Background: Although Dzip1 positively influences Hedgehog signaling by regulating ciliogenesis, it inhibits the Hedgehog pathway through an unclear mechanism.
Results: Dzip1 knockdown destabilizes Spop E3 ubiquitin ligase, leading to increased Gli transcription factor levels and phenotypes resembling Hedgehog signaling activation.
Conclusion: Dzip1 inhibits Hedgehog signaling through stabilizing Spop.
Significance: We uncover a novel Gli regulatory mechanism.
Keywords: Cilia, Development, Hedgehog, Protein Degradation, Xenopus, Dzip1, Gli, Spop
Abstract
The Hedgehog (Hh) pathway is essential for embryonic development and adult tissue homeostasis. The Gli/Cubitus interruptus (Ci) family of transcription factors acts at the downstream end of the pathway to mediate Hh signaling. Both Hh-dependent and -independent Gli regulatory mechanisms are important for the output of Hh signaling. Daz interacting protein 1 (Dzip1) has bipartite positive and negative functions in the Hh pathway. The positive Hh regulatory function appears to be attributed to a requirement for Dzip1 during ciliogenesis. The mechanism by which Dzip1 inhibits Hh signaling, however, remains largely unclear. We recently found that Dzip1 is required for Gli turnover, which may account for its inhibitory function in Hh signaling. Here, we report that Dzip1 regulates Gli/Ci turnover by preventing degradation of speckle-type POZ protein (Spop), a protein that promotes proteasome-dependent turnover of Gli proteins. We provide evidence that Dzip1 regulates the stability of Spop independent of its function in ciliogenesis. Partial knockdown of Dzip1 to levels insufficient for perturbing ciliogenesis, sensitized Xenopus embryos to Hh signaling, leading to phenotypes that resemble activation of Hh signaling. Importantly, overexpression of Spop was able to restore proper Gli protein turnover and rescue phenotypes in Dzip1-depleted embryos. Consistently, depletion of Dzip1 in Drosophila S2 cells destabilized Hh-induced BTB protein (HIB), the Drosophila homolog of Spop, and increased the level of Ci. Thus, Dzip1-dependent stabilization of Spop/HIB is evolutionarily conserved and essential for proper regulation of Gli/Ci proteins in the Hh pathway.
Introduction
Members of the highly conserved Hedgehog (Hh)2 family of secreted proteins are essential in a variety of developmental processes. In many cases Hh ligands function as morphogens to regulate cell fate determination in a multicellular target field or as mitogens to control cellular proliferation and survival. Understanding the many orchestrated events that transduce the Hh signal is of considerable interest, as perturbations to the Hh pathway lead to a variety of developmental disorders (1–3). In addition, a wide range of cancers are caused by unchecked Hh signaling (4–6). At the molecular level the Cubitus interruptus (Ci)/Gli family of zinc finger transcription factors are key downstream effectors of Hh signaling. Before pathway activation, Ci/Gli are proteolytically processed, leading to truncated repressor forms that suppress the expression of Hh target genes (7–9). Hh proteins initiate the signaling by binding to its receptor Patched (Ptc) (10–12) and relieving the inhibition of Smoothened (Smo) by Ptc (13, 14). Activation of Smo leads to a series of intracellular signal transduction events that ultimately prevent proteolytic processing of Ci/Gli and convert Ci/Gli to transcriptional activators that, in turn, promote Hh target gene expression. In vertebrates, activation of Smo requires translocation of Smo into the primary cilium. Interestingly, the cilium is dispensable for Drosophila Hh signaling, wherein the pathway becomes activated once Smo is accumulated on the cell membrane (15–20).
In addition to Hh-dependent regulation of Ci/Gli processing, Hh-independent Ci/Gli regulatory mechanisms also influence the output of Hh signaling. It has been shown that Speckle-type POZ protein (Spop) and its Drosophila homolog Hh-induced BTB protein (HIB, also known as Roadkill (Rdx)), substrate binding adaptors of cullin3 E3 ubiquitin ligase (21), directly interact with Ci/Gli and promote their degradation by the proteasome (22–24). In vertebrates, the function of Spop/Cullin3 ubiquitin ligase is antagonized by Suppressor of Fused (Sufu), another Gli interacting protein (25–27). It is believed that Sufu competes with Spop for Gli binding and protects Gli2 and Gli3 from proteasome degradation (22, 23). In Drosophila, HIB functions to promote Ci degradation (24, 28, 29). Loss of Hib leads to increased Ci and elevated Hh signaling, whereas its overexpression inhibits Hh signaling by decreasing Ci levels (28, 29). Thus, the function of HIB/Spop in Ci/Gli turnover is conserved between vertebrates and invertebrates. Nevertheless, Sufu is dispensable for Drosophila Hh signaling (30).
Daz interacting protein 1 (Dzip1) has been implicated in the Hh pathway. Dzip1 is a C2H2 zinc finger domain protein with coiled-coil domains and a nuclear localization signal. Requirements for Dzip1 during embryonic development have been best studied using zebrafish iguana mutants, which harbor specific mutations in the Dzip1 gene (31, 32). Iguana mutants exhibit midline and somitic muscle defects, both hallmark phenotypes of Hh loss of function in zebrafish. Interestingly, although global Hh signaling activity is reduced in iguana mutant embryos, low levels of ectopic expression of Hh-responsive genes, ptc1 and engrailed 1, were also observed. This suggests that Dzip1 can regulate the Hh signaling pathway in a positive and negative manner (31–33). Subsequent investigations revealed that the positive role of Dzip1 in the Hh pathway is due to its requirement for ciliogenesis (34–37). Regarding the inhibitory function of Dzip1, we recently provided evidence that Dzip1 is required for Gli turnover (38). In this study we extend our previous findings by demonstrating that Dzip1 regulates Gli turnover through stabilizing Spop. We provide evidence that Dzip1 regulates Spop independent of its role in ciliogenesis. We show that decreased Spop protein levels in response to knockdown of Dzip1 can account for elevated Gli protein levels and increased Hh target gene expression. Collectively, these findings provide mechanistic insight to explain the ectopic Hh pathway activation in Dzip1-deficient embryos. Furthermore, we show that Dzip1 stabilizes HIB and regulates Ci turnover in Drosophila S2 cells. Taken together, our work demonstrates that the novel regulatory mechanism involving Dzip1-Spop-Gli is evolutionarily conserved and essential for the output of Hh signaling.
EXPERIMENTAL PROCEDURES
Xenopus Embryos and Manipulations
Xenopus embryos were obtained and injected as described (39). Embryo sectioning and staining were performed as described (39). The dosage of RNA or morpholino for microinjection is indicated in the text or figure legend. For experiments in which RNA and morpholino was injected into the same embryo, injections were performed sequentially. Morpholinos against Dzip1 (DMO1 and DMO2) (38), Gli1 and -2 (40), and intraflagellar transport protein 88 (IFT88)/Polaris (41) were described. For lactacystin treatment, animal caps were dissected at the late blastula stage and cultured in 1× Marc's modified Ringers with or without 2 μg/ml lactacystin. Caps were harvested at stage 15 for Western blotting. For purmorphamine treatments, a 10 mm stock solution in 100% ethanol was diluted to 2.5–10 μm in 0.5× Marc's modified Ringers. Embryos were transferred into 0.5× Marc's modified Ringers containing purmorphamine or vehicle at stage 14 and harvested at stage 35.
Expression Constructs
Xenopus Sonic hedgehog (Shh) (42), Xenopus Gli1, Xenopus Gli2, and human Gli3 (38) expression constructs were described. The full-length Spop and Spop deletion constructs were constructed by standard PCR/cloning methods. These constructs were derived from mouse Spop (IMAGE: 6825425). The Gli21–799 mutant was generated by site-directed mutagenesis. FLAG-HIB and FLAG-dDzip were constructed by cloning individual cDNA sequence in-frame with the 2XFLAG-UAST backbone. FLAG-Ci has been described (43). Cloning details will be provided upon request.
Quantitative Real Time PCR, Whole Mount in Situ Hybridization, and Immunostaining
RNA extraction and RT-PCR method were described (44). Real-time PCR reactions were performed in triplicate using SYBR Green master mix (Applied Biosystems) on the Applied Biosystems 7500 Real-time PCR System. PCR primers for odc, ptc1, and gli1 were described (38). Whole mount in situ hybridization was performed as described (39). For sectioning embryos after whole-mount in situ hybridization, embryos were washed 3–5× in PBS, suspended in 4% low-melt agarose, 0.5× PBS, and sectioned at 100 μm using a Vibratome (Series 100, Electon Beam Sciences). Cilia were stained by labeling acetylated tubulin (anti-acetylated tubulin, Sigma). Superficial slow fibers and total muscle fibers were stained using monoclonal antibodies BA-F8 and 12/101 (Developmental Studies Hybridoma Bank), respectively.
