Background: The motif D loop in poliovirus RNA-dependent RNA polymerase is important for catalysis and fidelity.
Results: A vaccine-derived mutation in motif D decreases RdRp fidelity by changing motif D conformational dynamics.
Conclusion: Non-conserved residues of motif D can alter RdRp function.
Significance: Motif D of the RdRp may be a universal target permitting creation of enzymes with perturbed fidelity and viruses with reduced virulence.
Keywords: Enzyme Kinetics, NMR, Positive-strand RNA Viruses, Protein Dynamics, Viral Polymerase
Abstract
All viral RNA-dependent RNA polymerases (RdRps) have a conserved structural element termed motif D. Studies of the RdRp from poliovirus (PV) have shown that a conformational change of motif D leads to efficient and faithful nucleotide addition by bringing Lys-359 into the active site where it serves as a general acid. The RdRp of the Sabin I vaccine strain has Thr-362 changed to Ile. Such a drastic change so close to Lys-359 might alter RdRp function and contribute in some way to the attenuated phenotype of Sabin type I. Here we present our characterization of the T362I RdRp. We find that the T362I RdRp exhibits a mutator phenotype in biochemical experiments in vitro. Using NMR, we show that this change in nucleotide incorporation fidelity correlates with a change in the structural dynamics of motif D. A recombinant PV expressing the T362I RdRp exhibits normal growth properties in cell culture but expresses a mutator phenotype in cells. For example, the T362I-containing PV is more sensitive to the mutagenic activity of ribavirin than wild-type PV. Interestingly, the T362I change was sufficient to cause a statistically significant reduction in viral virulence. Collectively, these studies suggest that residues of motif D can be targeted when changes in nucleotide incorporation fidelity are desired. Given the observation that fidelity mutants can serve as vaccine candidates, it may be possible to use engineering of motif D for this purpose.
Introduction
Positive-strand RNA viruses cause a number of acute and chronic diseases, including the common cold, myocarditis, encephalitis, hepatitis, and paralytic poliomyelitis (1–7). The enzyme responsible for RNA genome replication is the virally encoded RNA-dependent RNA polymerase (RdRp).3 RdRps belong to a superfamily of template-directed nucleic acid polymerases, including DNA-dependent DNA polymerases, DNA-dependent RNA polymerase, and RNA/DNA-dependent DNA polymerases (i.e. reverse transcriptases) (8–11). A strategy to rationally design live, attenuated vaccine strains has been proposed based on modifying RdRps to change their nucleotide incorporation fidelity (12, 13); viral strains encoding RdRps with altered fidelity (either higher or lower fidelity) have been shown to lead to virus incapable of causing disease but capable of acting as vaccine strains to protect mice from a lethal challenge from wild-type virus (12, 13). A better understanding of the fidelity determinants of RdRps would thus provide a framework for rational vaccine design.
The structure of viral RdRps has been described as a “cupped right hand” with fingers, thumb, and palm subdomains (11) (see Fig. 1A). Most of the catalytic machinery resides in the palm region, which can be further divided into five major structural motifs (A–E). Motifs A and C contain absolutely conserved Asp residues critical for binding Mg2+ ions required for RdRp function, and conserved residues in motif B are important for interacting with the sugar of the incoming nucleoside triphosphate (NTP) (15–22). Structures of RdRps bound with RNA and/or nucleotide have highlighted structural rearrangements within these palm motifs that are necessary before nucleotide incorporation (23–27).
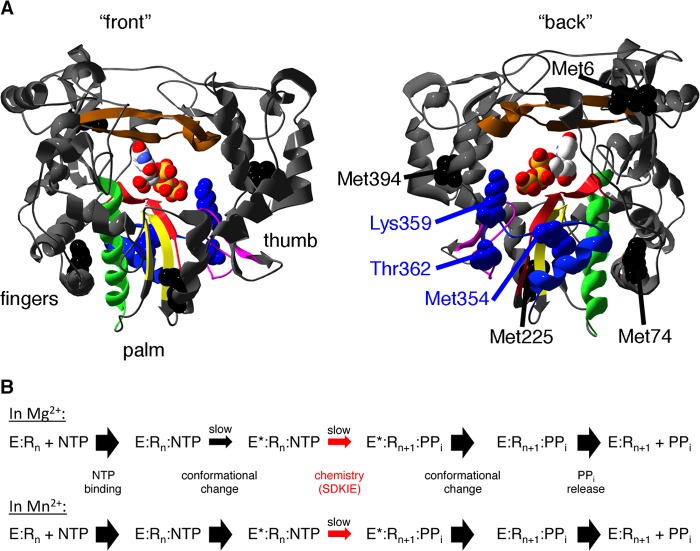
FIGURE 1.
The T362I substitution in motif D may impact the structural dynamics of motif D and surrounding regions to affect RdRp catalysis and fidelity. A, ribbon structure of PV RdRp bound with UTP (Protein Data Bank 2IM2; Ref. 24) with palm motifs colored (A, red; B, green; C, yellow; D, blue; E, purple; F, brown), highlighting the location of Thr-362, near the general acid Lys-359 (shown in blue). Useful NMR probes for the structure and dynamics of PV RdRp are also indicated (Met-354 in blue, Met-6, Met-74, Met-225, and Met-394 in black). B, kinetic mechanism of PV RdRp in the presence of Mg2+ (50) and Mn2+ (51). In the presence of Mg2+, both the pre-chemistry conformational change and the chemistry step itself are partially rate-determining. In contrast, the chemistry step (for correct nucleotide incorporation) is fully rate-determining in the presence of Mn2+. The SDKIE reports on the proton transfers occurring in the chemistry step.
Our recent kinetic and solution-state nuclear magnetic resonance (NMR) studies have also identified motif D as an important determinant of RdRp fidelity. Motif D contains an absolutely conserved Lys (Lys-359 in PV RdRp) that protonates the pyrophosphate leaving group in the nucleotide addition reaction (28, 29). NMR experiments have indicated that there are important structural rearrangements in motif D when correct, but not incorrect, nucleotide binds (30, 31), which might be responsible for repositioning Lys-359 for catalysis. Computational studies are also consistent with the motif D Lys acting as a general acid (32). Amino acid substitutions at Lys-359 (e.g. K359R) lead up to a 50-fold decrease in the maximum turnover rate constant (kpol), a 5-fold increase in RdRp fidelity (as determined by kpol,correct/kpol,incorrect), and lead to the severe attenuation of the virus (29, 30, 33). We have suggested that the role of motif D in RdRp and reverse transcriptase fidelity is analogous to the function of the O/P helices in A/B family DNA polymerases (30).
Amino acid substitutions in the O/P helices of DNA polymerases appear to be able to tune polymerase fidelity (34–38). Amino acid changes in motif D may likewise alter PV RdRp fidelity. One particularly noteworthy substitution is the T362I substitution that is encoded by the RdRp from the Sabin type I strain of PV. The T362I substitution may interfere with the structural dynamics of the motif D loop to disrupt RdRp catalytic function and may play a role in the attenuation of the Sabin vaccine.
In this article we demonstrate that the T362I substitution lowers RdRp fidelity by altering the conformational dynamics of motif D and other active-site regions for more efficient nucleotide misincorporation. The decrease in RdRp fidelity induced by the T362I substitution likely contributes to the reduced virulence of the strain carrying this genetic change. These studies highlight motif D as a potential, rational target for modifying viral RdRps to produce attenuated viruses for use as vaccine candidates.
