Abstract
Mass spectrometry based proteomics has facilitated sperm composition studies in several mammalian species but no studies have been undertaken in non-human primate species. Here we report the analysis of the 1247 proteins that comprise the Rhesus macaque (Macaca mulatta) sperm proteome (termed the MacSP). Comparative analysis with previously characterized mouse and human sperm proteomes reveals substantial levels of orthology (47% and 40% respectively) and widespread overlap of functional categories based on Gene Ontology analyses. Approximately 10% of macaque sperm genes (113/1247) are significantly under-expressed in the testis as compared with other tissues, which may reflect proteins specifically acquired during epididymal maturation. Phylogenetic and genomic analyses of three MacSP ADAMs (A-Disintegrin and Metalloprotease proteins), ADAM18-, 20- and 21-like, provides empirical support for sperm genes functioning in non-human primate taxa which have been subsequently lost in the lineages leading to humans. The MacSP contains proteasome proteins of the 20S core subunit, the 19S proteasome activator complex and an alternate proteasome activator PA200, raising the possibility that proteasome activity is present in mature sperm. Robust empirical characterization of the Rhesus sperm proteome should greatly expand the possibility for targeted molecular studies of spermatogenesis and fertilization in a commonly used model species for human infertility.
The application of mass spectrometry (MS) based proteomics, coupled with whole genome annotation of an increasing number of species, has greatly extended our knowledge of sperm composition. Traditional methods used to assess sperm composition, including the use of sperm-specific antibodies and 2D gel electrophoresis, have identified a limited number of sperm proteins. These traditional studies have been augmented in recent years by the use of high throughput and highly sensitive MS (shotgun proteomics) that have substantially increased the accuracy of peptide identification, resulting in a significant increase in proteome coverage. Indeed, advances in MS instrumentation, data acquisition, and the availability of genome annotations have, for example, increased sperm proteome coverage in Drosophila from 381 (1) to 1108 proteins (2) over a five year period.
Two main MS based methodologies have been applied to study sperm composition, including (i) 2D PAGE followed by spot excision and MS and (ii) digestion of proteins, followed by MS/MS analysis of the resulting peptides (3). Although each method has its own advantages and disadvantages, a far greater level of proteome coverage is obtained using MS/MS (4). A previous comparative study found that each method identified proteins not found in the other and vice versa, and therefore it has been suggested that these methods should be used to complement each other (5). Thus, although no single methodology yet exists capable of producing a complete whole cell proteome, MS/MS methods provide deeper and broader coverage and are therefore the current method of choice. Shotgun proteomics has characterized sperm proteomes in a variety of taxa including plants, invertebrates and mammals such as human, mouse, rat, and bull (3, 6–11). These studies achieve varying levels of proteome coverage as a result of several factors including the choice of MS equipment, sample acquisition, purification, solublization, and fractionation schemes. Although these different approaches make direct comparisons difficult they nevertheless have provided invaluable information regarding the composition of sperm and have helped to identify novel proteins that play important roles in sperm function and reproduction.
In this study we use MS based proteomics to elucidate the sperm proteome of a species of old world monkey, the Rhesus macaque (Macaca mulatta). Due primarily to their genetic and physiological similarities to humans, Rhesus macaques are the most widely used nonhuman primate model system for basic and applied biomedical research (12). Rhesus macaques are also used extensively as a model of human reproduction where numerous similarities at the molecular level have been observed between gametes of the two species, and why Rhesus macaques have become a useful model system for fertility and assisted reproductive technology research (13). A more complete knowledge of the sperm proteome will facilitate reproductive studies using the Rhesus macaque as a model organism. However, despite its widespread use in reproductive biology, the macaque sperm proteome (MacSP)1 has yet to be characterized.
Although insight into the MacSP will facilitate reproductive studies using the Rhesus macaque as a model organism, this knowledge can also be used to better understand the composition of human sperm. Sperm mature and gain fertilization competency as they traverse the epididymis, a specialized duct that connects the testis to the vas deferens (14). During the maturation process, sperm lose or modify a number of their surface proteins and gain additional transient or permanent surface proteins in a well-organized manner, and it is only after emerging from the cauda epididymis that sperm are motile and considered fertilization competent (14, 15).
Proteomic studies of human sperm have been undertaken (3, 6, 10), identifying between 98–1760 sperm proteins, however these studies used sperm from ejaculates which complicates sperm proteome analysis. A previous study identified 923 proteins present in human seminal plasma (16), which is likely to be only a fraction of the seminal plasma proteome. Human sperm proteome data sets derived from human ejaculates makes it difficult to differentiate which of the identified proteins are sperm or seminal plasma constituents. For example, a major seminal protein family, the semenogelins are not expressed in the testis but are found in sperm proteomes determined from ejaculates (6, 10). Such highly abundant seminal proteins may mask lower abundance integral sperm proteins and inhibit their identification by MS. In order to avoid these problems, we collected mature sperm directly from the cauda epididymis of the Rhesus macaque, thus avoiding contamination from seminal plasma proteins.
In the present study, sperm proteins were separated using 1D SDS-PAGE, digested and the resulting peptides analyzed by LC MS/MS. Using high stringency parameters for peptide identification, we conservatively identified 1247 proteins from purified samples of Rhesus macaque sperm. Given their close evolutionary relationship, the Rhesus macaque and human share 93% nucleotide homology (12). Data from this study can be used to complement what is currently known about the composition of human sperm and provides a more useful proxy of human sperm proteome composition than the proteomes of other non-primate mammals for which data is available. Studies of sperm composition, especially those in human, can be applied to develop novel molecular based clinical diagnostic tests of sperm quality, which is currently limited to evaluating parameters such as sperm count, morphology and motility. In addition, knowledge of sperm components can lead to the discovery of novel contraceptives and infertility treatments.
EXPERIMENTAL PROCEDURES
Tissue Preparation and Isolation of Mature Sperm
Intact epididymides were harvested from two euthanized adult macaques (Macaca mulatta) that had previously been used for a series of neurophysiological experiments. Each animal was euthanized and tissue processed successively over a 2 day period and stored in PBS at 4 °C until used. Each intact epididymis was removed and both proximal and distal ends ligated. All procedures were approved by the Institutional Animal Care and Use Committee of Northwestern University.
Mature and motile sperm were collected from the cauda epididymis by needle puncture and collected in 50 ml conical centrifuge tubes. Sperm pellets were obtained by differential low speed centrifugations using a tabletop centrifuge (Sorvall). Purified sperm samples were obtained by repeated cycles of centrifugation and pellet resuspension in standard Ringers solution. Following each pellet resuspension, sperm purity was assessed using a DNA-based fluorescence assay. Equal 5 μl aliquots of sperm were mounted on a microscope slide along with an equal volume of PBS containing 1.0 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and subsequently examined using a Zeiss Axioskop equipped with DIC and epifluorescence optics (Fig. 1). Digital images captured with an onboard ccd camera and exported to Photoshop.
Fig. 1.