Immunoprecipitation and Western Blots
Immunoprecipitations and Western blots were performed as described (45). To detect ubiquitinated Spop, an antigen retrieval step was added after proteins were transferred to the PVDF membrane. Membranes were washed 3× in TBS-Tween, placed protein side up on a water-soaked filter paper, wrapped in saran wrap, and heated in a pressure cooker for 25 min. After antigen retrieval, the membrane was washed 3× in TBS-Tween and used immediately for Western blotting. Antibodies were anti-Myc (9E10, Sigma, 1:1,000), anti-FLAG (M2, Sigma, 1:1,000), anti-Spop (Santa Cruz, 1:200), anti-ubiquitin (Santa Cruz, 1:500), and anti-β-tubulin (Sigma, 1:2,000).
Cell Culture, Transfection, RNAi, and Treatments
NIH3T3 cells were cultured and transfected as described (46). S2 cell culture and transfection were performed with standard protocols, and the use of dsRNA has been described (43). dDzip1 dsRNA was synthesized against the region of nucleotide 91–676 of dDzip1 coding sequence. MG132 (Calbiochem) was added to inhibit proteasome activity at a final concentration of 50 μm for 4 h before harvesting the cells.
RESULTS
Partial Knockdown of Dzip1 Sensitizes Xenopus Animal Caps to Shh
Gli regulatory mechanisms have an important impact on the output of Hh signaling. Dzip1 is required for Gli turnover (38) and ciliogenesis (34–36, 38). We previously reported that these functions of Dzip1 could be uncoupled. Stabilization of Gli proteins could be detected even when Dzip1 was partially knocked down. In contrast, ciliogenesis defects occurred only when the depletion of Dzip1 was more complete (38). Because different levels of Dzip1 are required for its functions in Gli turnover and ciliogenesis, we set out to investigate how the Dzip1-dependent Gli regulatory mechanism influences the output of Hh signaling by partially depleting Dzip1.
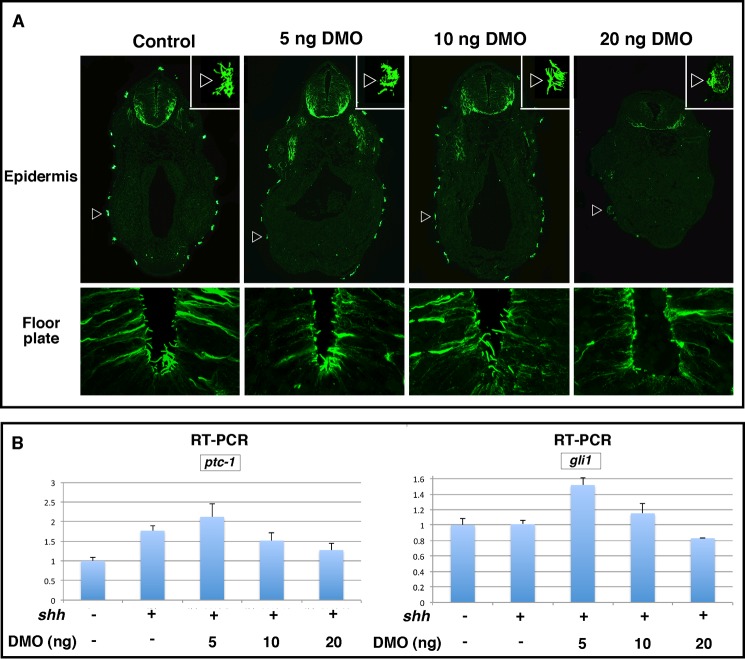
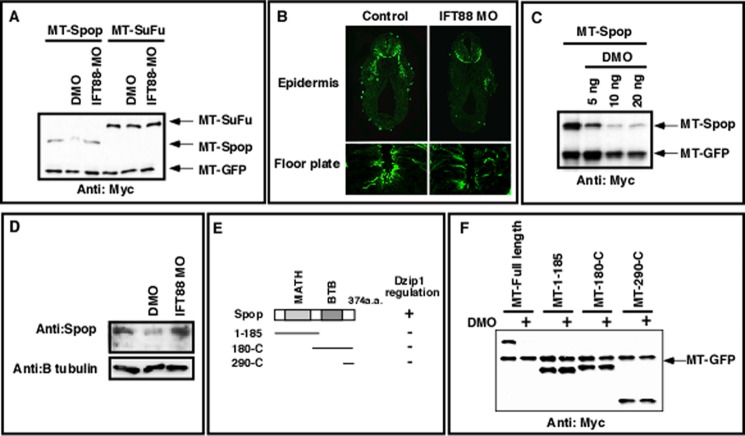
Using a combination of two Dzip1 morpholinos (DMO, described in Jin et al. (38)), we knocked down Dzip1 to various degrees and examined the response of cells to Shh in an animal cap assay. In parallel, we examined the formation of floor plate primary cilia and skin motile cilia in tadpoles. We found that injection of 5 ng of DMO, which had no effect on the formation of cilia (Fig. 1A), caused a weak but reproducible increase in Shh-induced expression of gli1 and ptc1 in animal caps (Fig. 1B). Embryos receiving 10 ng of DMO displayed intermediate phenotypes (Fig. 1, A and B). Injection of higher doses of DMO (20 ng) caused defective primary and motile cilia (Fig. 1B) and reduced Shh-induced expression of gli1 and ptc1 (Fig. 1A). These data indicate that partial depletion of Dzip1 sensitizes animal caps to Shh.
FIGURE 1.
Sensitization of Xenopus animal caps to Shh by partial knockdown of Dzip1. A, immunofluorescence showing primary cilia in the floor plate (bottom panel) and epidermis motile cilia (top panel, arrowheads) in control (left column) and embryos injected with 5, 10, or 20 ng of DMOs (from left to right). B, real-time RT-PCR showing the expression of ptc1 and gli1 in animal caps from controls and embryos injected with shh mRNA (500pg) alone or in combination with 5, 10, or 20 ng of DMOs. All samples were normalized to odc levels.
Partial Knockdown of Dzip1 Leads to Ectopic Hh Signaling in Whole Embryo
In light of the above findings, we next sought to perform a detailed phenotypic characterization of embryos after partial Dzip1 depletion. We speculated that partially depleting Dzip1 levels, below the threshold to perturb ciliogenesis, would result in phenotypes characteristic solely of Hh activation in the embryo. To do so, a low concentration (5 ng) of DMO was injected at the one cell stage, and phenotypic characterization was carried out carefully at the tadpole stage.