EXPERIMENTAL PROCEDURES
Materials
[γ-32P]ATP (>7000 Ci/mmol) was from VWR-MP Biomedical; nucleoside 5′-triphosphates and 2′-deoxynucleoside 5′-triphosphates (all nucleotides were ultrapure solutions) were from GE Healthcare; 3′-deoxyadenosine 5′-triphosphate (cordycepin) was from Trilink Biotechnologies. All RNA oligonucleotides were from Dharmacon Research, Inc. (Boulder, CO); T4 polynucleotide kinase was from New England Biolabs, Inc; [methyl-13C]methionine was from Cambridge Isotope Laboratories. All other reagents were of the highest grade available from Sigma or Fisher.
Site-directed Mutagenesis, Protein Overexpression, and Purification of PV RdRp
The T362I variant was generated using the QuikChange method (Stratagene) and appropriate primers. The mutation was confirmed by DNA sequencing (Nucleic Acid Facility, The Pennsylvania State University).
It should be noted that wild-type (WT) and T362I RdRp used in the kinetic and NMR studies also contain two interface I amino acid substitutions (L446D and R455D) to prevent polymerase dimerization. Overexpression and purification of PV RdRp were performed as described previously (31, 39–41). Both kinetic and NMR experiments involved protein samples labeled by [methyl-13C]Met (31). The incorporation of this isotope did not change RdRp kinetics (data not shown).
PV RdRp Kinetic Assays
Purifying, 5′-32P end labeling and reannealing of RNA templates 10-mer sym/subU (5′-GCAUGGGCCC-3′) and 11-mer sym/subUA (5′-GCAUGGGCCCA-3′) were carried out as described previously (31, 40).
Enzyme assays were conducted at 30 °C in reaction buffer consisting of 50 mm HEPES, pH 7.5, 10 mm 2-mercaptoethanol, 5 mm MgCl2, and 60 μm ZnCl2. Immediately before the reaction, RNA oligonucleotides were reannealed, and concentrated RdRps were diluted to the required concentration by reaction buffer (50 mm HEPES, pH 7.5, 10 mm 2-mercaptoethanol, 20% glycerol, and 60 μm ZnCl2). The volume of enzyme added into any reactions was never above of the total volume.
For stopped flow experiments, the RdRp-RNA binary complexes were formed by incubating RdRp (0.5 μm) with sym/sub RNA (0.25 μm duplex) for 2 min at room temperature. Experiments were initiated by rapidly mixing RdRp-RNA binary complexes in 50 mm HEPES, pH 7.5, 10 mm 2-mercaptoethanol, 5 mm MgCl2, and 60 μm ZnCl2 with the equal volume of NTP solution in the same buffer. The utilized RNA templates sym/subU and sym/subUA were substituted with 2-aminopurine and pyrrolo-cytosine at the 5′ side of the template RNA respectively. These fluorescent bases create fluorescence changes to monitor the chain elongation reaction. For 2-aminopurine-modified sym/sub-U RNA, excitation and emission wavelengths were 313 and 370 nm, respectively. For pyrrolo-cytosine-modified sym/sub-UA RNA, the excitation and emission wavelengths were 350 and 450 nm, respectively.
For benchtop assays, the RdRp-RNA binary complexes were formed through incubating RdRp (0.5 μm) with RNA (0.25 μm duplex) for 3 min at room temperature followed by 2 min at 30 °C. The RNA template was 32P end-labeled. After initiation, the reaction was quenched by adding an equal volume of quench buffer (50 mm EDTA, 85% formamide, 0.025% bromphenol blue, and 0.025% xylene cyanol). The quenched samples were analyzed by denaturing PAGE, and products were quantitated by phosphorimaging as previously described (40).
NMR Sample Preparation and Spectroscopy
NMR sample preparation followed procedures previously described using [methyl-13C]Met-labeled PV RdRp (30, 31). 13C,1H HSQC (heteronuclear single quantum correlation) NMR spectra were collected at 293 K on a Bruker Avance III 600 MHz spectrometer equipped with a 5-mm “inverse detection” triple-resonance (13C,1H,15N) single axis gradient TCI for the following samples: free RdRp, RdRp-RNA complex, RdRp-RNA complex, and excess 3′-dATP, RdRp-RNA-3′-blocked complex after passage over the desalting column (RdRp-RNA binary complex), RdRp-RNA-binary complex and excess of second nucleotide (i.e. RdRp-RNA-NTP ternary complex), and RdRp-RNA-NTP ternary complex after passage over a second de-salting column.
Molecular Dynamics Simulations
Atomistic molecular dynamics (MD) simulation for T362I PV RdRp (20 ns) was performed following the same method used in our previous MD simulations of picornaviral RdRps (42). Because there is no structure available for the T362I variant, the initial structural model of T362I RdRp was constructed based on the WT PV RdRp (Protein Data Bank code 1RA6) by substituting Thr-362 with an Ile residue using the TLEAP module of the AMBER package (43). The protein was immersed in a truncated octahedral box filled with TIP3P (44) water molecules, with a distance of at least 20 Å between any protein atom and the edge of the solvent box. MD simulation was carried out using SANDER/PMEMD in AMBER10 package (43) using the AMBER99SB force field. Initial minimization of the modeled structure was done in multiple steps using SANDER; all subsequent steps of the MD simulation were carried out using PMEMD. The solvated system was heated stepwise from 0 to 300 K with an increment of 50 K over a period of 10 ps under constant volume and temperature conditions (NVT) followed by 200 ps of NVT simulation. Finally, the simulation was switched to constant pressure and temperature dynamics for the remainder of the MD simulation. An integration time step of 1 fs was used, and the cutoff distance for nonbonded interactions was set to 9 Å. The neighbor pair list was updated every 10 steps. The Berendsen thermostat (45) was applied with temperature and pressure coupling constants of 1 ps (weak coupling). Electrostatic interactions were calculated with the Particle Mesh Ewald method (46), and all bond lengths involving hydrogen atoms were constrained using SHAKE algorithm (47).
Ribavirin Sensitivity Assay
50 plaque forming units of WT, G64S, or T362I PV were used to infect HeLa cell monolayers pretreated with the various concentrations of ribavirin and then washed and overlaid with agarose media containing the same concentration of ribavirin. Cells were incubated for 3–4 days at 37 °C before being stained with crystal violet (48).
Mouse Infection Studies
Mice were bred and housed in standard ventilated caging for all experiments. Protocols for animal studies were approved by The Pennsylvania State University Institutional Animal Care and Use Committee (IACUC). All experiments were performed in accordance to guidelines and regulations overseen by the IACUC. Viral stocks were generated in serum-free media and harvested, and genome copies were quantitated by quantitative PCR performed by the Genomics Core Facility of The Pennsylvania State University. 4–6-Week-old outbred (ICR) mice transgenic for the PV receptor (cPVR) were infected with PV at the indicated genome copy by intraperitoneal injection in 3 ml of serum-free media. Mice were observed for 10 days for signs of disease and were euthanized upon showing dual limb paralysis or paralysis such that their ability to obtain food and water was compromised; this was in accordance with approval by the IACUC at The Pennsylvania State University.
RESULTS
The T362I Substitution in Motif D Lowers RdRp Fidelity by Making the Pre-chemistry Conformational Change for Misincorporation More Efficient
The T362I substitution as derived from the PV Sabin I vaccine strain is located in the motif D loop of the palm subdomain (Fig. 1A). Motif D also contains the highly conserved Lys (for PV RdRp, Lys-359), which has been shown to behave as a general acid to protonate the pyrophosphate leaving group in the nucleotide incorporation reaction (28, 29). Pre-steady-state enzyme kinetics indicated that both a pre-chemistry conformational change and the chemistry step itself are partially rate-determining (in the presence of Mg2+) and thus are both important fidelity checkpoints (50) (Fig. 1B). Our recent NMR studies have implicated structural rearrangements in motif D, likely to re-position Lys-359, as an important component of the pre-chemistry conformational change(s) (30). The T362I substitution may thus interfere with the structural rearrangements in motif D necessary for RdRp function, including changes to RNA/NTP affinity, catalytic activity, and/or RdRp fidelity.