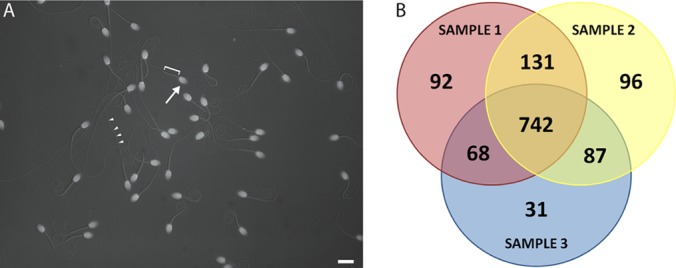
Sperm purification and overview of shotgun proteomics analyses. A, Epi-fluorescent/DIC image of purified macaque sperm. DAPI fluorescence in bright oblate spheroids (white, arrow) indicate the presence of DNA in morphologically normal sperm nuclei and head structures. Associated sperm midpiece (bracket) and flagella (arrowheads) were visualized simultaneously under Nomarski DIC optics also indicative of normal morphology for mature sperm. Bar = 10 microns. B, Venn diagram of protein overlap in three sperm samples identified using shotgun MS/MS.
Protein Solubilization and Quantitation
Sperm samples were solubilized in SDS and quantified using the EZQ® Protein Quantitation Kit (Invitrogen, Carlsbad, CA). Protein fluorescence was measured using a Typhoon Trio+ (Amersham Biosciences/GE Healthcare) equipped with a 488 nm laser and 610 nm bandpass filter. ImageQuant™ TL software was used to analyze fluorescence data. A standard curve was generated using fluorescence data from control samples of known concentration and used to determine sperm sample concentration.
1-Dimensional SDS-PAGE
A 1 mm 10% NuPAGE® Novex® Bis-Tris Mini Gel was set up using the XCell SureLock Mini-Cell system (Invitrogen) as per manufacturer's instructions for reduced samples. Fifty micrograms of Rhesus macaque sperm was loaded in triplicate and the gel was run for 35 min at a 200V constant. Following electrophoresis, the gel was stained using SimplyBlue™ SafeStain (Invitrogen) and destained as per manufacturer's instructions. The gel was transferred to a gel slicer (built in house) and each of the three lanes was separated from the rest of the gel by cutting vertically. Each lane was then cut into 16 equal sections and half of each section was transferred to a 96-well plate, resulting in 48 total gel slices. Gel slices were stored at −80 °C until needed.
In-Gel Digestion of Proteins/MS Analysis
Using a scalpel, each gel slice was cut into smaller (∼1 × 1 × 1 mm) pieces which were transferred to a 0.6 ml microcentrifuge tube (one tube per gel slice, for a total of 48 tubes). A standard in-gel digestion protocol was performed on each gel slice as described previously (17). Gel slices were further destained using 50 mm ammonium bicarbonate/50% acetonitrile and dehydrated with 100% acetonitrile. Proteins were reduced and alkylated by treatment with dithiothreitol and iodoacetamide and subsequently digested using 15 ng/μl Trypsin. Protein digests were extracted with 5% formic acid and 50% acetonitrile, and dried down using a vacuum centrifuge. Resulting peptides were reconstituted in 20 μl of 0.1% formic acid.
Liquid Chromatography Mass Spectrometry
Extracted peptides were analyzed by nanoflow reverse phase liquid chromatography using a nanoAcquity LC system (Waters, Milford, MA) coupled in-line with the linear trap quadrupole LTQ Orbitrap Velos instrument (Thermo Fisher Scientific). The nano LC system included a Symmetry C18 5 μm 180 μm × 20 mm trap column and a BEH130 C18 1.7 μm 100 μm × 100 mm analytical column (Waters). Mobile phases A and B consisted of 0.1% formic acid in water and 0.1% formic acid in acetonitrile, respectively. Samples were loaded onto the trap column for desalting and preconcentration with 99%/1% mobile phase A:B at a flow rate of 5 μl/min for 3 min. Peptides were separated on the analytical column at a flow rate of 500 nl/min with a two-step linear gradient consisting of 7% B to 25% B in 72 min and 25% B to 45% B in 10 min. The electrospray ion source consisted of a nanospray head (Thermo Fisher Scientific) coupled with a coated PicoTip fused silica spray tip with OD 360 μm, ID 20 μm, and 10 μm diameter emitter orifice (New Objective, Inc.). Samples were analyzed using positive ion spray voltage and heated capillary temperature of 1.9 kV and 220 °C, respectively.
Mass spectrometry data were collected with the instrument operating in data dependent MS/MS mode. MS survey scans (m/z 300–2000) were acquired in the Orbitrap analyzer with a resolution of 60,000 at m/z 400 and an accumulation target of 1 × 106. This was followed by the collection of MS/MS scans of the 15 most intense precursor ions with a charge state ≥2 and an intensity threshold above 500 in the LTQ with the accumulation target of 10,000, an isolation window of 2 Da, normalized collision energy of setting of 35%, and activation time of 30ms. Dynamic exclusion was used with repeat counts, repeat duration, and exclusion duration of 1, 30 s, and 60 s, respectively.
Peptide Identification and Protein Annotation
The mass spectra data files were analyzed using Sequest (Thermo Fisher Scientific, San Jose, CA, USA; version 1.2.0.208) and X!Tandem (The GPM; version 2007.01.01.1) searched against the NCBI Macaca mulatta protein fasta file (dated 10/14/10, with 27088 entries). SEQUEST and X!Tandem were searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 10.0PPM. Iodoacetamide derivative of cysteine was specified as a fixed modification, whereas oxidation of methionine was specified as a variable modification. Results were merged using Scaffold (Proteome Software) version 3.0.8 which calculated False Discovery Rates (FDRs) using a reverse concatenated decoy database. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (18) and protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least three identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (19). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Protein identifications from Scaffold after analysis with Sequest and X!Tandem consisted of Macaque RefSeq (NCBI) protein IDs. These protein IDs were entered into BioMart (biomart.org, version 0.7) using the Ensembl Genes 54 (Sanger UK) Macaca mulatta genes (MMUL_1.0) database and used to obtain cross-referenced Ensembl Gene IDs and associated gene names. The same method was used to obtain human orthologs. Macaque and human Ensembl numbers were matched to each RefSeq protein ID in a 1:1 fashion such that there was only one macaque and one human Ensembl number per RefSeq protein ID, thus the size of the original data set was maintained. A gene name was assigned to each of the 1247 proteins identified in this study. Macaque Ensembl IDs and Human Ensembl IDs were assigned to 94% (1170/1247) and 93% (1165/1247) of proteins, respectively.