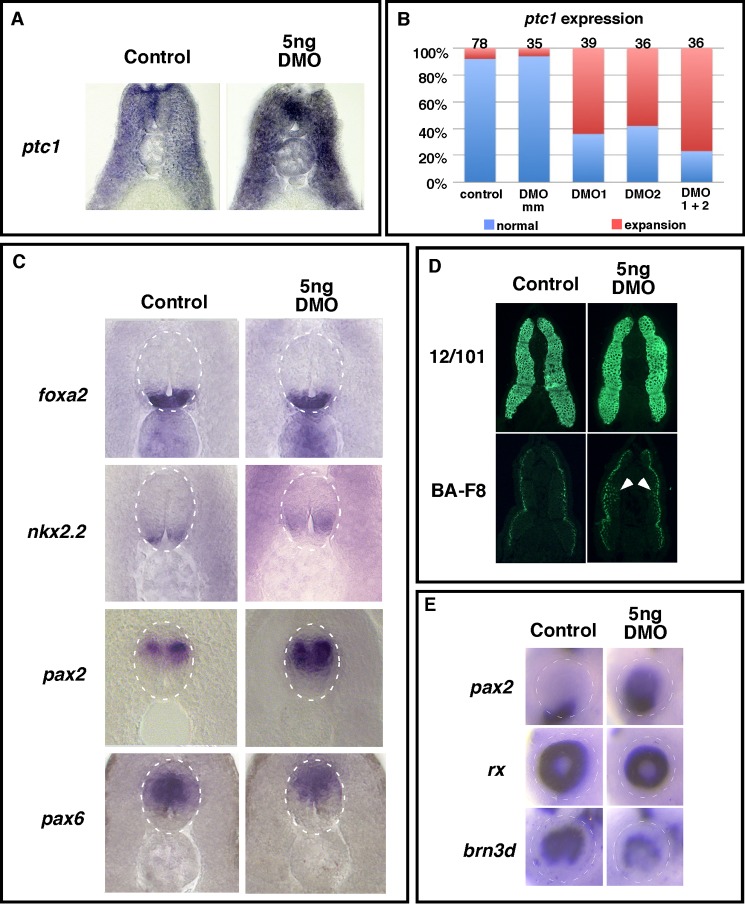
To both confirm results from the animal cap assay, which showed that a low DMO dose could enhance Shh-induced target gene expression, and investigate how Hh signaling was spatially regulated in DMO injected embryos, we examined ptc1 expression in the neural tube and somitic mesoderm, two tissues whose development is regulated by Hh signaling. As shown in Fig. 2A, ptc1 was markedly enhanced throughout the neural tube and somitic tissues after DMO injection (78%, n = 36) compared with controls, indicating that Hh signaling is enhanced throughout both tissues. Importantly, the enhanced ptc1 expression phenotype was visualized when DMO1 or DMO2, which target different sequences of Dzip1 mRNA, was injected individually or in combination, indicating that increased ptc1 expression was not an off-target effect. In addition, injection of a five-base pair (bp)-mismatch control morpholino yielded ptc1 staining patterns that were indistinguishable from uninjected controls (Fig. 2B), further supporting the specificity of Dzip1 knockdown by DMOs.
FIGURE 2.
Partial depletion of Dzip1 increased the expression of Hh target genes in embryos. A, in situ hybridization showing that partial Dzip1 depletion (right) markedly increased the expression of ptc1 in the neural tube and somites at stage 35. B, summary of the ptc1 expression phenotypes in control and morpholino-injected embryos. DMO mm, 5-bp mismatch control morpholino of DMO2. C, in situ hybridization showing the expression of floor plate markers (foxa2 and nkx2.2) and markers for more dorsal neural tube cells (pax2 and pax6) in controls (left) and embryos injected with DMO (right). The white dotted lines outline the neural tube borders. D, immunofluorescence showing total muscle (12/101 reactivity; top panel) and slow muscle cells (BA-F8 reactivity; bottom panel) in controls (left column) and DMO-injected embryos (right column). Arrowheads denote ectopic BA-F8 reactive slow muscle cells evident in DMO injected embryos. E, in situ hybridization showing eye patterning in controls (left) and DMO injected (right) embryos. Markers for optic stalk (pax2, top panel), total retina (rx, middle panel), and dorsal retina (brn3d, bottom panel) were analyzed. Dotted lines outline the eye.
To determine how partial Dzip1 depletion impacts Hh pathway-dependent developmental processes, we analyzed markers of neural tube, somite, and eye in DMO-injected tadpoles. In the neural tube, proper expression of foxa2 and nkx2.2, markers for medial and lateral floor plate respectively, is dependent on Hh signals from the underlying notochord (47–49). In Xenopus, these markers are differentially sensitive to increased Hh signaling, with only nkx2.2 being affected after up-regulation of Hh signaling (50). We found that there was no distinguishable difference in foxa2 expression between DMO injected (n = 30) and control embryos (n = 30). The expression of nkx2.2, however, was expanded dorsally in DMO-injected embryos (58%, n = 36) compared with controls (n = 33). The expression of pax2, which is localized to an adjacent dorsal region to the nkx2.2 domain and is positively regulated by Hh signaling (50), was enhanced and expanded dorsally in DMO injected embryos (59%, n = 32) compared with controls (n = 25). The expression of pax6, a gene that is restricted from the ventral neural tube and negatively regulated by Hh signaling (47, 51, 52), was reduced and shifted dorsally in DMO-injected embryos (50%, n = 42) compared with controls (n = 27) (Fig. 2C). Collectively, marker expression in the neural tubes of DMO injected embryos is suggestive of enhanced Hh signaling, in accordance with the increased ptc1 expression in morphant neural tubes.
Hh signaling regulates cell fate determination in the somitic mesoderm. In Xenopus and zebrafish embryos, superficial slow fibers (SSF) arise at the lateral border of the somite. Specification of SSF depends on low levels of Hh signals from the notochord (53–55). Ectopic Hh signaling in Xenopus embryos results in a mis-positioning and an increased incidence of SSF within more medial positions of the posterior somite (55). To determine how partial Dzip1 depletion affected somitic mesoderm development, we examined total muscle fiber and SSF formation in posterior trunk sections of DMO-injected embryos. The monoclonal antibodies, BA-F8 and 12/101, that react with SSF and all muscle fibers (55), respectively, were utilized for this analysis. In controls, BA-F8 reactivity was confined to a single layer of cells at the lateral edge of 12/101 staining field (n = 20). In contrast, DMO-injected embryos contained more slow muscle fibers (57%, n = 14), with ectopic BA-F8-reactive cells arising in more medial layers of the 12/101 staining field (arrowheads) (Fig. 2D). The SSF expansion in DMO-injected embryos is reminiscent of Hh activation phenotypes, consistent with enhanced ptc1 expression throughout the somitic mesoderm in these embryos.
Hh signaling regulates multiple aspects of eye development, including dorsoventral patterning in the eye. During eye patterning, the optic vesicle becomes specified into a ventral domain, which gives rise to the optic nerve, and a dorsal domain that forms the retina. During zebrafish, Xenopus, and chick development, elevated Hh signaling represses eye field transcription factors and activates forebrain differentiation markers. This results in an expansion of the ventral forebrain, which includes the optic nerve, at the expense of retinal primordia, leading to a small eye phenotype with reduced numbers of retinal cells (51, 56–62). We assessed eye-patterning phenotypes in DMO-injected embryos by examining expression of pax2, a ventral optic stalk marker, as well as retinal markers rx and brn3d. In controls, pax2 expression was confined to the ventral extent of the eye (n = 67), whereas rx and brn3d were expressed throughout the dorsoventral extent of the eyefield (n = 23, rx; n = 47, brn3d). Partial Dzip1 depletion disrupted the dorsoventral organization of the eye. In DMO-injected eyes, pax2 expression was expanded dorsally (59%, n = 90), and the ventral expression domain of brn3d was significantly reduced (54%, n = 48). Partial knockdown of Dzip1 was not sufficient to down-regulate rx expression. However, the size of the rx expression domain was smaller than that in controls (57%, n = 28) (Fig. 2E), indicating that DMO-injected embryos had smaller eyes than their siblings. These observations are again consistent with the notion that partial knockdown of Dzip1 elevates Hh signaling. Collectively, partial knockdown of Dzip1 resulted in phenotypes reminiscent of Hh activation during neural tube, somite, and eye patterning.
Partial Knockdown of Dzip1 Sensitizes Embryos to Smo Agonist Purmorphamine
To further strengthen the conclusion that partial depletion of Dzip1 sensitizes cells to Hh signaling, we determined whether partial Dzip1 knockdown enhances the response of embryos to purmorphamine, a Smo agonist. Thus, uninjected and DMO-injected embryos were treated with various doses of purmorphamine. The expression of ptc1and pax6 in the neural tube and that of pax2 during eye development was analyzed at the tadpole stage.