To determine if the T362I substitution has an effect on RNA affinity and ternary RdRp-RNA-NTP complex formation, we compared the formation and stability of pre-chemistry RdRp-RNA reactant complexes by enzyme assembly and dissociation assays respectively (40) (Fig. 2). For our kinetic assays, we used two RNA templates (Fig. 2A), which contain a six (or seven)-base pair duplex flanked by two-, four-, (or three)-nucleotide 5′ overhangs. We have previously used these symmetrical primer/template substrate (sym/sub) RNA templates in enzyme kinetic and solution-state NMR studies (30, 31, 40, 48, 50, 51). The rate and yield of competent RdRp-RNA complexes (Fig. 2, B and C) and the stability of the assembled RdRp-RNA complexes (Fig. 2, D and E) were essentially identical for WT and T362I RdRp using either RNA template. These results indicate that the T362I substitution does not substantially affect RNA affinity or formation of catalytically competent RdRp-RNA-NTP complexes.
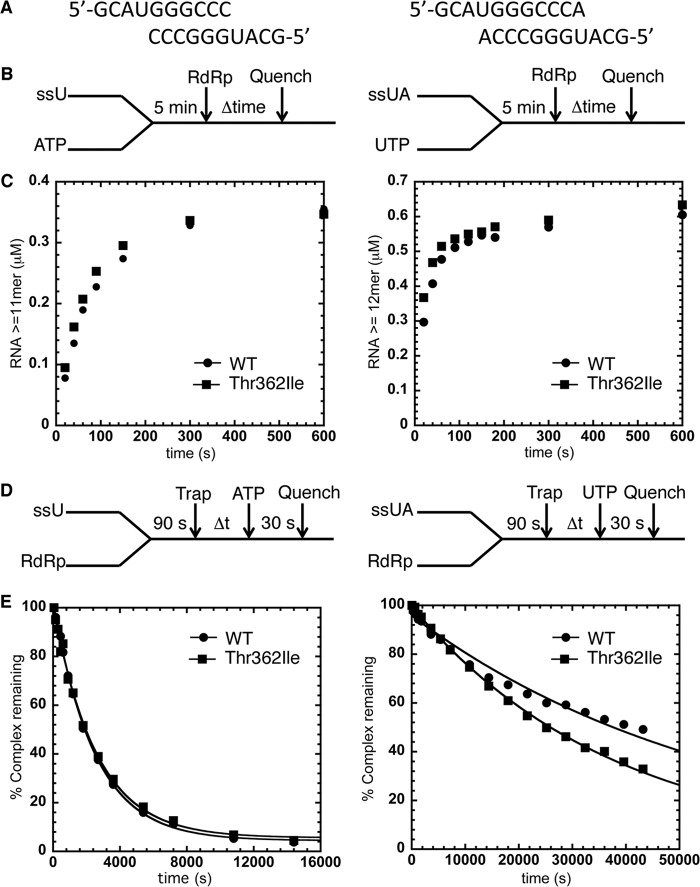
FIGURE 2.
Formation and stability of RdRp-RNA complexes is not affected by the T362I substitution in motif D. A, RNA sequences for sym/subU (left) and sym/subUA (right). B, experimental design for RdRp-RNA-NTP assembly assay. Reactions were initiated by adding RdRp (1 μm) into sym/subU or sym/subUA (0.5 μm duplex) and its corresponding ATP or UTP (500 μm), which were incubated at 30 °C for 5 min. At the indicated times, reactions were quenched by adding 25 mm EDTA. C, the RdRp-RNA-NTP assembly assay for WT RdRp (●) and T362I RdRp (■) using both sym/subU and sym/subUA. There is essentially no difference in this assay between WT and T362I RdRp. D, experimental design for the RdRp-RNA dissociation assay. RdRp and RNA were incubated at 30 °C for 90 s at which time trap (100 μm unlabeled RNA) was added to the reaction buffer. After the indicated times, the reaction buffer was mixed with ATP or UTP (500 μm) and then quenched after 30 s by adding EDTA (25 mm). E, the RdRp-RNA dissociation assay for WT RdRp (●) and T362I RdRp (■) using both sym/subU and sym/subUA RNA. The solid line is the data fit to a single exponential function. The kdiss(obs) for sym/subU (sym/subUA) was 4.00 ± 0.09 × 10−4 s−1 (1.70 ± 0.07 × 10−5 s−1) for WT RdRp and 3.90 ± 0.02 × 10−4 s−1 (2.70 ± 0.03 × 10−5 s−1) for T362I RdRp, indicating that the RNA binding characteristics of WT and T362I RdRp are nearly identical. Each data point is from the averages of at least two separate experiments on different days.
We also performed kinetics experiments to compare the maximal rate constant for nucleotide incorporation, kpol, and the apparent dissociation constant for NTP, Kd,app, for WT and T362I RdRp (Fig. 3, Table 1). The T362I substitution did not substantially change kpol or Kd,app values for correct NTP incorporation using ATP for sym/subU RNA (Fig. 3B, Table 1) and UTP for sym/subUA (Table 1). However, the T362I substitution led to more efficient misincorporation of incorrect NTP either with 2′-dNTP (Fig. 3C) or incorrect nucleobase (Fig. 3D). This change resulted in lowered RdRp fidelity for both nucleobase and sugar selection (as evaluated by either kpol,correct/kpol,incorrect or (kpol/Kd,app)correct/(kpol/Kd,app)incorrect). For example, 2′-OH selection was decreased up to 2-fold for the T362I variant relative to WT RdRp (Table 1).
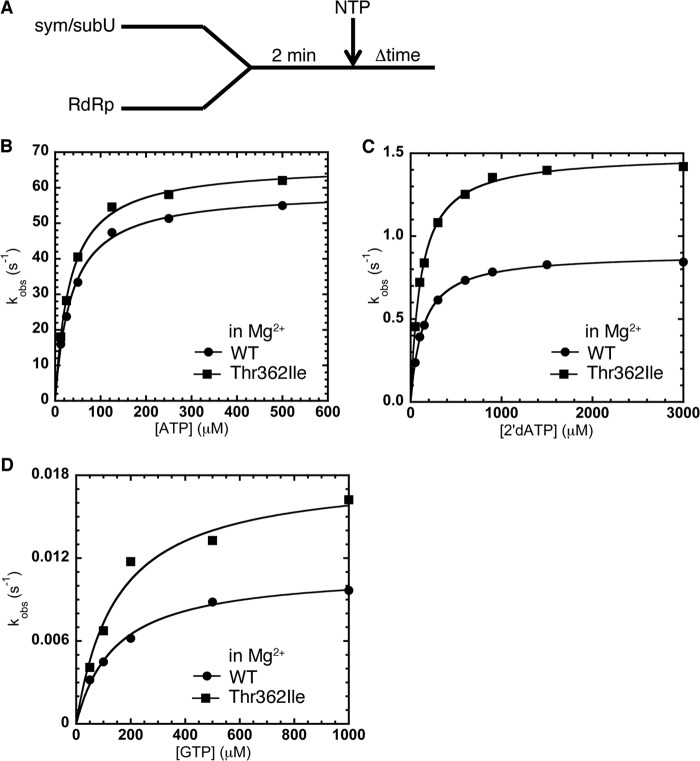
FIGURE 3.