Gene Ontology, Sperm Phenotype and Genomic Analysis
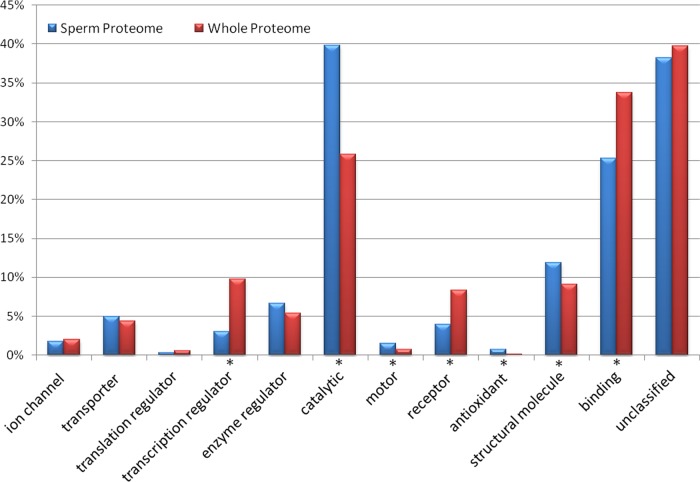
The gene group functional profiling (g:GOSt) analysis tool from g:Profiler (20) was used to conduct Gene Ontology (GO) analyses. Macaque Ensembl Gene IDs (1166) for the genes expressed in the MacSP were queried against the Macaca mulatta data set. Significance of over-represented GO categories, as compared with the whole macaque proteome, was determined using the Benjamini-Hochberg FDR significance threshold. The expected number of proteins in each functional category in the whole macaque proteome was calculated for comparison with the MacSP. The distribution of functional categories in the MacSP as compared with the whole proteome is shown in Fig. 2. GO category enrichment is indicated by an asterisk (p ≤ 0.05). A description of each of the GO terms can be found at www.geneontology.org.
Fig. 2.
Gene Ontology (GO) molecular function analysis for the MacSP and whole proteome. Bars represent the number of hits (proteins) in the functional category (note that some proteins are placed in multiple functional GO categories). The unannotated category represents the number of proteins with no annotated molecular function. (*) indicates significant (p ≤ 0.05) difference between the number of entries for each category in the MacSP and whole proteome.
Mammalian phenotype information was obtained from the MGI mammalian phenotype database (http://www.informatics.jax.org/) using annotated orthology relationships with the mouse and human. These results were further curated to obtain a comprehensive list of abnormal sperm morphology or process phenotypes. GO category enrichment was conducted using g:Profiler. The chromosomal distribution of macaque sperm genes was obtained from gene map coordinate file available from NCBI (Cyto_gene mapview 1.2). A Chi-squared test was used to compare the proportion of X- linked and autosomal macaque sperm genes to the genome-wide distribution of genes.
Mouse and Human Sperm Proteome Comparisons
To compare the MacSP to other published proteomes, we obtained the MouseSP and HumanSP IDs from the published literature (6, 9) and converted them to their corresponding Ensembl IDs using Biomart (Biomart.org). The resulting 934 mouse and 925 Human Ensembl gene IDs were then used to identify the corresponding orthologs present in the MacSP data set.
Network analyses were conducted on the genes found to be common to the macaque, mouse, and humanSPs and on the genes found to be common to both the macaque and mouse, and the macaque and human sperm proteomes. Networks were created using GO Biological Process Annotations (downloaded 23.05.2013) as implemented in the ClueGo plugin v2.0.5 (21) of Cytoscape v3.0.0 (22). For the macaque-, mouse-, and humanSP overlap, an enrichment (right-sided hypergeometric) test was used with a Benjamini-Hochberg multiple test correction. GO Tree Levels (min = 1; max = 2) and GO term restriction (min#genes = 3, min% = 1%) were set and terms were grouped using a Kappa Score Threshold of 0.3. For the macaque/mouse and macaque/humanSP overlap networks, an enrichment (right-sided hypergeometric) test was used with a Benjamini-Hochberg multiple test correction. GO Tree Levels (min = 1; max = 3) and GO term restriction (min#genes = 5,min% = 4%) were set and terms were grouped using a Kappa Score Threshold of 0.3. For all networks, resulting groups consisted of a minimum of two terms, and groups sharing >50% of terms were merged.
Phylogenetic Analysis
The MacSP contains 3 ADAM-like proteins annotated as ADAM18, 20, and 21-like. Protein BLAST queries of these proteins identified higher similarity to other ADAMs, in particular ADAMs 3, 4, and 6. To further examine this, all available macaque, human, chimpanzee, mouse, and rat ADAM sequences (101 total) were obtained from NCBI. Alignment and phylogenetic analysis of these protein sequences were conducted in MEGA5 (23) as described below with 50 bootstrap replicates to investigate placement of the ADAM-like proteins in the tree (not shown).
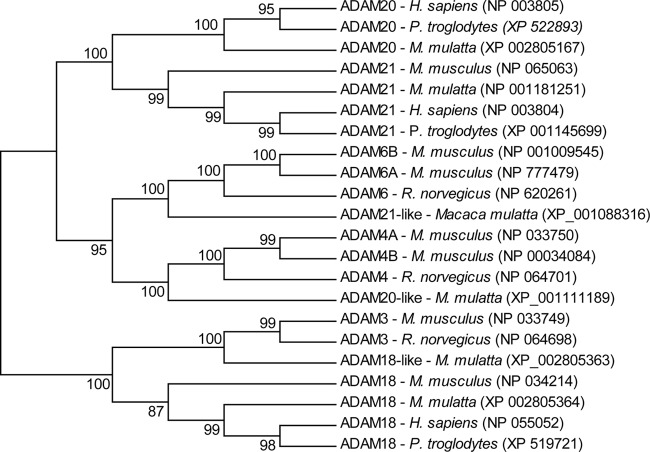
For subsequent analyses, a subset of 22 amino acid sequences (supplemental Table S1) from the macaque, human, chimpanzee, mouse, and rat for ADAMs 3, 4, 6, 18, 20, and 21 were aligned by ClustalW as implemented by MEGA5. All ambiguous positions were removed for each sequence pair and there were a total of 939 positions in the final data set. Evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (22) and the bootstrap consensus tree, as shown in Fig. 3, was inferred from 1000 replicates (24). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically as follows. When the number of common sites was < 100 or less than one fourth of the total number of sites, the maximum parsimony method was used; otherwise BIONJ method with MCL distance matrix was used. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA5. Orthology was determined using reciprocal best protein BLAST queries of ADAM18, 20, and 21-like against mouse (Mus musculus) and rat (Rattus norvegicus) proteins.
Fig. 3.
Maximum-likelihood phylogenetic analysis of Macaque ADAM-like proteins. Evolutionary history was inferred by using the Maximum Parsimony method. The bootstrap consensus tree (shown) was inferred from 1000 replicates (24). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The analysis involved 22 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 939 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (23).
Expression Analysis
Gene expression data for five rhesus macaque tissues (cerebral cortex, pancreas, thymus, testis, and an immortalized fibroblast cell line) obtained from three different research centers (25) was downloaded from the NCBI Gene Expression Omnibus (GEO, series GSE7094). The data set from the Yerkes National Primate Research Center was omitted from our analysis because their total intensities were markedly higher than the other two. Affymetrix Probeset Identifiers were matched to Ensembl macaque sperm gene IDs using BioMart. In total, there were 1774 Affymetrix probeset matches to macaque sperm genes, some represented by multiple probes on the array. Multiple matches were collated using the similarity of gene expression intensities across samples implemented in the Inferno software program (https://code.google.com/p/inferno4proteomics/), resulting in 1041 MacSP genes for which expression data was available.
The average expression of the MacSP data set in the testis was compared with other tissues (cerebral cortex, pancreas, thymus, fibroblast). The ratio of average expression of sperm proteome genes to the averages in the other tissues was log2 transformed to determine the fold change of that particular gene in the testis as compared with all other tissues (supplemental Table S2).