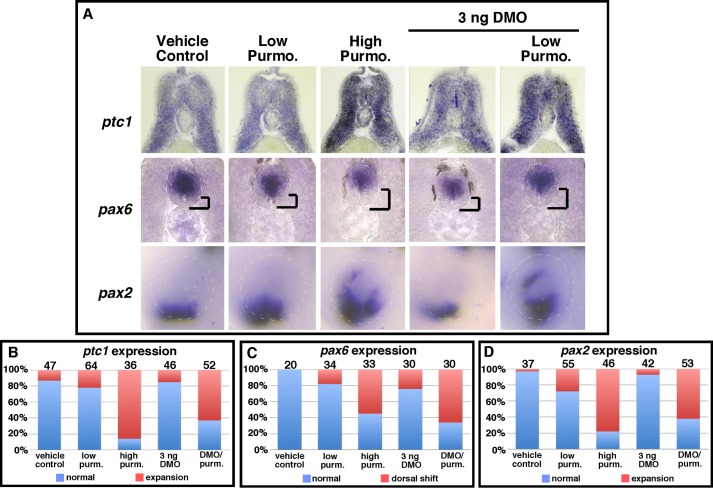
As expected, in embryos treated with a high dose of purmorphamine (10 μm), we observed an expansion of ptc1 (86%, n = 36), a reduction and dorsal shift in the pax6 expression domain in the neural tube (57%, n = 33), and dorsally expanded pax2 expression in the eye (78%, n = 46) (Fig. 3A) in a similar manner to DMO-injected embryos (Fig. 2). By comparison, the expression of ptc1 (n = 47), pax6 (n = 20), or pax2 (n = 37) in vehicle-treated (ethanol) controls (Fig. 3A) was similar to nontreated controls (Fig. 2). To test if partial knockdown of Dzip1 sensitizes embryos to purmorphamine, we performed titration experiments to determine doses of both purmorphamine and DMO that were below the threshold capable of altering marker gene expression when delivered on their own. We found that the majority of 3 ng of DMO injections were indistinguishable from controls with respect to ptc1 (85%, n = 46), pax6 (78%, n = 30), and pax2 (93%, n = 42) expression. Similarly, a purmorphamine dose of 2.5–5.0 μm often led to normal phenotypes with respect to the expression of ptc1 (78%, n = 64), pax6 (82%, n = 34), and pax2 (72%, n = 55) (Fig. 3A). Notably, low doses of DMO (3 ng) and purmorphamine (2.5–5.0 μm) synergized when delivered in combination, resulting in enhanced expression of ptc1 (63%, n = 52), reduction and dorsal expansion of pax6 (65%, n = 30), and elevated pax2 (62%, n = 53) expression, in a similar fashion to higher doses of purmorphamine or Dzip1 knockdown (Fig. 3A). The gene expression changes for all treatments/injection groups are summarized in Fig. 3, B--D. Taken together, our results demonstrate that partial depletion of Dzip1 to levels insufficient for perturbing ciliogenesis sensitizes cells to Hh signaling.
FIGURE 3.
Partial depletion of Dzip1 sensitizes embryos to the Smo agonist purmorphamine. A, whole mount in situ hybridization of ptc1 (top), pax6 (middle), and pax2 (bottom) in vehicle-treated (ethanol) controls and embryos treated with either a low dose of purmorphamine (2.5–5.0 μm), a high dose of purmorphamine (10 μm), injected with a low concentration of DMO (3 ng), or receiving a combination of a low dose of purmorphamine (Purmo. or purm.) and an injection with a low concentration of DMO. In the middle panels, the distance between the floor plate and the ventral border of the pax6 expression domain was highlighted. B–D, summary of ptc1 expression phenotypes (B), pax6 expression phenotypes (C), or pax2 expression phenotypes (D) for vehicle-treated (ethanol) controls and the different groups of embryos subjected to purmorphamine, DMO, or both.
Knockdown of Gli1 or Gli2 Rescues Phenotypes Induced by Partial Knockdown of Dzip1
Dzip1 regulates Gli turnover (38). To determine if partial knockdown of Dzip1 sensitizes cells to Hh signaling by stabilizing Gli proteins, we performed double knockdown experiments in which morpholino against Gli1 or Gli2 was injected together with DMO. We then assayed the expression of ptc1, pax6, and pax2 in these embryos.
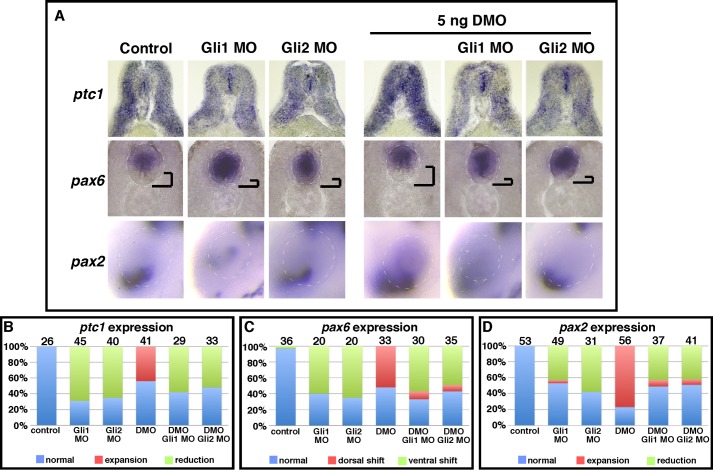
As expected, knockdown of either Gli1 or Gli2 alone suppressed Hh signaling, as evidenced by reduced ptc1 expression throughout the neural tube and somites (Gli1-MO, 69%, n = 45; Gli2-MO, 65%, n = 40), ventral expansion of pax6 in the neural tube (Gli1-MO, 60%, n = 20; Gli2-MO, 65%, n = 20), and reduction of pax2 in the ventral eye (Gli1-MO, 43%, n = 49; Gli2-MO, 58%, n = 31). Next, we compared double knockdown embryos to DMO-injected embryos. Consistent with findings presented in Fig. 2, injection of DMO alone enhanced ptc1 expression (44%, n = 41) (Fig. 4A). In contrast, injection of DMO/Gli1-MO or DMO/Gli2-MO reduced ptc1 expression in 58% (n = 29) and 52% (n = 33) of embryos (Fig. 4A), respectively. In accordance with this, knockdown of Gli1 or Gli2 in combination with DMO was capable of rescuing the dorsal shift of pax6 expression that is evident in the neural tube after DMO injection alone (52%, n = 33) (Fig. 4A). Moreover, double knockdown often caused a reversal of the phenotype, leading to ventral expansion of pax6 within the neural tube after either DMO/Gli1-MO (57%, n = 30) or DMO/Gli2-MO (49%, n = 35) injection (Fig. 4A). As previously reported in Fig. 2, DMO-injected embryos display enhanced pax2 expression throughout the eye (77%, n = 56) (Fig. 4A). By comparison, knockdown of Gli1 or Gli2 in DMO-injected embryos prevented DMO-induced dorsal expansion of pax2. Moreover, double knockdown often led to an overall reduction of pax2 expression in the eye (43%, n = 37, DMO/Gli1-MO; 41%, n = 41, DMO/Gli2-MO). These results are summarized in Fig. 4, B--D. Collectively, these data support the notion that Dzip1 influences the sensitivity of cells to Hh signaling by stabilizing Gli proteins.
FIGURE 4.
Knockdown of Gli1 or Gli2 rescues phenotypes induced by partial depletion of Dzip1. A, whole mount in situ hybridization of ptc1 (top), pax6 (middle), and pax2 (bottom) in controls and embryos receiving either a single injection of Gli1 MO, Gli2 MO, or DMO and double knockdown embryos injected with Gli1 MO/DMO or Gli2 MO/DMO. In the middle panels, the distance between the floor plate and the ventral border of the pax6 expression domain was highlighted. B--D, summary of ptc1 expression phenotypes (B), pax6 expression phenotypes (C), or pax2 expression phenotypes (D) in controls and the different groups of embryos receiving either a single or double morpholino injection against Gli1, Gli2, or Dzip1.
Dzip1 Regulates Spop Stability in a Cilia-independent Manner
The Spop/HIB-dependent regulation of Gli/Ci is one of the best-characterized Gli/Ci regulatory mechanisms. Spop/HIB promotes Gli/Ci ubiquitination and degradation through recruiting Gli/Ci to the Cullin3 E3 ligase (24, 28, 29). In vertebrates, the function of Spop is antagonized by Sufu, which stabilizes Gli by competing with Spop for Gli binding (22, 23).
Our motivation to understand the mechanism by which Dzip1 regulates Gli turnover led us to examine whether Dzip1 regulates Gli turnover by controlling Spop or Sufu. Strikingly, we found that the stability of overexpressed Spop was decreased dramatically in DMO-injected embryos. By contrast, Sufu remained unaltered in Dzip1-depleted embryos (Fig. 5A). Interestingly, the level of Spop remained unaffected in IFT88/Polaris-depleted embryos (Fig. 5A), which exhibited severe ciliogenesis defects (Fig. 5B). Moreover, reduced levels of Spop were detected in embryos injected with 5 ng of DMO (Fig. 5C), a dose of DMO insufficient for disturbing ciliogenesis. Importantly, we found that endogenous Spop was destabilized after knockdown of Dzip1 but was unaffected by IFT88 knockdown (Fig. 5D). These results demonstrate that Dzip1-mediated regulation of Spop occurs in a cilia-independent fashion.
FIGURE 5.