The T362I substitution in motif D lowers RdRp nucleotide incorporation fidelity in the presence of Mg2+. A, experimental design for nucleotide incorporation assay. RdRp (0.5 μm) was incubated with sym/subU RNA (0.5 μm) at room temperature for 2 min and quickly mixed with various concentrations of NTP. The reactions were monitored by fluorescence changes with time. The solid line represents the data fit to a hyperbola function to give an apparent dissociation constant (Kd,app) and a maximal rate constant for nucleotide incorporation (kpol). B, kinetics of correct ATP incorporation into sym/subU RNA for WT RdRp (●) and T362I RdRp (■) were very similar in the presence of Mg2+. Misincorporation (in Mg2+) of 2′dATP (C) and GTP (D) into sym/subU RNA was more efficient for T362I RdRp (■) compared with WT RdRp (●). Kinetic parameters are located in Table 1.
TABLE 1.
Comparison of the kinetic constants of nucleotide incorporation for WT and T362I RdRp reveals that the T362I substitution decreases polymerase fidelity
| Variant | NTP | kpol | KD,app | kpol/KD,app | kpol,correct/kpol,incorrect | (kpol/KD)correct/(kpol/KD)incorrect |
|---|---|---|---|---|---|---|
| s−1 | μm | μm−1s−1 | ||||
| Correct NTP | ||||||
| Sym/Sub-U RNA | ||||||
| WT | ATP | 59 ± 1 | 36 ± 2 | 1.6 | ||
| T362I | ATP | 67 ± 1 | 33 ± 2 | 2.0 | ||
| Sym/Sub-UA RNA | ||||||
| WT | UTP | 130 ± 7 | 142 ± 36 | 0.91 | ||
| T362I | UTP | 131 ± 2 | 114 ± 9 | 1.1 | ||
| Incorrect sugar | ||||||
| Sym/sub-U RNA | ||||||
| WT | 2'-dATP | 0.89 ± 0.01 | 134 ± 4 | 6.7 × 10−3 | 66 | 240 |
| T362I | 2'-dATP | 1.5 ± 0.0 | 112 ± 4 | 1.3 × 10−2 | 45 | 150 |
| Sym/Sub-UA RNA | ||||||
| WT | 2'-dUTP | 8.8 ± 0.3 | 925 ± 85 | 9.5 × 10−3 | 15 | 96 |
| T362I | 2'-dUTP | 12 ± 0.4 | 748 ± 77 | 1.6 × 10−2 | 11 | 69 |
| Incorrect nucleobase | ||||||
| Sym/sub-U RNA | ||||||
| WT | GTP | 0.011 ± 0.000 | 142 ± 15 | 7.7 × 10−5 | 5400 | 21,000 |
| T362I | GTP | 0.018 ± 0.001 | 149 ± 25 | 1.2 × 10−4 | 3700 | 17,000 |
| WT | RTPa | 0.019 ± 0.002 | 292 ± 86 | 6.5 × 10−5 | 3100 | 25,000 |
| T362I | RTP | 0.025 ± 0.003 | 224 ± 67 | 1.1 × 10−4 | 2700 | 18,000 |
a RTP, ribavirin triphosphate.
The T362I substitution may alter the conformational rearrangements necessary to re-position Lys-359 and/or alter the final positioning of Lys-359 to affect nucleotide addition kinetics. The effect of Mn2+ and D2O on nucleotide incorporation kinetics can help to distinguish changes to the pre-chemistry conformational change and/or the chemistry step (Fig. 1B). In the presence of Mn2+, the pre-chemistry conformational change becomes more efficient, and kpol is then fully rate-determined by the chemistry step (51). Solvent deuterium kinetic isotope effects (SDKIE; kpol,H2O/kpol,D2O), which arise from proton transfers within the chemistry step (28, 29), also report on the relative contribution of the chemistry step to kpol.
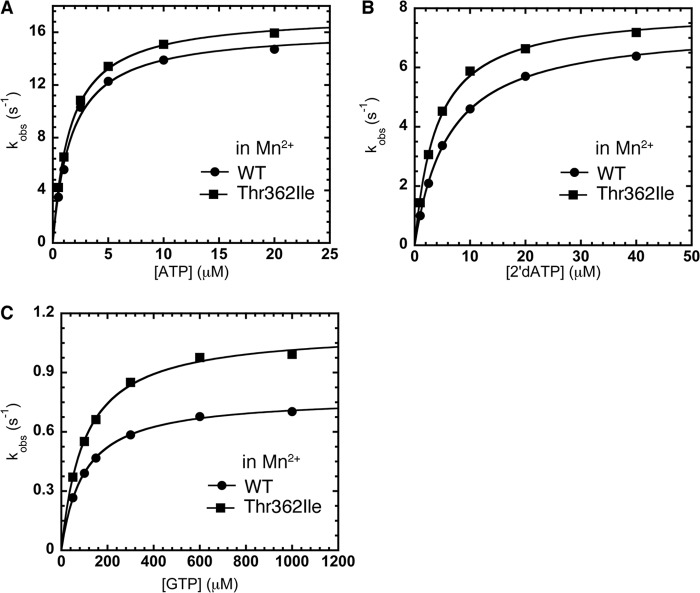
In the presence of Mn2+, the T362I substitution did not substantially alter correct NTP incorporation or misincorporation of the 2′-dNTP compared with WT RdRp (Fig. 4, A and B); the RdRp fidelity for sugar selection in Mn2+ is identical for WT (kpol,correct/kpol,incorrect = 2.2 ± 0.1) and T362I RdRp (kpol,correct/kpol,incorrect = 2.2 ± 0.1). This result implies that the T362I alteration primarily impacts the step made more efficient by the presence of Mn2+. The pre-chemistry conformational change is more efficient with Mn2+, such that it is no longer rate-determining (51), and thus introduction of the T362I substitution to further enhance the pre-chemistry conformational change has no additional impact on the kinetic parameters as governed by the rate-determining step (i.e. in this case, the chemistry step).
FIGURE 4.
The T362I substitution in motif D lowers sugar and nucleobase selection by making the pre-chemistry conformational change more efficient. A, kinetics of correct ATP incorporation into sym/subU RNA in the presence of Mn2+ is very similar for WT (●) and T362I (■) RdRp. B, in contrast to what occurs in the presence of Mg2+ (see Fig. 3), misincorporation of 2′-dATP into sym/subU RNA in the presence of Mn2+ is similar between WT (●) and T362I (■) RdRp. The metal dependence of the RdRp fidelity is consistent with the T362I substitution primarily impacting the pre-chemistry conformational change in 2′dATP misincorporation. C, the T362I substitution may also impact the chemistry step itself for misincorporation of NTP with incorrect nucleobase. The T362I (■) substitution leads to more efficient GTP misincorporation into sym/subU RNA even in the presence of Mn2+ compared with WT RdRp (●). Kinetic parameters are located in Table 1.
For misincorporation of NTP with incorrect nucleobase in Mn2+, the T362I variant (kpol = 1.1 ± 0.0 s−1) was still more efficient than WT RdRp (kpol = 0.78 ± 0.00 s−1) (Fig. 4C). In the case of GMP misincorporation, the pre-chemistry conformational change is primarily rate-determining in Mg2+ (30). The introduction of Mn2+ and the T362I substitution may both enhance the rate of the pre-chemistry conformational change but not sufficiently to make the chemistry step rate-determining for GTP misincorporation. To further test which step(s) is being affected, we also determined SDKIEs (i.e. kpol,H2O/kpol,D2O) for WT and T362I RdRp for both correct and incorrect incorporation. The SDKIEs for T362I RdRp for correct (2.39 ± 0.02) and incorrect (1.80 ± 0.07) NTP incorporation were both higher than the corresponding SDKIEs for WT RdRp (correct, 2.18 ± 0.02; incorrect, 1.51 ± 0.06). These findings suggest that chemistry is more rate-determining for the T362I variant compared with WT RdRp, implying that the pre-chemistry conformational change is more efficient for T362I RdRp.