Statistical tests were performed to determine if any sperm proteome genes were over- or underexpressed in the testis compared with other tissues and cell types. Gene expression intensities in the testis samples were compared with a background data set that consisted of the four averaged sets taken from the GSE7094 series. A standard t test, as implemented in the Inferno program, identified a set of 242 genes differentially expressed in the testis (p < 0.05) (supplemental Table S2).
Network Analysis
A network analysis was conducted on the 242 genes expressed in macaque sperm that are over- or underexpressed significantly in the testis. The network was created by comparing GO Molecular Function Annotations (downloaded 05.01.2012) for over- and underexpressed genes as implemented in the ClueGo plugin v1.4 (21) of Cytoscape v2.8.2 (22). A two-sided hypergeometric enrichment and depletion test was used with a Benjamini-Hochberg multiple test correction. GO Tree Levels (min = 1; max = 3) and GO term restriction (min#genes = 5, min% = 1%) were set and terms were grouped using a Kappa Score Threshold of 0.1. Resulting groups consisted of a minimum of two terms, and groups sharing >50% of terms were merged.
RESULTS
Sperm Protein Identification by MS
Shotgun MS/MS identified 1247 proteins in purified macaque sperm samples using strict criteria mass spectra analysis as well as protein identification (supplemental Table S3). False Discovery Rates (FDRs) using a reverse concatenated decoy database resulted in estimates of peptide and protein FDR of <1%, supporting the high quality of the data set. Three replicate sperm samples were analyzed and each sample resulted in the identification of between 5449 and 7523 unique peptides that identified between 928–1056 proteins per sample (Table I). The average coverage of identified proteins ranged from 17.8% to 24.0% with an average across all samples of 20.7% (Table I, Supplemental Table S3). Although the cutoff for the minimum number of peptides identified per protein was set to 3, the average number of peptide hits per protein was over twice this value (6.46) reflecting the depth of the MS data. In addition, 59.5% (742/1247) of proteins in our data set were identified in all three samples whereas 82.4% (1028/1247) of proteins were identified in at least two samples (Fig. 1).
Table I. Summary of MS results.
| Sample 1 | Sample 2 | Sample 3 | Total | |
|---|---|---|---|---|
| Total proteins identified | 1033 | 1056 | 928 | 1247 |
| Total peptides identified | 7523 | 6578 | 5449 | 19550 |
| Sample 1 | Sample 2 | Sample 3 | Average | |
|---|---|---|---|---|
| Average percent protein coverage | 23.97 | 20.35 | 17.82 | 20.71 |
| Average peptide hits per protein | 7.28 | 6.23 | 5.87 | 6.46 |
Abundant proteins in the MacSP were ranked by spectral counts providing a semiquantitive measure of relative protein levels (Table II, supplemental Table S3) (26). By far, the most highly abundant protein was a-kinase anchor protein 4-like (AKAP4), a known constituent of mammalian sperm. Orthology searches of the corresponding gene found that it shares one-to-one orthology with AKAP4 of 7 primate species, including human, as well as rat and mouse. AKAP4 was also identified as the most abundant protein in the rat sperm proteome (7) and is the most abundant protein constituent of the sperm fibrous sheath (27). Several keratins, including KRT1, 2, 5, 6a, 10, and 16 were also found to be among the 20 most abundant proteins identified in the MacSP. Although keratins are often associated with sample contamination, these keratins have previously been demonstrated to be present in the human sperm nucleus (5). Additional keratins have also been identified in the rat, human and mouse sperm proteomes (2, 6, 8, 10) suggesting that these proteins play important functional roles in sperm. LOC703932, the 13th most abundant protein, has high sequence homology with keratin 9 from several species including human, orangutan (Pongo abelii), and marmoset (Callithrix jacchus). Keratin 9, along with other proteins, has been found to comprise the perinuclear ring of the manchette, which nucleates extending microtubules in the developing spermatid (28).
Table II. Top 50 abundant macaque sperm proteins by spectral count.
| Rank | Macaque sperm protein | Human ortholog (Ensembl) | Total spectral counts | Average percent coverage (%) |
|---|---|---|---|---|
| 1 | LOC694888 | AKAP4 | 7901 | 76 |
| 2 | LOC702153 | KRT10 | 2866 | 51 |
| 3 | LOC699320 | KRT1 | 2551 | 48 |
| 4 | ODF2 | ODF2 | 2226 | 44 |
| 5 | GAPDHS | GAPDHS | 2182 | 76 |
| 6 | TUBB2C | TUBB4B | 2123 | 78 |
| 7 | AKAP3 | AKAP3 | 2047 | 63 |
| 8 | ODF1 | ODF1 | 2018 | 61 |
| 9 | LOC707215 | TUBA3E | 1658 | 70 |
| 10 | LOC699567 | KRT2 | 1639 | 46 |
| 11 | GSTM3 | GSTM3 | 1524 | 77 |
| 12 | TUBB2B | TUBB2B/TUBB2Aa | 1495 | 65 |
| 13 | LOC703932 | KRT9a | 1211 | 32 |
| 14 | ALDOA | ALDOA | 1151 | 43 |
| 15 | LOC100424400 | KRT5 | 1062 | 45 |
| 16 | ROPN1B | ROPN1B | 1053 | 72 |
| 17 | DNAH2 | DNAH2 | 1030 | 4 |
| 18 | GPX4 | GPX4 | 992 | 53 |
| 19 | KRT14 | KRT16 | 988 | 49 |
| 20 | LOC100424162 | KRT6A | 969 | 56 |
| 21 | LDHAL6B | LDHAL6B | 955 | 57 |
| 22 | LOC704257 | VDAC2a | 948 | 60 |
| 23 | LOC707236 | KRT16 | 909 | 20 |
| 24 | ATP5B | ATP5B | 865 | 60 |
| 25 | LOC705465 | ATP5B | 861 | 60 |
| 26 | HK1 | HK1 | 851 | 47 |
| 27 | LDHA | LDHA | 852 | 66 |
| 28 | ACO2 | ACO2 | 735 | 56 |
| 29 | PDHB | PDHB | 705 | 55 |
| 30 | PKM2 | PKM2 | 705 | 53 |
| 31 | LDHC | LDHC | 624 | 64 |
| 32 | ACR | ACR | 608 | 40 |
| 33 | KRT17 | KRT17 | 604 | 29 |
| 34 | DNAH8 | DNAH8 | 600 | 20 |
| 35 | LOC714425 | LELP1 | 537 | 47 |
| 36 | HSPD1 | HSPD1 | 535 | 51 |
| 37 | LOC706830 | LELP1a | 535 | 10 |
| 38 | LAP3 | LAP3 | 528 | 65 |
| 39 | TPI1 | TPI1 | 519 | 82 |
| 40 | PDHA2 | PDHA2 | 516 | 51 |
| 41 | OAZ3 | OAZ3 | 516 | 16 |
| 42 | ZPBP | ZPBP | 513 | 45 |
| 43 | MTCH2 | MTCH2a | 502 | 51 |
| 44 | ENO1 | ENO1 | 491 | 55 |
| 45 | HADHA | HADHA | 491 | 49 |
| 46 | LRRC37A | - | 467 | 18 |
| 47 | PGK2 | PGK2 | 451 | 65 |
| 48 | LOC10402 | TEKT5 | 440 | 58 |
| 49 | ALB | ALB | 438 | 40 |
| 50 | ZAN | ZAN | 427 | 22 |
a Likely orthologs.