Dzip1 regulates the stability of Spop independent of its role in ciliogenesis. A, Western blot showing that the stability of Spop, but not that of Sufu, was decreased in Dzip1-depleted embryos. RNAs encoding MT-Spop (250 pg) or MT-Sufu (250 pg) were co-injected with MT-GFP (50 pg) into controls or embryos that had received a prior injection of DMO or IFT88 morpholino (MO). Embryos were harvested at the neurula stage. MT-GFP serves as injection and loading control. B, immunofluorescence showing a significant reduction of motile cilia on the epidermis (top panel) and primary cilia in the floor plate (bottom panel) within embryos injected with 40 ng IFT88 morpholino (right column) compared with controls (left column). C, DMO injection negatively impacted Spop stability in a dose-dependent fashion. MT-Spop was injected with MT-GFP into controls or embryos that had received increasing concentrations of DMO (5–20 ng). Expression of Spop was assessed by Western blotting. D, endogenous Spop protein was reduced in Dzip1-depleted embryos and unchanged in IFT88-depleted embryos. Embryos were harvested at stage 18 and subjected to Western blot with the anti-Spop antibody to detect endogenous Spop levels. Detection of β-tubulin levels serves as a loading control. E, schematic presentation of Spop deletion constructs. All deletion constructs were N-terminally myc-tagged. F, Western blot results showing the effects of Dzip1 knockdown on the stability of full-length and deletions of Spop. All constructs were co-injected with MT-GFP. Embryo lysates were subjected to Western blot with the anti-myc antibody.
Spop has two functional domains, an N-terminal MATH domain and a C-terminal BTB (Bric-a-brac/Tramtrack/Broad complex) domain (63). We sought to determine whether individual functional domains mediate Dzip1-dependent regulation of Spop. Several Spop deletion constructs (Fig. 5E) were generated and expressed in embryos. We found that the expression of all deletion constructs was unchanged after Dzip1 depletion (Fig. 5F), suggesting that both MATH and BTB domains are required for Spop to be regulated by Dzip1. In full, these data indicate a novel function of Dzip1 in controlling Spop stability.
Dzip1 Protects Spop/HIB from Proteasome-dependent Degradation
To better understand how Spop protein stability is regulated, we examined whether Spop is ubiquitinated. Thus, we purified myc-tagged (MT)-Spop from embryo lysate by immunoprecipitation using an anti-myc antibody. Purified Spop was separated on SDS-PAGE and analyzed by immunoblotting using anti-myc and anti-ubiquitin antibodies.
The predicted size of MT-Spop is ∼68 kDa. As shown in the anti-myc immunoblot in Fig. 6A, the majority of MT-Spop proteins are ∼68 kDa. Interestingly, we could detect the presence of slower migrating bands ranging in size from 85 to 170 kDa, well above the unmodified MT-Spop protein at 68 kDa, indicating that Spop protein becomes modified in embryos. Consistent with this, myc immunoprecipitates probed with an anti-ubiquitin antibody revealed a smear of high molecular weight forms, ranging in size from 85 to 170 kDa (Fig. 6A). Importantly, these bands were only present in immunoprecipitates from embryos expressing MT-Spop, indicating that they represent ubiquitinated Spop. This suggests that Spop undergoes ubiquitin/proteasome-dependent degradation.
FIGURE 6.
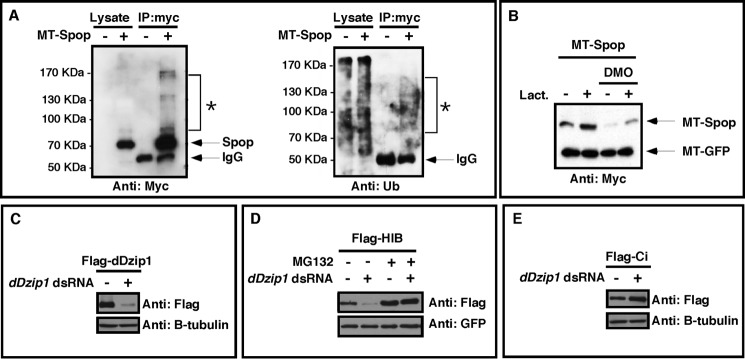
Dzip1 prevents ubiquitin/proteasome-dependent degradation of Spop/HIB. A, Spop is ubiquitinated in vivo. MT-Spop was injected into Xenopus embryos. Spop protein was purified by immunoprecipitation using an anti-myc antibody. Lysates and immunoprecipitates (IP) from injected and uninjected embryos were subjected to Western blot with the anti-myc antibody (left panel) and anti-ubiquitin antibody (right panel). The unmodified MT-Spop band migrated to 68 kDa. A smear of high molecular weight proteins (*) migrating slower than unmodified MT-Spop could be detected in both myc and ubiquitin Westerns. B, MT-Spop was injected into control or embryos that had received a prior injection of DMO. Animal caps were dissected at stage 9. Subsequently, caps were cultured in media with or without 2 μg of lactacystin (Lact.). Animal caps were harvested at stage 15 and subjected to Western blot to detect the levels of Spop. C, Western blot results showing severe reduction of FLAG-dDzip1 by dDzip1 dsRNA in S2 cells. β-Tubulin serves as a loading control. D, Dzip1 knockdown promoted proteasome-dependent degradation of HIB in S2 cells. S2 cells were cotransfected with FLAG-HIB and GFP constructs and treated with dDzip1 dsRNA and/or MG132. Cell extracts were subjected to Western blot with the anti-FLAG antibody to detect the levels of HIB. GFP serves as the transfection and loading control. E, Dzip1 knockdown promotes Ci stability in S2 cells. S2 cells were transfected with FLAG-Ci and treated with dDzip1 dsRNA. Western blots were carried out with the cell lysates to determine the levels of Ci. Tubulin serves as a loading control.
To determine if Dzip1 prevents proteasome-dependent degradation of Spop, we examined the effect of the proteasome inhibitor lactacystin on Spop protein levels in an animal cap assay. By dissecting animal caps from embryos, the vitelline membrane was removed, allowing MT-Spop-expressing cells to become exposed to lactacystin in the culture media. As shown in Fig. 6B, DMO-induced degradation of Spop was blocked by treatment of lactacystin. These data indicate that Dzip1 prevents Spop from proteasome-dependent degradation.
We next addressed whether regulation of Spop by Dzip1 is evolutionarily conserved. To achieve this, we knocked down dDzip1 in Drosophila S2 cells by RNAi (Fig. 6C) and examined the stability of HIB, the fly homolog of Spop. Indeed, knockdown of dDzip in S2 cells decreased the levels of HIB protein. The effect of dDzip1 RNAi on HIB was blocked by MG-132, a proteasome inhibitor (Fig. 6D), demonstrating that dDzip1 prevents proteasome-dependent turnover of HIB protein. As predicted, the level of Ci protein was increased in dDzip1 RNAi-treated cells (Fig. 6E). It appears that Dzip1-dependent regulation of Spop is evolutionarily conserved and essential for controlling the stability of Ci/Gli.
Overexpression of Spop Rescues Phenotypes Induced by Partial Knockdown of Dzip1
Knowing Dzip1 prevents proteasome-dependent turnover of Spop, we set out to determine whether phenotypes induced by partial Dzip1 knockdown could be rescued by overexpression of Spop.
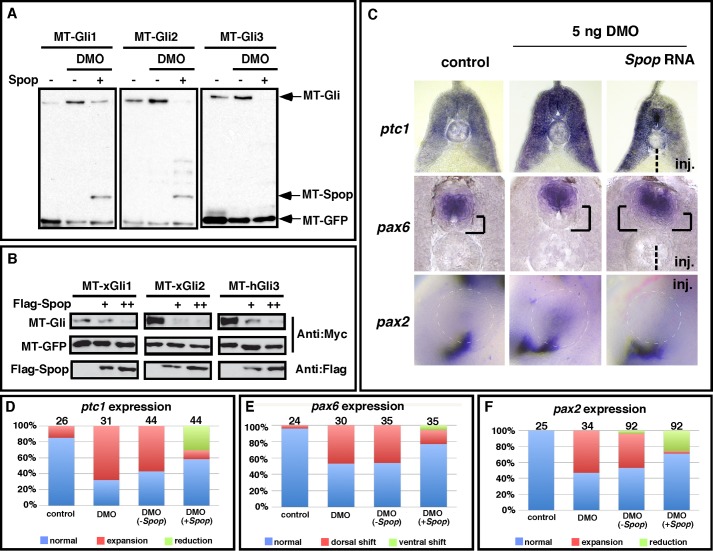
First, we tested the potential of Spop to rescue Gli turnover in Dzip1 knockdown embryos. As shown in Fig. 7A, forced Spop expression in Dzip1 morphants abolished the DMO-induced stabilization of xGli1, xGli2, and hGli3. Previous studies reveal that the stability of mammalian Gli1 is not sensitive to Spop (23, 24). Because Spop prevented DMO-induced stabilization of Xenopus Gli1 in our experiments, we tested the effects of Spop overexpression on xGli1, xGli2, and hGli3 in NIH3T3 cells. We found that xGli1, xGli2, and hGli3 were all destabilized upon Spop expression (Fig. 7B). This suggests that loss of Spop activity in DMO-injected embryos accounts for stabilization of all three Glis.