Solution-state NMR Indicates That T362I Induces Structural Rearrangements within Motif D to Lower RdRp Fidelity
Our previous NMR experiments have been able to identify key chemical shift changes associated with Met residues in PV RdRp induced by the binding of RNA and the formation of ternary RdRp-RNA-NTP complexes (30, 31). In particular, we identified a large chemical shift change to the Met-354 resonance and smaller chemical shift changes to the Met-6, Met-74, and Met-225 resonances upon formation of the correct RdRp-RNA-NTP complex for the WT enzyme (30, 31). We have suggested that these chemical shift patterns correspond to a “closed” conformation. These chemical shift changes do not occur in the presence of the incorrect NTP, either with incorrect 2′-deoxy sugar (Fig. 5A) or with incorrect nucleobase (Fig. 5B). In fact, the resonances for WT RdRp bound with RNA and 2′-deoxy sugar or incorrect nucleobase essentially overlap (Fig. 5C), and we have suggested that these complexes are in an “open” conformation.
FIGURE 5.
The T362I substitution in motif D alters the conformational equilibrium between open and closed states of PV RdRp as revealed by NMR spectra of [methyl-13C]Met-labeled RdRp. A–C, comparisons of HSQC spectra between WT RdRp bound with RNA lacking a 3′-OH and correct UTP nucleotide (black), incorrect 2′-dUTP nucleotide (red), or incorrect ATP nucleotide (blue). The chemical shift patterns for the RdRp-RNA complexes bound with 2′-dUTP and ATP are strikingly similar (C), representing an open conformation, and are different from the chemical shift pattern when bound to UTP, representing a closed conformation (A and B). D–F, comparisons of HSQC spectra between T362I RdRp bound with RNA lacking 3′-OH and correct UTP nucleotide (black), incorrect 2′-dUTP (red), or incorrect ATP nucleotide (blue). T362I RdRp bound with RNA and 2′-dUTP displays two sets of resonances (D and F), where one set overlays with that from T362I RdRp bound with RNA and UTP (D) and the other with that from T362I RdRp bound with RNA and ATP (F). These results indicate that T362I RdRp bound with RNA and 2′-dUTP accesses both the open and closed conformations. This finding is consistent with the results presented in Figs. 3 and 4, which suggests that the T362I substitution makes the pre-chemistry conformational change (open to closed) more efficient. Spectra were collected at 293 K with 245 μm RdRp, 500 μm RNA, and 4 mm UTP, 10 mm 2′-dUTP or 8 mm ATP.
With the T362I substitution, there is a small chemical shift change to the Met-354 resonance in the absence (data not shown) and presence of RNA and NTP (Fig. 5). The most substantial change with the T362I RdRp spectra occurs in the presence of 2′-dNTP (red spectrum in Fig. 5D). In this case two sets of resonances were observed, especially for Met-354, indicating that the enzyme is exchanging between two conformations on the slow NMR chemical shift timescale. This behavior is not observed in the presence of NTP with incorrect nucleobase (blue spectrum in Fig. 5E). It should be kept in mind that the concentrations of NTP (2′-dUTP = 10 mm, CTP = 8 mm) are identical for WT and T362I RdRp, and we would expect binding saturation given the Kd,app of the incorrect NTP (Table 1), which is borne out by the small chemical shift changes observed compared with the enzymes bound with RNA alone (30) (i.e. incorrect NTP is bound to RdRp).
One set of resonances for the T362I variant bound to RNA and 2′-dUTP (red spectrum in Fig. 5F) overlaps that of the T362I variant bound to RNA and ATP (blue spectrum in Fig. 5F). The other set of resonances for the T362I variant bound to RNA and 2′-dNTP (red spectrum in Fig. 5D) overlaps the spectrum of the T362I variant bound to RNA and correct UTP (black spectrum in Fig. 5D). These findings suggest that T362I RdRp exchanges between the closed and open conformations of RdRp, such that the closed conformation is accessible even in the presence of the 2′-dNTP. This result is consistent with the enzyme kinetic results, which also suggest that the T362I substitution increases 2′-dNTP misincorporation rates through a more efficient pre-chemistry conformational change. The “open-closed” conformational equilibrium when RdRp is bound with NTP with incorrect nucleobase may also be altered, but the effect is unlikely to be observable in the NMR spectrum considering that the conformational equilibrium would still likely heavily favor the open state even in the T362I variant. This statement is consistent with the Mn2+ effects and the diminished SDKIE (Table 1), which suggest that the pre-chemistry conformational change from open to closed largely governs kpol for incorrect nucleobase incorporation.
The T362I Substitution in Motif D Results in Long-range Structural Changes That May Allow for More Efficient Misincorporation
To gain more insight into how the T362I substitution changes the structure and/or internal dynamics of RdRp to impact catalytic function and fidelity, we constructed an in silico model for the T362I variant, as there is no crystal structure available for this protein, and conducted 20 ns of all-atom MD simulations. The intention of these studies was to focus on interactions that may be perturbed by the T362I substitution. The MD simulation of the T362I variant was compared with that of the WT enzyme from our previous study (42). It should be emphasized that these MD simulations are for RdRp enzymes without RNA and NTP bound. Nonetheless, previous MD simulations have given insight into the function and fidelity of RdRp enzymes and have been consistent with NMR studies (42).
MD simulations revealed that substitution of the solvent-exposed residue Thr-362 by a hydrophobic Ile in the T362I variant resulted in conformational changes nearby and distant from the mutation site (Fig. 6A). In the simulated T362I structure, the side chain of Ile-362 is stabilized by van der Waals interactions with the side chain of the nearby Phe-363; the average distance between the two side chains is 4.6 Å. The simulation revealed that the substitution at position 362 disrupted the local environment observed in the WT enzyme, and the most perturbed residues were those of the motif D loop (i.e. amino acids 359–365) (Fig. 6B). The side chain of Glu-364 adopts a completely different conformation in the T362I structure relative to WT RdRp; in the WT enzyme, the Glu-364 side chain is engaged in electrostatic interactions with the Lys-228 preceding motif A (amino acids 229–240), whereas in the T362I variant the side chain of Glu-364 is swung away from the WT position to a new position where it can form H-bonding interactions with Asn-370 and transient H-bonding interactions with Thr-367. The negatively charged Glu-364 in the T362I variant is partly stabilized by the nearby side chain of Lys-359. Furthermore, side chains of residues Thr-365, Glu-337, Asp-339, and Ser-341, all belonging to motif D and located in the vicinity of Glu-364, are affected by the conformational change of Glu-364 side chain. Of note, the local conformational changes around the mutation site can be sensed by the nearby Met-354 as shown by NMR (Fig. 5). In addition to the local conformational changes in the vicinity of Ile-362, MD simulations showed propagation of the conformational changes from the mutation site to other, more distant residues at the active-site (Fig. 6, C and D). These conformational differences include residues Ser-288 of the loop 283–293, Asp-238 of motif A (both known to play a role in recognition of the sugar moiety of the nucleotide substrate), and the region encompassing the metal binding aspartates Asp-328 and Asp-329 of motif C.
FIGURE 6.