Mammalian Sperm Phenotypes
We identified 74 proteins in the MacSP which, when mutated or defective in mice or humans, result in abnormal sperm development or function (Table III). This includes 37 proteins associated with abnormal sperm morphology and 50 associated with abnormal physiological, motility, capacitation, or fertilization processes. Statistical analysis of functional over-representation among these genes found that abnormal sperm morphology phenotypes are enriched for proteins that localize to the flagellum (p = 2.50 e-8; GO:19861) and cilium (p = 2.00 e-4; GO:5929), including the structural genes TEKT2, 3 and 4 and ODF2. As expected, abnormal sperm process genes are significantly enriched for genes involved in fertilization (p = 8.04 e-16; GO: 9566), sperm-egg recognition (p = 1.64 e-4; GO: 35036) and those which localize to the acrosomal vesicle (p = 4.68 e-11; GO:1669). Notably this group includes a number of highly studied sperm proteins involved in sperm-egg interaction, including IZUMO1, ADAM1, AKAP4, CRISP1, ACR, ZPBP1, and ZPBP2.
Table III. Macaque sperm proteins with mammalian sperm phenotypes.
| Phenotype | Mammalian phenotype IDa | Total genes in genome | Genes identified in the MacSP |
|---|---|---|---|
| Abnormal morphology | |||
| Axoneme | 9838 | 13 | ODF2, SPAG6, THEG, TEKT2, VDAC3 |
| Flagellumb | 8892, 8545, 9238, 8893, 9243, 9237, 9839, 9239 | 104 | AKAP4, ATP1A4, CSNK2A1, CSNK2A2, HK1, OAZ3, ODF2, PPP1CC, PRSS21, SPAG6, TCTE3, TEKT2, TEKT3, TEKT4, THEG, ZPBP, ZPBP2 |
| Head | 9230 | 122 | ACR, CSNK2A1, CSNK2A2, CD59, H1FNT, PPP1CC, PRSS21, SMCP, TCTE3, TSSK4, SPAG6 |
| Nucleus | 9232 | 26 | CSNK2A1, CSNK2A2, GPX5, H1FNT, HK1, THEG, ZPBP, ZPBP2 |
| Principle piece | 9836 | 8 | AKAP4, PPP1CC, SPAG6, TEKT4 |
| Midpiece | 9831 | 27 | NUP210L, ODF2, PPP1CC, TEKT4, SPAG6 |
| Mitochondrial sheath | 9832, 9833 | 19 | AKAP4, PPP1CC, VDAC3 |
| Spermatogonia | 6378 | 32 | ALPL |
| Spermatid | 6380 | 125 | ATM, CSNK2A1, CSNK2A2, CSTF2T, HERC2, HK1, NPEPPS, PPP1CC, PRKDC, PSME4, RSPH1, THEG, TSSK2, ZPBP, ZPBP2 |
| Spermatocyte | 6379 | 59 | ATM, DDX4, PLEKHA1, PSME4, SLC25A31 |
| Abnormal Process | |||
| Spermatogenesis | 1156, 8261 | 386 | ALMS1, BSG, GNPAT, H1FNT, HRG, HSP90AA1, HSPA4L, IMMP2L, KLHL10, LMNA, MLH3, NR2C2, RET, RPS27A, STX2, SUV39H1, TDRD6 |
| Physiology | 4543 | 215 | AQP7, ATP1A4, ATP2B4, CRISP1, KCNU1, LDHC, PPP1CC, PRSS21, RSPH1, SPAM1, SPESP1, TEKT4, ZAN |
| Capacitation | 3666 | 36 | ADCY3, ATP1A4, KCNU1 |
| Motility | 2674, 2675 | 187 | ADAM2, CAPZA3, CHDH, CSNK2A1, CSNK2A2, DNAH1, GAPDHS, HSPA4L, PGK2, PRKACA, TEKT2 |
| Fertilization | 242, 5410, 5411 | 96 | ACR, ACRBP, ADAM1, ADCY3, ATP1A4, CD46, IZUMO1, KCNU1, LDHC, PRKACA, PRSS21, SERPINA5, SMCP, SPAM1, SPESP1, ZPBP2 |
a Mammalian phenotypes were obtained from the Mouse Genome Informatics database.
b Defects include abnormal flagellum morphology (coiled, detached, hairpin, kinked, and short), flagellum absence and multiflagellate sperm.
Gene Ontology (GO) Functional Analysis
Annotated molecular functions were obtained for 72% (896/1247) of the genes in the data set, which were placed among 13 broad functional categories (Fig. 2). Three of the categories, catalytic, antioxidant, and electron transport were statistically enriched in the MacSP (p < 0.05) as compared with the entire genome. Further analyses of the two most abundant annotated categories in the MacSP, catalytic and binding functions (described below), revealed a higher proportion of proteins with a catalytic function, but a slightly lower proportion of proteins with binding function in the MacSP (Fig. 2). With regards to catalytic function, the MacSP contains an overabundance of proteins with oxidoreductase- (116 proteins, p = 3.44 e-17), hydrolase- (210 proteins, p = 4.83 e-12), and isomerase (20 proteins, p = 1.69 e-2) activity as compared with the whole proteome. The MacSP is enriched for oxidoreductases that act on NADH and NADPH (58 proteins, p = 3.90 e-08), including dehydrogenases that act on NADH (26 proteins, p = 9.78 e-08). The MacSP also contains hydrolases enriched in peptidase activity (65 proteins, p = 1.75 e-05), particularly threonine-type peptidases/endopeptidases (14 proteins, p = 1.77 e-10), and hydrolases acting on acid anhydrides (92 proteins, p = 5.55 e-09).
The MacSP is enriched in a number of binding proteins including small molecule binding (205 proteins, p = 2.30 e-14), cofactor binding (33 proteins, p = 2.63 e-05), and metal cluster binding (12 proteins, p = 2.63 e-03) proteins. Small molecule binding proteins are enriched for nucleotide binding proteins (194 proteins, p = 4.90 e-14) whereas cofactor binding proteins are enriched for coenzyme (26 proteins, p = 4.68 e-05) and NAD binding (13 proteins, p = 6.54 e-06) proteins. Metal cluster binding proteins are enriched for proteins involved in unfolded protein (21 proteins, p = 1.00 e-06), cytoskeletal protein (36 proteins, p = 2.14 e-0.3) and calcium ion (54 proteins, p = 3.5 e-03) binding.
Overall, 59% (733/1247) of the MacSP has annotated GO biological processes such as metabolic processes (539, p = 8.79 e-04), including small molecule metabolic processes (158 proteins, p = 2.89 e-10), oxidation-reduction process (101 proteins, p = 3.03 e-18), and catabolic process (140 proteins, p = 2.84e-15). In addition, the MacSP is enriched for proteins involved in particular cellular processes including response to oxidative stress (26 proteins, p = 2.21 e-06), generation of precursor metabolites and energy (50 proteins, p = 1.23 e-19), protein folding (29 proteins, p = 1.31 e-05), and microtubule based processes (40 proteins, p = 3.21 e-06), which includes sperm motility (8 proteins, p = 2.93 e-04).