FIGURE 7.
Overexpression of Spop rescues phenotypes caused by partial knockdown of Dzip1. A, Western blot results showing that increases to Gli stability after Dzip1 depletion were reversed by Spop overexpression. RNAs encoding MT-Gli1–3 (2 ng) and MT-GFP (50 pg) were injected alone or in combination with MT-Spop (100 pg) into controls or embryos that had received a prior injection of DMO. Embryo were harvested at the neurula stage and subjected to Western blot with the anti-myc antibody to detect the levels of Spop. B, forced Spop expression decreases the stability of xGli1, xGli2, and hGli3 in NIH3T3 cells. NIH3T3 cells were transfected with MT-Glis and MT-GFP alone or in combination with increasing levels of FLAG-Spop. Western blots were carried out with the cell lysates to determine the levels of Gli1, -2, and -3. GFP serves as a transfection and loading control. C, in situ hybridization results showing the expression of ptc1, pax6, and pax2 in controls or embryos injected with DMO alone or DMO and Spop RNA. inj. indicates the side that received Spop RNA injection. D–F, summary of the ptc1 (D), pax6 (E), and pax2 (F) expression phenotypes in controls and injected embryos.
Next we sought to determine whether restoration of Spop expression in DMO-injected embryos was sufficient to rescue phenotypes induced by partial Dzip1 depletion. To achieve this, embryos were injected with DMO at the one-cell stage. Subsequently, RNAs encoding Spop and the lineage tracer were injected into one side of DMO-injected embryos. Embryos were harvested at the tadpole stage and subjected to in situ hybridization for ptc1, pax6, and pax2. The marker gene expression was compared between the Spop-injected and -uninjected sides.
As seen in Fig. 7C, ptc1 expression on the Spop-injected side was markedly reduced compared with the side that did not receive an injection of Spop mRNA. Regarding pax6, overexpression of Spop was sufficient to reverse dorsal expansion of pax6 in the neural tube. Pax6 expression on the Spop-injected side often resembled control embryos and occasionally displayed a ventral shift, similar to Gli1- and Gli2-depleted embryos. Consistent with these data, the expansion of pax2 in the eye field of DMO-injected embryos was reversed by Spop overexpression (Fig. 7C). These results are summarized in Fig. 7, D--F. Collectively, these data indicate that Dzip1 exerts its inhibitory effect on Hh signaling through stabilizing Spop, which targets Gli for proteasome-dependent degradation.
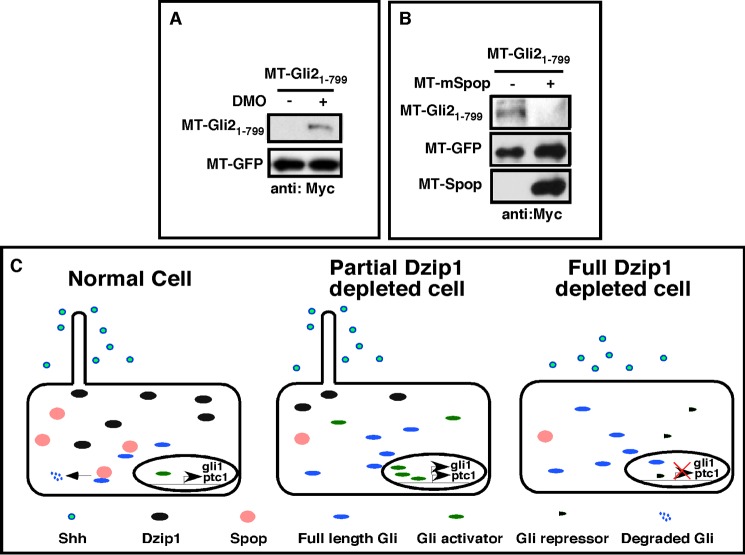
In zebrafish, the Hh signaling inhibitory effect of the you-too (yot) allele, which encodes a dominant negative mutant of Gli2 due to a nonsense mutation at Arg-929 (64), is markedly enhanced in iguana mutants (31, 32). The molecular mechanisms underlying this perplexing finding remain unclear. In light of our findings above, we hypothesize that loss of Dzip1 enhances the dominant negative function of the yot allele through stabilizing the mutated Gli2 protein. We thus generated Gli21–799, which is analogous to Yot119 (64). We found that the stability of Gli21–799 was markedly enhanced after depletion of Dzip1 in Xenopus embryos (Fig. 8A). Consistently, overexpression of Spop destabilized Gli21–799 in NIH3T3 cells (Fig. 8B). These findings thus provide a mechanistic explanation for the synergistic relationship between iguana and yot mutant alleles that negatively impacts Hh signaling. They also provide further evidence that Dzip1 promotes the turnover of Gli transcription factors through a Spop-dependent mechanism.
FIGURE 8.
Regulation of Gli21–799, an analog Gli2 mutant of zebrafish Yot, by Dzip1 and Spop. A, Western blot results showing that knockdown of Dzip1 stabilized Gli21–799 in Xenopus embryos. RNAs encoding Gli21–799 and MT-GFP were injected into controls and embryos that had received a prior injection of DMO. Embryos were harvested at the neurula stage and subjected to Western blotting. MT-GFP served as an injection and loading control. B, forced Spop expression decreased the protein stability of Gli21–799. MT-Gli21–799 and MT-GFP were transfected alone or in combination with MT-Spop into NIH3T3 cells. Cell lysates were subjected to Western blot with the anti-myc antibody to detect the levels of Spop. C, model of Dzip1 functions during Hh signaling. Dzip1 supports ciliogenesis and maintains the proper expression level of Gli proteins through controlling the stability of Spop (left panel). Partial knockdown of Dzip1 has no effect on the formation of cilia but induces degradation of Spop, leading to increased levels of Gli proteins. When exposed to Shh, these cells produce more Gli activators, resulting in a more robust Hh target gene expression (middle panel). When Dzip1 was depleted more completely, ciliogenesis was disrupted. Although these cells have increased levels of Gli, they cannot receive Shh. This prevents the formation of Gli activators. Consequently, gene expression induced by Shh were inhibited (right panel). Under normal conditions, cells located at a distance from the source of Shh are not exposed to Shh. Whether these cells form cilia or not are not important. However, when Dizp1 is depleted, destabilization of Spop will induce accumulation of full-length Gli proteins (not Gli activators). This will lead to weak ectopic Hh target gene expression in embryos.
DISCUSSION
Phenotypic characterization of zebrafish iguana mutants reveals that Dzip1 is required for cells to respond to Shh. In iguana mutants, there is a global reduction of Hh target gene expression (31, 32). More recent studies indicate that the reduced Hh signaling in iguana mutants is likely a consequence of impaired ciliogenesis (34–36). Strikingly, in iguana mutants, the expression of some Hh target genes is increased in cells located at a distance from the source of Shh, for example, cells in somites (31, 32). This argues for an inhibitory role of Dzip1 in the Hh pathway. We recently reported that, in addition to controlling ciliogenesis, Dzip1 is required for Gli turnover in Xenopus embryos (38), providing important clues to the mechanism through which Dzip1 inhibits Hh signaling. Because we were able to disrupt Dzip1-dependent Gli turnover without perturbing ciliogenesis by injection of low concentrations of DMO, we knocked down Dzip1 partially and specifically investigated the inhibitory function of Dzip1 in the Hh pathway.