The T362I substitution in motif D leads to structural and dynamics changes consistent with changes to RdRp function as revealed by MD simulation. A, the average structures calculated from the MD simulations of WT (gray) from our previous study (42) and T362I variant (black) are shown. The two structures were superimposed using Cα atoms; the proteins are depicted as ribbons, and the mutated residues Thr-362/Ile-362 are shown as sticks and labeled. Other residues that are altered in the Sabin variant (Asp-53, Tyr-73, and Lys-250) are also shown as sticks and labeled. The Thr to Ile substitution at position 362 resulted in conformational changes, both local and remote from the site of mutation. B, zoom-in view of the circled region around the mutation site T362I variant in A; selected residues in that region surrounding the mutation site are shown as sticks. The substitution at position 362 disrupted the local interactions, which is most pronounced for residue Glu-364. C, zoom-in view of the circled region in A surrounding the loop 283–293. Residues of the loop 283–293 and selected residues in the surrounding environment belonging to the functional motifs are shown as sticks and labeled (motif A, Asp-238; motif C, Asp-328 and Asp-329; motif D, Lys-359). The T362I substitution resulted in conformational changes of key active-site residues known to be important for the RdRp function. D, the per-residue root mean square deviation (per-RMSD) calculated from the last 10 ns of the WT (gray line) and T362I (black line) trajectories are plotted. In general, the per-root mean square deviation structural changes for the T362I variant were smaller compared with WT RdRp. The points in the per-root mean square deviation plot corresponding to the other Sabin sites that are displayed in panel A are indicated by a dark red diamond (WT) and a dark red triangle (T362I). Similar to the surrounding environment of T362I, the local region encompassing Asp-53 (amino acid residues 46–58) showed reduced nanosecond dynamics in the T362I variant compared with WT RdRp.
T362I PV Expresses a Mutator Phenotype in Vivo and Has Reduced Virulence in the Mouse Model
To determine how the biophysical and biochemical changes associated with the T362I substitution impact virus multiplication in cell culture, we constructed a PV genome encoding the T362I RdRp by changing the ACA codon encoding Thr to the ATC codon encoding Ile. Passage of T362I PV at a low multiplicity of infection showed no evidence of reversion to the wild-type codon even after four consecutive passages, suggesting that there is no strong fitness consequence for this mutation in cell culture.
Ribavirin is a nucleoside analog that is a mutagen to PV and other RNA viruses (52–54). A high fidelity PV variant, G64S, exhibiting increased nucleotide incorporation fidelity exhibits reduced sensitivity to the antiviral nucleoside, ribavirin (48). Therefore, to assess T362I fidelity in cells, we evaluated the sensitivity of T362I PV to ribavirin. By plaque assay, there was an increase in sensitivity of T362I PV to ribavirin when compared with WT PV in cells (Fig. 7A); a more than 2-fold decrease in the concentration of ribavirin needed to reduce the number of plaques by half compared with the untreated population. This result is consistent with the kinetic results indicating that the T362I substitution decreases RdRp fidelity and would, therefore, more readily incorporate the antiviral nucleotide (Table 1).
FIGURE 7.

PV encoding the T362I substitution in motif D expresses a mutator phenotype in cell culture and is attenuated in vivo. A, ribavirin (RBV) sensitivity assay. 50 plaque forming units of WT, G64S, or T362I PV were used to infect HeLa cell monolayers pretreated with various concentrations of ribavirin. Plaque numbers were plotted against ribavirin concentration normalized to untreated (0 mm) control. The solid line represents the fit of the data to a sigmoidal dose response equation (four parameter logistic model), yielding IC50 values of 0.67 ± 0.06, 2.15 ± 0.75, and 0.30 ± 0.02 mm for WT, G64S, and T362I respectively. Data are the means from three independent experiments. Error bars represent ± S.E. B and C, T362I PV is attenuated in the cPVR mouse model. cPVR mice (4–6 weeks old) were infected via the intraperitoneal route with serum-free media containing WT or T362I PV at the indicated genome copy and observed for 10 days. Mice that developed the symptoms of two-limb paralysis or paralysis that compromised the ability to get food and water were euthanized. The number of mice per group was n = 3.
The increased ribavirin sensitivity of T362I PV suggests that this mutant may be more attenuated in animals than WT PV, similar to other mutants that lead to lower fidelity RdRps (55). To test this proposal, we compared the virulence of T362I and WT PVs in a mouse transgenic for the PV receptor (56). In this system, infection by WT PV is generally lethal. In the case of infection at the highest dose (1.6 × 1011 genomes), T362I PV resulted in the delay of onset of double-limb paralysis by 1 day compared with WT PV (Fig. 7, B and C). At the lower dose (1.6 × 1010 genomes), onset of double-limb paralysis in all animals showed an even more gradual progression. These results are consistent with T362I PV having reduced virulence in mice.
DISCUSSION
Previous studies provided evidence that mutations in the RdRp gene contribute to the attenuation of the Sabin I vaccine strain (57–62). Here we have shown that the Sabin-derived T362I substitution in PV RdRp induces a decrease in polymerase fidelity (Figs. 3 and 4) by making the pre-chemistry conformational change more efficient (Fig. 5), likely through disrupting interactions in motif D and other regions of the active-site (Fig. 6), and PV strains expressing this variant have reduced virulence in mice (Fig. 7). These studies further highlight the functional roles for motif D, which until recently (29, 30) were only believed to play a structural role in maintaining the integrity of the palm region (16, 63). We have proposed that motif D in RdRps and reverse transcriptases play an analogous role to helix O/P in family A/B of DNA polymerases (30); both regions contain a conserved Lys that acts as a general acid in phosphodiester bond formation (29), and both regions undergo fidelity-governing conformational changes (30, 64–70). We have now demonstrated that amino acid substitutions elsewhere in motif D impact RdRp fidelity just as amino acid substitutions in helix O/P can lower polymerase accuracy (34–38).
One way for RNA viruses to “tune” their fidelity may be to alter the conformational dynamics of motif D. In fact, it was previously shown that motif D in cystoviral φ6 RdRp undergoes conformational exchange on the same timescale as catalysis (71, 72). It has also been previously demonstrated that RdRp fidelity has a major impact on viral virulence; viral strains carrying RdRp with reduced or enhanced fidelity are attenuated in mice (12–14, 33, 48, 55, 73). Thus, the general concept of modifying the conformational dynamics of motif D to impact RdRp fidelity and virus biology may be a guiding principle in rational vaccine design. The highly conserved motif D Lys has already been identified as an important residue to modify to attenuate the virus and provide immunoprotective effects almost to the same level as Sabin I (33); viral attenuation is likely caused by the change in RdRp fidelity as proposed for other RdRp mutants (12, 13, 33, 48, 55).
In picornaviruses, similar interactions as those highlighted by the current MD simulations may govern the motion of motif D in other RdRps and hence be important for this fidelity checkpoint. Although Thr-362 is not conserved among picornoviral RdRps, the amino acid equivalent to this position is polar in coxsackievirus virus (CVB3; Cys-363), foot-and-mouth disease virus (Lys-372), and human rhinovirus serotype 16 (HRV16; Glu-361) RdRps (Fig. 8). Modifying these positions to something more non-polar may allow them to interact with the well conserved motif D Phe and lead to similar structure and/or dynamic changes that we have proposed for T362I PV RdRp. The MD simulations predict that the T362I substitution perturbs the interaction between Glu-364 and Lys-228, but a new interaction forms between Glu-364 and Asn-370. Again, these residues are not highly conserved, but similar interactions between equivalent positions (may) form in other RdRps. For example, there is an electrostatic interaction in HRV16 RdRp between Asp-228/Lys-363 comparable to the Lys-228/Glu-364 interaction in PV RdRp. Substituting Glu-361 in HRV16 RdRp to something more non-polar may perturb the Asp-228/Lys-363 interaction but then allow Lys-363 to interact with Asn-369. Based on our studies with T362I PV RdRp, we would predict that such changes would decrease HRV16 RdRp fidelity and attenuate the virus.