Comparison to the Mouse and Human Sperm Proteomes
Previous studies have characterized the mouse and human sperm proteome but used fundamentally different sperm isolation techniques. The mouse sperm proteome was determined using sperm purified from the cauda epididymis, consistent with the methods used in this study, whereas the human analysis used sperm purified from total ejaculates. To assess the possible impact of nonsperm ejaculate contamination within the human sperm proteome, we calculated the number of orthologous proteins within each proteome (supplemental Table S4). Based on sperm proteome size and expected overlap based on total genome orthology, the MouseSP and MacSP are expected to share 589 orthologs whereas the HumanSP and MacSP are expected to share 692 orthologs. Contrary to what would be expected based on their evolutionary relationship, we identified a significantly higher (p = 0.0037) proportion of orthology relationships between the macaque and mouse sperm proteomes (436 proteins; 47% of total proteome, 74% of expected) than between the MacSP and HumanSP (368 proteins; 40% of total proteome, 53% of expected). This finding is likely attributed to differences in sample preparation, MS techniques and the potential for the human sperm proteome to contain a number of contaminating seminal proteins that are not conserved components of sperm. This illustrates the difficulty in comparing sperm proteome data across species and across studies where varying methods are employed.
To identify similarities and potential functional differences between the sperm proteomes, we conducted GO cluster network analyses on the 436 proteins common to the MacSP/MouseSP and the 368 proteins common to the MacSP/HumanSP (supplemental Fig. S1). Pair-wise comparison highlights several functional similarities in processes related to energy generation and metabolism and processes closely related to sperm biology including response to oxidative stress, sperm motility, and sexual reproduction. These common biological processes are also reflected in the triple overlap between the MacSP, HumanSP and MouseSP as detailed below.
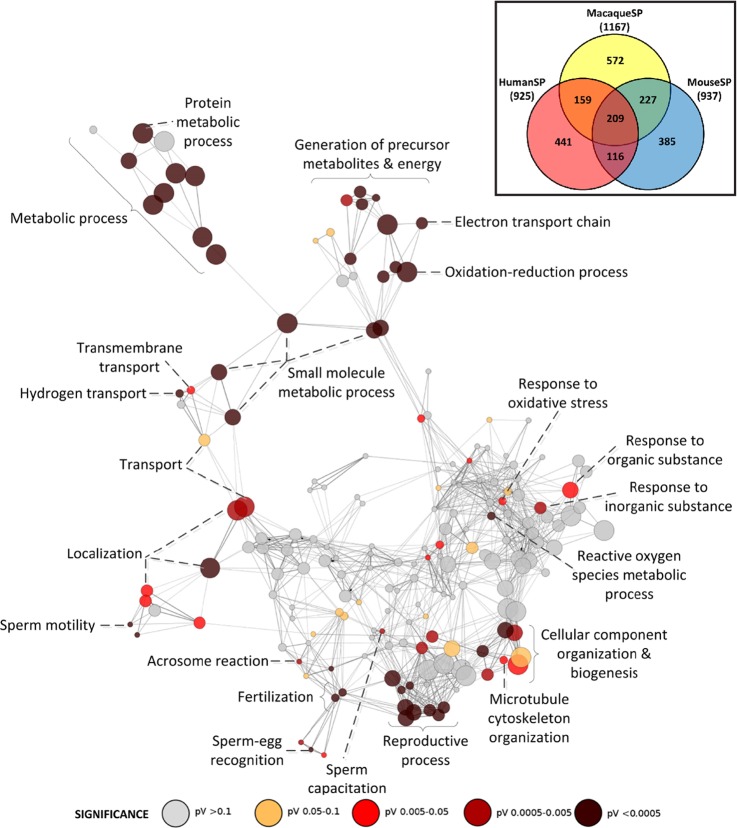
Overall, 209 proteins comprise a core data set common to all three sperm proteomes (supplemental Table S4). We analyzed the pattern of enrichment of the GO functional categories statistically over-represented by the triple overlap data set using Cytoscape (Fig. 4). As expected from the pair-wise comparisons, a number of functional categories related to reproduction, fertilization, and sperm-egg recognition are represented, along with metabolic and energy production processes. The high level of functional coherence observed reflects a significant coverage of the proteomes by our MS approach, however, a number of differences between the three sperm proteomes were also apparent (see Discussion).
Fig. 4.
Biological process network showing enriched functional categories for proteins found in the macaque, mouse and human sperm proteomes. Inset shows a venn diagram illustrating the overlap between the macaque, mouse and human sperm proteomes.
Phylogenetic Analysis of Macaque ADAM-like Proteins
Consistent with biochemical studies of mammalian sperm and functional studies of sperm-egg fusion (29, 30) our analysis identified eight members of the ADAM family in the MacSP. Among these, three proteins were annotated as ADAM-like, including ADAM18-, 20-, and 21-like, indicating high sequence similarity with the annotated ADAM18, 20, and 21 genes. The ADAM family of proteins has undergone significant expansion in the mouse lineage, whereas many of the human orthologs are pseudogenes. Because many ADAM proteins have evolved key roles in sperm biology, we were interested in determining whether these ADAM-like proteins (iii) were mis-annotated, and represent known mammalian ADAMs or (ii) represented a functional expansion of ADAM proteins in the primate lineage. Frequent gene turnover and pseudogenization among the ADAM gene family of proteins led us to suspect that these genes may be mis-annotated because of gene gain/loss during primate evolution. Construction and examination of the ADAM family phylogenies using all available ADAM sequences for the macaque, human, chimpanzee, mouse, and rat robustly groups ADAM18, 20 and 21 within a monophyletic group, exclusive of the ADAM18, 20 and 21-like proteins. Further phylogenetic and reciprocal BLAST analysis demonstrates a close evolutionary relationship and phylogenetic cluster between ADAM18-like and ADAM3-, ADAM20-like and ADAM4- and ADAM21-like and ADAM6-groups (Fig. 3). The identification of orthologs of ADAM3, 4, and 6 in the MacSP provides empirical evidence of sperm genes functioning in primate taxa, which have been subsequently lost in the human lineage.
Genomic Analysis of the MacSP
The X chromosome contains an underrepresentation of genes with male biased expression or function because (1) the X chromosome spends two-thirds of its life history in females, and may therefore be a selectively unfavorable location for such genes, and (2) the mammalian X chromosome is inactivated following the meiotic stages of spermatogenesis. These predictions have been supported in the analysis of male-biased expressed genes in Drosophila and mouse (31, 32) and in the documented patterns of gene movement between the autosomes and the X chromosome (33, 34). An analysis of the macaque sperm proteome further supports these predictions as the chromosomal distribution of sperm proteome genes is not uniform and displays a 28.3% underrepresentation on the X chromosome (χ2 = 4.91; p < 0.05).