Results presented here indicate that partial knockdown of Dzip1 sensitizes Xenopus embryos to Hh signaling (Figs. 1 and 3). We found that partial knockdown of Dizp1-induced phenotypes indicative of elevated Hh signaling, including ventralized neural tubes and eyes, expanded slow muscle fibers, and ectopic ptc1 expression in multiple groups of tissues (Fig. 2). These phenotypes were rescued by depletion of Gli1 or Gli2 (Fig. 4). Thus, the phenotypes associated with partial Dzip1 depletion are likely attributed to elevated Hh signaling as a direct consequence of increased Gli stability.
Intriguingly, some phenotypes observed in Dzip1-depleted Xenopus embryos were in stark contrast to that of zebrafish iguana mutants. Partial knockdown of Dzip1 in Xenopus embryos results in ventralization of the neural tube, whereas the iguana mutant displays neural tube patterning defects indicative of an Hh signaling reduction, including the absence of lateral floor plate specification and reduced ptc1 expression within the floor plate and ventrally located inter- and motoneurons (31). We argue that the phenotypic differences between the Dzip1 morphants in this study and those reported in iguana mutants lies in the differential outcome to cilia formation. In our studies we chose to knock down Dzip1 partially, allowing normal ciliogenesis in embryos. Due to stabilization of Gli proteins, neural tube cells become sensitized and generated a more robust response to Shh from the notochord and floor plate. In iguana, however, although the levels of Gli proteins are likely increased, cells in embryos are deficient in primary cilium (34–36), the organelle essential for cells to receive Shh. Consequently, Hh signaling is impaired, and the neural tube of iguana becomes dorsalized (31).
Our results further reveal that Dzip1 regulates Gli turnover by preventing proteasome-dependent degradation of Spop/HIB, adaptors of Cullin3 E3 ligase that promotes Gli/Ci turnover. We were able to show that forced expression of Spop could prevent DMO-induced Gli stabilization and rescue phenotypes caused by partial knockdown of Dzip1 in Xenopus embryos. This demonstrates that Dzip1 exerts its inhibitory effect on the Hh pathway by preventing degradation of Spop/HIB.
In agreement with the notion that Dzip1 regulates Gli turnover independent of its role in ciliogenesis, we found that Dzip1 stabilizes Spop independent of its function in ciliogenesis. We showed that knockdown of IFT88, which disrupted ciliogenesis in embryos, had no effect on the stability of Spop. Furthermore, reduction of Spop was detected in embryos injected with 5 ng of DMO, a dose of DMO insufficient for causing ciliogenesis defects. It is worth noting that Dzip1 overexpression did not lead to any discernable effects on Spop protein stability. In addition, we could not detect physical interaction between Dzip1 and Spop proteins (data not shown). Thus, further analysis will be needed to fully elucidate the mechanism by which Dzip1 prevents proteasome-dependent degradation of Spop/HIB.
Based on our studies and works from others (31, 32, 34–37), we now propose that vertebrate Dzip1 has at least two independent functions during vertebrate Hh signaling (Fig. 8C). First, Dzip1 plays critical roles during ciliogenesis, which in turn positively influences the Hh pathway. Additionally, Dzip1 is required for stabilizing Spop. Because Spop promotes degradation of Gli proteins, the latter function of Dzip1 acts negatively to regulate Hh signaling. Collectively, these dual actions likely account for the contradictory combination of activation and depletion of Hh signaling in zebrafish and Xenopus embryos. In Drosophila, in which cilia are dispensable for Hh signaling, Dzip1 action is likely restricted to its regulation of HIB stability. In the absence of Dzip1, HIB becomes destabilized, leading to an increased level of Ci. Our work presented here demonstrates that regulation of the stability of Spop/HIB by Dzip1 is an evolutionarily conserved mechanism important for the output of Hh signaling.
Acknowledgments
We thank Drs. A. Liu and R. Harland for plasmids. BA-F8 and 12/101 antibodies were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD, National Institutes of Health and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM093217 (to J. Y.), R01GM079684 (to J. J.), and F32EY021708 (to T. S.).
- Hh
- Hedgehog
- Ci
- Cubitus interruptus
- Ptc
- Patched
- Smo
- Smoothened
- Dzip1
- Daz interacting protein 1
- Spop
- Speckle-type POZ protein
- BTB
- Bric-a brac/Tramtrack/Broad complex protein domain
- HIB
- Hh induced BTB protein
- Sufu
- Suppressor of Fused
- DMO
- Dzip1 morpholino
- Shh
- Sonic Hedgehog
- SSF
- superficial slow fibers
- IFT88
- intraflagellar transport protein 88 homolog
- MT
- myc-tagged
- yot
- you-too allele.
REFERENCES
- 1. Dellovade T., Romer J. T., Curran T., Rubin L. L. (2006) The hedgehog pathway and neurological disorders. Annu. Rev. Neurosci. 29, 539–563 [DOI] [PubMed] [Google Scholar]
- 2. Ingham P. W., McMahon A. P. (2001) Hedgehog signaling in animal development. Paradigms and principles. Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 3. Mullor J. L., Sánchez P., Ruiz i Altaba A. (2002) Pathways and consequences. Hedgehog signaling in human disease. Trends Cell Biol. 12, 562–569 [DOI] [PubMed] [Google Scholar]
- 4. Kasper M., Regl G., Frischauf A. M., Aberger F. (2006) GLI transcription factors. Mediators of oncogenic Hedgehog signalling. Eur. J. Cancer 42, 437–445 [DOI] [PubMed] [Google Scholar]
- 5. Taipale J., Beachy P. A. (2001) The Hedgehog and Wnt signalling pathways in cancer. Nature 411, 349–354 [DOI] [PubMed] [Google Scholar]
- 6. Jiang J., Hui C. C. (2008) Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang G., Amanai K., Wang B., Jiang J. (2000) Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 14, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pan Y., Bai C. B., Joyner A. L., Wang B. (2006) Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 26, 3365–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aza-Blanc P., Ramírez-Weber F. A., Laget M. P., Schwartz C., Kornberg T. B. (1997) Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 10. Fuse N., Maiti T., Wang B., Porter J. A., Hall T. M., Leahy D. J., Beachy P. A. (1999) Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc. Natl. Acad. Sci. U.S.A. 96, 10992–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hooper J. E., Scott M. P. (2005) Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6, 306–317 [DOI] [PubMed] [Google Scholar]
- 12. Marigo V., Davey R. A., Zuo Y., Cunningham J. M., Tabin C. J. (1996) Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176–179 [DOI] [PubMed] [Google Scholar]
- 13. Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H., Noll M., Hooper J. E., de Sauvage F., Rosenthal A. (1996) The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134 [DOI] [PubMed] [Google Scholar]
- 14. Chen Y., Struhl G. (1996) Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563 [DOI] [PubMed] [Google Scholar]
- 15. Eggenschwiler J. T., Anderson K. V. (2007) Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huangfu D., Anderson K. V. (2006) Signaling from Smo to Ci/Gli. Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 133, 3–14 [DOI] [PubMed] [Google Scholar]
- 17. Oro A. E. (2007) The primary cilia, a “Rab-id” transit system for hedgehog signaling. Curr. Opin. Cell Biol. 19, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scholey J. M., Anderson K. V. (2006) IntraFLAGellar transport and cilium-based signaling. Cell 125, 439–442 [DOI] [PubMed] [Google Scholar]
- 19. Wong S. Y., Reiter J. F. (2008) The primary cilium at the crossroads of mammalian hedgehog signaling. Curr. Top. Dev. Biol. 85, 225–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goetz S. C., Anderson K. V. (2010) The primary cilium. A signalling centre during vertebrate development. Nature Rev. Genetics 11, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwon J. E., La M., Oh K. H., Oh Y. M., Kim G. R., Seol J. H., Baek S. H., Chiba T., Tanaka K., Bang O. S., Joe C. O., Chung C. H. (2006) BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem. 281, 12664–12672 [DOI] [PubMed] [Google Scholar]
- 22. Chen M. H., Wilson C. W., Li Y. J., Law K. K., Lu C. S., Gacayan R., Zhang X., Hui C. C., Chuang P. T. (2009) Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 23, 1910–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang C., Pan Y., Wang B. (2010) Suppressor of fused and Spop regulate the stability, processing, and function of Gli2 and Gli3 full-length activators but not their repressors. Development 137, 2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Q., Shi Q., Chen Y., Yue T., Li S., Wang B., Jiang J. (2009) Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 106, 21191–21196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dunaeva M., Michelson P., Kogerman P., Toftgard R. (2003) Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 278, 5116–5122 [DOI] [PubMed] [Google Scholar]