FIGURE 8.
Comparison of motif D conformations in picornaviral RdRps. The region encompassing motif D and the surrounding environment in the average MD simulated structures of the different RdRps is shown in different colors (PV, gray; CVB3, magenta; foot-and-mouth disease virus, green; HRV16, orange) (42). The four RdRp structures were superimposed using PV RdRp as a reference. Equivalent residues involved in key interactions of motif D are shown as sticks and labeled. Thr-362 in PV RdRp is not conserved, but there is a polar residue equivalent to this position in the other picornaviral RdRps (CVB3, Cys-363; foot-and-mouth disease virus, Lys-372; HRV16, Glu-361). Although sequence similarity is low in this region, non-covalent interactions (i.e. electrostatic, H-bonding, van der Waals interactions) occur between equivalent positions. Of note is the conservation of interactions between residues occupying the equivalent positions of Lys-228 and Glu337 in PV RdRp.
One intriguing feature of PV RdRp is that amino acid substitutions remote from the active site can alter polymerase catalysis and fidelity (48). This finding suggests that there are long-range interaction networks important for modulating RdRp function (31, 42). The T362I substitution also leads to long-range structural and dynamics changes (Fig. 6D), including a reduction in nanosecond dynamics around residues 46–58, which encompasses the site of another Sabin substitution, D53N. This finding suggests that there may be communication pathways linking the Sabin sites, and modifications at these sites may act cooperatively to change RdRp function; previous MD simulations of WT RdRp have also indicated that the motions of the α-helix containing Tyr-73 are anti-correlated with the motions of motif D containing Thr-362 (42). The Sabin amino acid substitutions may also contribute to different RdRp functions; the Y73H substitution has been shown to interfere with RdRp initiation (62), K250E may interfere with protein structure and/or long-range interactions with RNA through changes to electrostatic interactions, and T362I, in addition to its role in RdRp fidelity shown here, has been shown to contribute to temperature sensitivity of the Sabin I vaccine (59). The other Sabin substitutions may even compensate for the effects observed here with T362I. Teasing apart the functions of the other Sabin amino acid substitutions in PV RdRp may lead to a more thorough understanding of RdRp mutations in viral attenuation and immunoprotective effects and provide a framework for predicting similar mutations in other viral RdRps for rational vaccine design.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI45818 (to C. E. C.). This work was also supported by startup funds from Pennsylvania State University (to D. D. B.).
- RdRp
- RNA-dependent RNA polymerase
- cPVR
- poliovirus receptor
- HSQC
- heteronuclear single quantum correlation
- IACUC
- Institutional Animal Care and Use Committee
- MD
- molecular dynamics
- PV
- poliovirus
- SDKIE
- solvent deuterium kinetic isotope effect
- sym/sub
- symmetrical primer-template substrate
- HRV16
- human rhinovirus serotype 16.
REFERENCES
- 1. Hayden F. G. (1999) Update on influenza and rhinovirus infections. Adv. Exp. Med. Biol. 458, 55–67 [DOI] [PubMed] [Google Scholar]
- 2. Hayden F. G. (2006) Antivirals for influenza. Historical perspectives and lessons learned. Antiviral Res 71, 372–378 [DOI] [PubMed] [Google Scholar]
- 3. Hayden F. G. (2006) Respiratory viral threats. Curr. Opin. Infect. Dis. 19, 169–178 [DOI] [PubMed] [Google Scholar]
- 4. Kim W. R. (2002) The burden of hepatitis C in the United States. Hepatology 36, S30–S34 [DOI] [PubMed] [Google Scholar]
- 5. Weiss S. R., Navas-Martin S. (2005) Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69, 635–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Racaniello V. R. (2006) One hundred years of poliovirus pathogenesis. Virology 344, 9–16 [DOI] [PubMed] [Google Scholar]
- 7. Howard R. S. (2005) Poliomyelitis and the postpolio syndrome. BMJ 330, 1314–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Svarovskaia E. S., Cheslock S. R., Zhang W. H., Hu W. S., Pathak V. K. (2003) Retroviral mutation rates and reverse transcriptase fidelity. Front. Biosci. 8, d117–d134 [DOI] [PubMed] [Google Scholar]
- 9. Joyce C. M., Benkovic S. J. (2004) DNA polymerase fidelity. Kinetics, structure, and checkpoints. Biochemistry 43, 14317–14324 [DOI] [PubMed] [Google Scholar]
- 10. Berdis A. J. (2009) Mechanisms of DNA polymerases. Chem. Rev. 109, 2862–2879 [DOI] [PubMed] [Google Scholar]
- 11. Ng K. K., Arnold J. J., Cameron C. E. (2008) Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 320, 137–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vignuzzi M., Stone J. K., Arnold J. J., Cameron C. E., Andino R. (2006) Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439, 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vignuzzi M., Wendt E., Andino R. (2008) Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 14, 154–161 [DOI] [PubMed] [Google Scholar]
- 14. Pfeiffer J. K., Kirkegaard K. (2005) Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 1, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poch O., Sauvaget I., Delarue M., Tordo N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Reilly E. K., Kao C. C. (1998) Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252, 287–303 [DOI] [PubMed] [Google Scholar]
- 17. Lang D. M., Zemla A. T., Zhou C. L. (2013) Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res. 41, 1464–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zemla A. T., Lang D. M., Kostova T., Andino R., Ecale Zhou C. L. (2011) StralSV. Assessment of sequence variability within similar 3D structures and application to polio RNA-dependent RNA polymerase. BMC Bioinformatics 12, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gohara D. W., Arnold J. J., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol). Kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 43, 5149–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gohara D. W., Crotty S., Arnold J. J., Yoder J. D., Andino R., Cameron C. E. (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol). Structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275, 25523–25532 [DOI] [PubMed] [Google Scholar]
- 21. Korneeva V. S., Cameron C. E. (2007) Structure-function relationships of the viral RNA-dependent RNA polymerase. Fidelity, replication speed, and initiation mechanism determined by a residue in the ribose-binding pocket. J. Biol. Chem. 282, 16135–16145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garriga D., Ferrer-Orta C., Querol-Audí J., Oliva B., Verdaguer N. (2013) Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J. Mol. Biol. 425, 2279–2287 [DOI] [PubMed] [Google Scholar]
- 23. Gong P., Peersen O. B. (2010) Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thompson A. A., Albertini R. A., Peersen O. B. (2007) Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J. Mol. Biol. 366, 1459–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson A. A., Peersen O. B. (2004) Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23, 3462–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferrer-Orta C., Arias A., Agudo R., Pérez-Luque R., Escarmís C., Domingo E., Verdaguer N. (2006) The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 25, 880–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zamyatkin D. F., Parra F., Machín A., Grochulski P., Ng K. K. (2009) Binding of 2′-amino-2′-deoxycytidine-5′-triphosphate to norovirus polymerase induces rearrangement of the active site. J. Mol. Biol. 390, 10–16 [DOI] [PubMed] [Google Scholar]