Expression Analysis
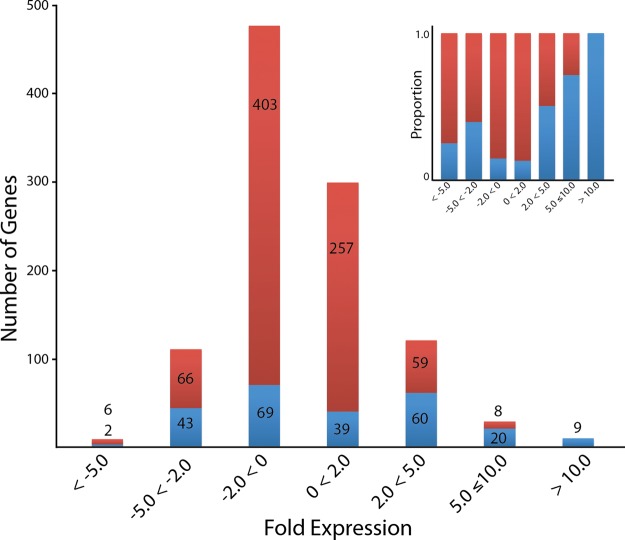
The testis expression of 1041 MacSP genes were extracted from previous macaque expression data sets (supplemental Table S2) and compared with the average expression of these genes in other tissues. The relative fold change distribution (Fig. 5) of the majority of testis-expressed genes (74%, 768/1041) was expressed in the interval between ± 2 fold and thus not significantly different from the relative expression in other tissues. A minority of genes were found to be under-expressed by more than twofold (117/1041; 11%), and less than 1% (8/1041) under-expressed fivefold or more. Only a modest fraction, (156/1041; 15%) were found to be overexpressed by >twofold, with only ∼4% (37/1041) overexpressed by >fivefold compared with other tissues. Interestingly, only nine genes (Table IV) were over-expressed by >10-fold. Functional analysis of the 37 genes overexpressed by >fivefold revealed that, five genes (SLC25A31, L1TD1, ZPBP, PACRG and REEP6) are unannotated for molecular function. However, GO annotates ZPBP for the biological process: binding of sperm to the zona pellucida, and two genes, SLC25A31 and PACRG, are annotated for cellular components: mitochondria. In humans, SLC25A31 is further annotated as having transporter activity and being involved in trans-membrane transport, and as a component of the flagellum. The remaining 32 genes were annotated for binding functions, including nucleotide, metal ion, and protein binding. Nearly half (15/32) also possess catalytic activity, and are annotated as hydrolases and transferases. Of the 32 genes, a number were also annotated as having structural molecule activity (5/32) and enzyme regulator activity (5/32).
Fig. 5.
Fold change distribution of the expression of macaque sperm genes in the testis as compared with the average expression in other tissues. The majority of genes are between twofold under- and overexpressed whereas a small subset show significant under or overexpression. Blue color indicates number of genes also found in the set of 242 unique genes whose testis expression is significantly different from expression in all other tissues. Inset: Comparison of the 242 statistically significant MacSP genes (blue) to all other MacSP genes as a proportion of the total.
Table IV. GO molecular function(s) and biological process(es) for macaque sperm genes found to be overexpressed in the testis by more than 10 fold as compared to other tissues.
| Gene ID (alternate gene ID) | Gene name | Fold change | GO molecular function(s) | GO biological process(es) |
|---|---|---|---|---|
| SERPINA5 (Q28500) | Plasma serine protease inhibitor | 17 | Protein binding; acrosin binding; peptidase inhibitor activity | Proteolysis, regulation of metabolic process, sexual reproduction, spermatogenesis |
| GSTM3 (Q6UIP7) | Glutathione S-transferase Mu 3 | 15 | Transferase activity; small molecule binding | Amine metabolic process, cellular catabolic process, response to stimulus |
| STK31 (TDRD8) | Serine/threonine-protein kinase 31 | 14 | Nucleotide binding; nucleic acid binding; kinase activity | Protein metabolic process, phosphorylation |
| MYH11 | Myosin heavy chain 11 | 13 | Nucleic acid binding transcription factor activity; hydrolase activity; nucleotide binding; nucleic acid binding | Gene expression, biosynthetic process, cell communication, cellular metabolic process, transcription, DNA dependent; response to stimulus, signaling, regulation of biological process |
| TUBA3E (LOC707215) | Loc707215 tubulin alpha-3C/D chain-like | 11 | GTPase activity, GTP binding, nucleotide binding | Catabolic process, microtubule-based process, nucleotide metabolic process, cellular component organization/biogenesis, |
| SLC25A31 | Solute carrier family 25, member 31 | 10 | Un-annotated | Un-annotated |
| DDX4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | 10 | Helicase activity; nucleotide binding; nucleic acid binding | Locomotion, protein localization, sperm motility, cell cycle process, meiosis, sexual reproduction, spermatogenesis |
| USP13 | Ubiquitin carboxyl-terminal hydrolase 13 | 10 | Ubiquitin thiolesterase activity; metal ion binding; protein binding | Catabolic process, proteolysis |
| MYLK | Myosin light chain kinase | 10 | Kinase activity; nucleotide binding, ATP binding, enzyme binding | Protein metabolic process, phosphorylation, developmental process, reproduction |
Table IV details the annotated molecular functions and biological processes of the nine genes found to be more than 10-fold overexpressed in the testis. Despite being present in sperm and highly overexpressed in the testis, three, SERPINA5, DDX4, and MYLK, are annotated as being involved with reproduction and only SERPINA5 and DDX4 are annotated as being involved in spermatogenesis. Five genes are annotated as having nucleotide binding molecular functions, three of which are also annotated as nucleic acid binding proteins. The remaining two genes, TUBA3E and MYLK, bind GTP and ATP, respectively. Overall, the nine highly overexpressed genes possess a range of molecular functions and are involved in diverse biological processes.
Our analyses further identified 242 genes expressed in macaque sperm that are significantly over- or underexpressed in the testis relative to other tissues (supplemental Table S2). Interestingly, these genes were about evenly split; 128 over- and 113 underexpressed. Despite the fact that sperm originate in the testis, this result illustrates that studying testis tissue expression is not necessarily informative with regards to sperm composition. Sperm generally acquire proteins in two different tissues, the testis during spermatogenesis and the epididymis during the post-testicular maturation process. This analysis can, however, provide some information with regards to the tissue of origin of sperm proteins. For example, the 128 significantly overexpressed testis genes represent a subclass of proteins with sperm-specific functions. Conversely, genes underexpressed in the testis, but whose protein products have been identified in the MacSP, may be translated and added to sperm during epididymal maturation. Additional functional genomic and bioinformatics analyses are needed to determine the overall significance of these protein classes.
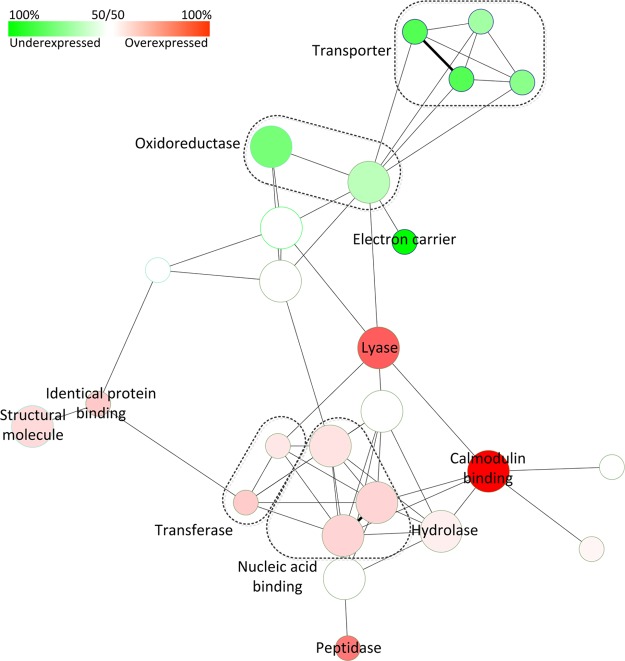
Fig. 6 shows a molecular function network analysis for the significantly over- and underexpressed genes. Nodes enriched with overexpressed genes are shown in red, and underexpressed genes shown in green. The most abundant annotated functional categories of over- and underexpressed genes include catalytic, binding, and structural molecules (data not shown), all of which are represented in the network analysis. However, genes with oxidoreductase activity have a tendency to be underexpressed in the testis as compared with other tissues, whereas genes with lyase, transferase, hydrolase, and peptidase activity are overexpressed. A similar number of genes also have a binding function, however nucleotide binding proteins, including ribonucleotide and purine nucleotide binding proteins tend to be overexpressed along with calmodulin binding proteins. Genes with an un-annotated molecular function comprise 34% of both overexpressed (44/128) of and underexpressed (39/114) genes, similar in proportion to all genes of the MacSP.