- 26. Kogerman P., Grimm T., Kogerman L., Krause D., Undén A. B., Sandstedt B., Toftgård R., Zaphiropoulos P. G. (1999) Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1, 312–319 [DOI] [PubMed] [Google Scholar]
- 27. Pearse R. V., 2nd, Collier L. S., Scott M. P., Tabin C. J. (1999) Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev. Biol. 212, 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Q., Zhang L., Wang B., Ou C. Y., Chien C. T., Jiang J. (2006) A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell 10, 719–729 [DOI] [PubMed] [Google Scholar]
- 29. Kent D., Bush E. W., Hooper J. E. (2006) Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133, 2001–2010 [DOI] [PubMed] [Google Scholar]
- 30. Préat T. (1992) Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sekimizu K., Nishioka N., Sasaki H., Takeda H., Karlstrom R. O., Kawakami A. (2004) The zebrafish iguana locus encodes Dzip1, a novel zinc-finger protein required for proper regulation of Hedgehog signaling. Development 131, 2521–2532 [DOI] [PubMed] [Google Scholar]
- 32. Wolff C., Roy S., Lewis K. E., Schauerte H., Joerg-Rauch G., Kirn A., Weiler C., Geisler R., Haffter P., Ingham P. W. (2004) Iguana encodes a novel zinc-finger protein with coiled-coil domains essential for Hedgehog signal transduction in the zebrafish embryo. Genes Dev. 18, 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vokes S. A., McMahon A. P. (2004) Hedgehog signaling. Iguana debuts as a nuclear gatekeeper. Curr. Biol. 14, R668–R670 [DOI] [PubMed] [Google Scholar]
- 34. Glazer A. M., Wilkinson A. W., Backer C. B., Lapan S. W., Gutzman J. H., Cheeseman I. M., Reddien P. W. (2010) The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev. Biol. 337, 148–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim H. R., Richardson J., van Eeden F., Ingham P. W. (2010) Gli2a protein localization reveals a role for Iguana/DZIP1 in primary ciliogenesis and a dependence of Hedgehog signal transduction on primary cilia in the zebrafish. BMC Biol. 8, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tay S. Y., Yu X., Wong K. N., Panse P., Ng C. P., Roy S. (2010) The iguana/DZIP1 protein is a novel component of the ciliogenic pathway essential for axonemal biogenesis. Dev. Dyn 239, 527–534 [DOI] [PubMed] [Google Scholar]
- 37. Wang C., Low W. C., Liu A., Wang B. (2013) Centrosomal protein Dzip1 regulates hedgehog signaling by promoting cytoplasmic retention of transcription factor Gli3 and affecting ciliogenesis. J. Biol. Chem. 288, 29518–29529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jin Z., Mei W., Strack S., Jia J., Yang J. (2011) The antagonistic action of B56-containing protein phosphatase 2As and casein kinase 2 controls the phosphorylation and Gli turnover function of Daz interacting protein 1. J. Biol. Chem. 286, 36171–36179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sive H., Grainger R., Harland R. (2000) Early Development of Xenopus laevis. A Laboratory Manual, 1st Ed., pp. 91–141, Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Nguyen V., Chokas A. L., Stecca B., Ruiz i Altaba A. (2005) Cooperative requirement of the Gli proteins in neurogenesis. Development 132, 3267–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dammermann A., Pemble H., Mitchell B. J., McLeod I., Yates J. R., 3rd, Kintner C., Desai A. B., Oegema K. (2009) The hydrolethalus syndrome protein HYLS-1 links core centriole structure to cilia formation. Genes Dev. 23, 2046–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rorick A. M., Mei W., Liette N. L., Phiel C., El-Hodiri H. M., Yang J. (2007) PP2A:B56ϵ is required for eye induction and eye field separation. Dev. Biol. 302, 477–493 [DOI] [PubMed] [Google Scholar]
- 43. Jia H., Liu Y., Yan W., Jia J. (2009) PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development 136, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J., Wu J., Tan C., Klein P. S. (2003) PP2A:B56ϵ is required for Wnt/β-catenin signaling during embryonic development. Development 130, 5569–5578 [DOI] [PubMed] [Google Scholar]
- 45. Jin Z., Shi J., Saraf A., Mei W., Zhu G. Z., Strack S., Yang J. (2009) The 48-kDa alternative translation isoform of PP2A:B56ϵ is required for Wnt signaling during midbrain-hindbrain boundary formation. J. Biol. Chem. 284, 7190–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jin Z., Wallace L., Harper S. Q., Yang J. (2010) PP2A:B56ϵ, a substrate of caspase-3, regulates p53-dependent and p53-independent apoptosis during development. J. Biol. Chem. 285, 34493–34502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chiang C., Litingtung Y., Lee E., Young K. E., Corden J. L., Westphal H., Beachy P. A. (1996) Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413 [DOI] [PubMed] [Google Scholar]
- 48. Ding Q., Motoyama J., Gasca S., Mo R., Sasaki H., Rossant J., Hui C. C. (1998) Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125, 2533–2543 [DOI] [PubMed] [Google Scholar]
- 49. Matise M. P., Epstein D. J., Park H. L., Platt K. A., Joyner A. L. (1998) Gli2 is required for induction of floor plate and adjacent cells but not most ventral neurons in the mouse central nervous system. Development 125, 2759–2770 [DOI] [PubMed] [Google Scholar]
- 50. Peyrot S. M., Wallingford J. B., Harland R. M. (2011) A revised model of Xenopus dorsal midline development. Differential and separable requirements for Notch and Shh signaling. Dev. Biol. 352, 254–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ekker S. C., McGrew L. L., Lai C. J., Lee J. J., von Kessler D. P., Moon R. T., Beachy P. A. (1995) Distinct expression and shared activities of members of the hedgehog gene family of Xenopus laevis. Development 121, 2337–2347 [DOI] [PubMed] [Google Scholar]
- 52. Ericson J., Rashbass P., Schedl A., Brenner-Morton S., Kawakami A., van Heyningen V., Jessell T. M., Briscoe J. (1997) Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90, 169–180 [DOI] [PubMed] [Google Scholar]
- 53. Du S. J., Devoto S. H., Westerfield M., Moon R. T. (1997) Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-β gene families. J. Cell Biol. 139, 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blagden C. S., Currie P. D., Ingham P. W., Hughes S. M. (1997) Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes Dev. 11, 2163–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grimaldi A., Tettamanti G., Martin B. L., Gaffield W., Pownall M. E., Hughes S. M. (2004) Hedgehog regulation of superficial slow muscle fibres in Xenopus and the evolution of tetrapod trunk myogenesis. Development 131, 3249–3262 [DOI] [PubMed] [Google Scholar]
- 56. Barth K. A., Wilson S. W. (1995) Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121, 1755–1768 [DOI] [PubMed] [Google Scholar]
- 57. Macdonald R., Barth K. A., Xu Q., Holder N., Mikkola I., Wilson S. W. (1995) Midline signalling is required for Pax gene regulation and patterning of the eyes. Development 121, 3267–3278 [DOI] [PubMed] [Google Scholar]
- 58. Zhang X. M., Yang X. J. (2001) Temporal and spatial effects of Sonic hedgehog signaling in chick eye morphogenesis. Dev. Biol. 233, 271–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang X. M., Yang X. J. (2001) Regulation of retinal ganglion cell production by Sonic hedgehog. Development 128, 943–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lupo G., Liu Y., Qiu R., Chandraratna R. A., Barsacchi G., He R. Q., Harris W. A. (2005) Dorsoventral patterning of the Xenopus eye. A collaboration of retinoid, hedgehog and FGF receptor signaling. Development 132, 1737–1748 [DOI] [PubMed] [Google Scholar]
- 61. Hallonet M., Hollemann T., Pieler T., Gruss P. (1999) Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev. 13, 3106–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Perron M., Boy S., Amato M. A., Viczian A., Koebernick K., Pieler T., Harris W. A. (2003) A novel function for Hedgehog signalling in retinal pigment epithelium differentiation. Development 130, 1565–1577 [DOI] [PubMed] [Google Scholar]
- 63. Nagai Y., Kojima T., Muro Y., Hachiya T., Nishizawa Y., Wakabayashi T., Hagiwara M. (1997) Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 418, 23–26 [DOI] [PubMed] [Google Scholar]
- 64. Karlstrom R. O., Talbot W. S., Schier A. F. (1999) Comparative synteny cloning of zebrafish you-too. Mutations in the Hedgehog target gli2 affect ventral forebrain patterning. Genes Dev. 13, 388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]