- 28. Castro C., Smidansky E., Maksimchuk K. R., Arnold J. J., Korneeva V. S., Götte M., Konigsberg W., Cameron C. E. (2007) Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 104, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C. E. (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang X., Smidansky E. D., Maksimchuk K. R., Lum D., Welch J. L., Arnold J. J., Cameron C. E., Boehr D. D. (2012) Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 20, 1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang X., Welch J. L., Arnold J. J., Boehr D. D. (2010) Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry 49, 9361–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen H., Sun H., Li G. (2012) What is the role of motif D in the nucleotide incorporation catalyzed by the RNA-dependent RNA polymerase from poliovirus? PLoS Comput. Biol. 8, e1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weeks S. A., Lee C. A., Zhao Y., Smidansky E. D., August A., Arnold J. J., Cameron C. E. (2012) A polymerase mechanism-based strategy for viral attenuation and vaccine development. J. Biol. Chem. 287, 31618–31622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bell J. B., Eckert K. A., Joyce C. M., Kunkel T. A. (1997) Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J. Biol. Chem. 272, 7345–7351 [DOI] [PubMed] [Google Scholar]
- 35. Kaushik N., Pandey V. N., Modak M. J. (1996) Significance of the O-helix residues of Escherichia coli DNA polymerase I in DNA synthesis. Dynamics of the dNTP binding pocket. Biochemistry 35, 7256–7266 [DOI] [PubMed] [Google Scholar]
- 36. Ogawa M., Tosaka A., Ito Y., Yoshida S., Suzuki M. (2001) Enhanced ribonucleotide incorporation by an O-helix mutant of Thermus aquaticus DNA polymerase I. Mutat. Res. 485, 197–207 [DOI] [PubMed] [Google Scholar]
- 37. Suzuki M., Avicola A. K., Hood L., Loeb L. A. (1997) Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J. Biol. Chem. 272, 11228–11235 [DOI] [PubMed] [Google Scholar]
- 38. Suzuki M., Yoshida S., Adman E. T., Blank A., Loeb L. A. (2000) Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J. Biol. Chem. 275, 32728–32735 [DOI] [PubMed] [Google Scholar]
- 39. Arnold J. J., Bernal A., Uche U., Sterner D. E., Butt T. R., Cameron C. E., Mattern M. R. (2006) Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity. Anal. Biochem. 350, 214–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arnold J. J., Cameron C. E. (2000) Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275, 5329–5336 [DOI] [PubMed] [Google Scholar]
- 41. Gohara D. W., Ha C. S., Kumar S., Ghosh B., Arnold J. J., Wisniewski T. J., Cameron C. E. (1999) Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17, 128–138 [DOI] [PubMed] [Google Scholar]
- 42. Moustafa I. M., Shen H., Morton B., Colina C. M., Cameron C. E. (2011) Molecular dynamics simulations of viral RNA-dependent RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 410, 159–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 45. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 46. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald. An N [center-dot] log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 47. Ryckaert J.-P., Ciccotti G., Berendsen H. J. C. (1977) Numerical integration of the cartesian equations of motion of a system with constraints. Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 48. Arnold J. J., Vignuzzi M., Stone J. K., Andino R., Cameron C. E. (2005) Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 280, 25706–25716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deleted in proof [Google Scholar]
- 50. Arnold J. J., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol). Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43, 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arnold J. J., Gohara D. W., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol). Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 43, 5138–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cameron C. E., Castro C. (2001) The mechanism of action of ribavirin. Lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Curr. Opin. Infect. Dis. 14, 757–764 [DOI] [PubMed] [Google Scholar]
- 53. Crotty S., Cameron C., Andino R. (2002) Ribavirin's antiviral mechanism of action. Lethal mutagenesis? J. Mol. Med. 80, 86–95 [DOI] [PubMed] [Google Scholar]
- 54. Crotty S., Maag D., Arnold J. J., Zhong W., Lau J. Y., Hong Z., Andino R., Cameron C. E. (2000) The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6, 1375–1379 [DOI] [PubMed] [Google Scholar]
- 55. Gnädig N. F., Beaucourt S., Campagnola G., Bordería A. V., Sanz-Ramos M., Gong P., Blanc H., Peersen O. B., Vignuzzi M. (2012) Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, E2294–E2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crotty S., Hix L., Sigal L. J., Andino R. (2002) Poliovirus pathogenesis in a new poliovirus receptor transgenic mouse model. Age-dependent paralysis and a mucosal route of infection. J. Gen. Virol. 83, 1707–1720 [DOI] [PubMed] [Google Scholar]
- 57. Christodoulou C., Colbere-Garapin F., Macadam A., Taffs L. F., Marsden S., Minor P., Horaud F. (1990) Mapping of mutations associated with neurovirulence in monkeys infected with Sabin 1 poliovirus revertants selected at high temperature. J. Virol. 64, 4922–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tardy-Panit M., Blondel B., Martin A., Tekaia F., Horaud F., Delpeyroux F. (1993) A mutation in the RNA polymerase of poliovirus type 1 contributes to attenuation in mice. J. Virol. 67, 4630–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Georgescu M. M., Tardy-Panit M., Guillot S., Crainic R., Delpeyroux F. (1995) Mapping of mutations contributing to the temperature sensitivity of the Sabin 1 vaccine strain of poliovirus. J. Virol. 69, 5278–5286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McGoldrick A., Macadam A. J., Dunn G., Rowe A., Burlison J., Minor P. D., Meredith J., Evans D. J., Almond J. W. (1995) Role of mutations G-480 and C-6203 in the attenuation phenotype of Sabin type 1 poliovirus. J. Virol. 69, 7601–7605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baker S., Richards O. C., Ehrenfeld E. (1995) Elongation activity of poliovirus RNA polymerase derived from Sabin type 1 sequence is not temperature-sensitive. J. Gen. Virol. 76, 2081–2084 [DOI] [PubMed] [Google Scholar]
- 62. Paul A. V., Mugavero J., Yin J., Hobson S., Schultz S., van Boom J. H., Wimmer E. (2000) Studies on the attenuation phenotype of polio vaccines. Poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature-sensitive in the uridylylation of VPg. Virology 272, 72–84 [DOI] [PubMed] [Google Scholar]
- 63. Hansen J. L., Long A. M., Schultz S. C. (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 64. Beard W. A., Wilson S. H. (2003) Structural insights into the origins of DNA polymerase fidelity. Structure 11, 489–496 [DOI] [PubMed] [Google Scholar]
- 65. Franklin M. C., Wang J., Steitz T. A. (2001) Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105, 657–667 [DOI] [PubMed] [Google Scholar]
- 66. Johnson S. J., Beese L. S. (2004) Structures of mismatch replication errors observed in a DNA polymerase. Cell 116, 803–816 [DOI] [PubMed] [Google Scholar]
- 67. Kiefer J. R., Mao C., Braman J. C., Beese L. S. (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature 391, 304–307 [DOI] [PubMed] [Google Scholar]
- 68. Wu E. Y., Beese L. S. (2011) The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 286, 19758–19767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yin Y. W., Steitz T. A. (2004) The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116, 393–404 [DOI] [PubMed] [Google Scholar]
- 70. Kirmizialtin S., Nguyen V., Johnson K. A., Elber R. (2012) How conformational dynamics of DNA polymerase select correct substrates. Experiments and simulations. Structure 20, 618–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ren Z., Ghose R. (2011) Slow conformational dynamics in the cystoviral RNA-directed RNA polymerase P2. Influence of substrate nucleotides and template RNA. Biochemistry 50, 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ren Z., Wang H., Ghose R. (2010) Dynamics on multiple timescales in the RNA-directed RNA polymerase from the cystovirus phi6. Nucleic Acids Res. 38, 5105–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pfeiffer J. K., Kirkegaard K. (2003) A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U.S.A. 100, 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]