Fig. 6.
Network molecular function analysis for genes found to be expressed in macaque sperm that are significantly over- or under-expressed in the testis at p ≤ 0. 05 (242 genes total). Functional category nodes enriched for overexpressed genes are shown in red, and under-expressed shown in green.
DISCUSSION
We used strict parameters for peptide and protein inclusion to generate a robust Rhesus macaque sperm proteome containing 1247 proteins. These criteria required scores for peptide identifications at greater than 95.0% probability as specified by the Peptide Prophet algorithm (18) and protein identifications were only accepted if they could be established at greater than 99.0% probability and contained at least 3 identified peptides. Previous sperm proteomics studies have used different search parameters including proteins identified by two, or in some cases even one peptide (2, 6, 7). We chose our parameters to ensure the accuracy of the identified proteins and to reduce false positives, which is particularly crucial for comparative studies of sperm proteome composition designed to gain evolutionary insights of sperm function and origin.
Relevance to Studies of Human Fertility
Despite a number of similarities, a number of functional differences can be observed between the macaque/mouse and macaque/human SP overlap networks (supplemental Fig. S1). Interestingly, the MacSP and MouseSP contains a group related to complement activation, whereas this node is absent in the MacSP and HumanSP network. Indeed, the MacSP and MouseSP contain several proteins involved in complement activation including C3, CD46, and CD55 not identified in the HumanSP. CD46 is a rapidly evolving protein in the mouse, and is associated with the sperm acrosome, although its precise role in acrosome biology is unknown, whereas CD55 is thought to play a role in modulating the complement pathway (35, 36). In the sexual reproduction group, proteins involved in sperm–egg recognition are enriched in the MacSP and MouseSP but not in the MacSP and HumanSP network. Further examination revealed three proteins present in the MacSP and MouseSPs absent from the HumanSP and include ADAM3 (a pseudogene in humans), ZAN and ZPBP2, all of which have previously characterized sperm phenotypes (see Table III). These differences could reflect the unique evolutionary trajectory of these proteins in the human lineage, and therefore provide targets for future studies into their functional significance.
The MacSP and HumanSP network analysis reveals a number of functional categories that are not present in the MacSP and MouseSP overlap networks including homeostasis, negative regulation of cell growth, cell cycle regulation, antigen processing and presentation, membrane organization, and maintenance of localization. Several proteins involved in membrane organization were identified in the MacSP and HumanSP not identified in the MouseSP and include ANXA1, ANXA2, CACNA1A, HSPA4, RPS27A, and SYNE2. Two proteins involved in antigen processing and presentation, RPS27A and NPEPPS, were also absent from the MouseSP but present in the MacSP and HumanSP. Finally, the MacSP and HumanSP contains several proteins involved in maintenance of localization, which were not identified in the MouseSP including ARHGAP21, FLNA, GAA, JUP, SYNE1, SYNE2, and TLN1. Overall, the proteins common to the MacSP and HumanSP fall into a much broader range of functional categories than the proteins identified in the MacSP and MouseSP. A deeper understanding and the significance of these differences await further MS verification and functional studies.
Several previous studies have used MS to investigate the proteomic composition of human ejaculates, a combination of sperm mixed with seminal fluids. A previous study identified 923 proteins in seminal fluid and noted that a few proteins (e.g. semenogelins) present at high concentrations complicated identification of lower abundance proteins (16). Likewise, proteomic analyses of sperm from ejaculates could similarly mask low abundance proteins and reduce the number of sperm proteins identified while at the same time making it difficult to distinguish sperm from seminal fluid proteins. For example, a previous study identified heat shock proteins HSPA5 and HSPD1 on the surface of ejaculated human sperm, however it was unclear whether these proteins are components of sperm or simply adsorbed to the sperm surface following ejaculation (37). The identification of HSPA5 and HSPD1 in macaque epididymal sperm suggests that these proteins are sperm components before ejaculation. This finding is further supported by the identification of HSPA5 and HSPD1 in mouse epididymal sperm (9). Because this study used epididymal sperm devoid of seminal fluid, the data from this study can, by inference, be extended to the human sperm proteome.
A study by Johnston et al. identified 1760 proteins in human sperm for which the resulting list of sperm proteins was not published (10). However, this study acknowledged that many of the proteins found on ejaculated sperm are synthesized in the accessory sex glands, and acquired long after the morphological differentiation of the spermatozoa, potentially indicating that a large portion of their data set was composed of seminal proteins, rather than sperm proteins. A subsequent study, using proteins separated into Triton X-100 soluble and insoluble fractions, identified 1053 proteins in human sperm (6). This data set contained semenogelins, which are a predominant component of semen (38–40). GO functional analysis also revealed that the most prevalent proteins are those with binding and catalytic functions (6), consistent with our analysis of the MacSP. Additionally, we observed that oxidoreductases, including oxidoreductases acting on NADH and NADPH, and proteins with electron carrier and antioxidant activity are highly enriched in the macaque, mouse and human sperm proteomes. The abundance of these GO categories presumably reflects the demand on sperm to generate energy for motility.
We compared the human sperm proteome (925/1053 proteins for which we were able to identify macaque orthologs) and MacSP and found 368 proteins common to both proteomes. Based on their evolutionary relationship, it was expected that the macaque and human sperm proteomes would share a higher degree of overlap than the macaque (iii) and mouse sperm proteomes. Interestingly, we observed a higher degree of overlap between the macaque and mouse sperm proteomes. This observation may be because of a number of factors including (i) contamination of the HumanSP with seminal proteins leading to problems discussed above (ii) differences in sample preparation and MS, leading to selection or enrichment of particular types of proteins, or (iii) significant evolutionary differences between sperm composition in humans and primates, reducing the amount of overlap observed between the Rhesus macaque and human sperm proteomes.
Sperm Proteasome
The sperm proteasome has recently become of interest because of its likely role in facilitating penetration through the egg vitelline coat during fertilization, sperm capacitation and the acrosome reaction (41, 42). GO analysis revealed that the MacSP, MouseSP, and HumanSP are all highly enriched for proteins in the proteasome complex as determined using g:Profiler. (p = 5.86 e-10, p = 2.08 e-28 and p = 3.93 e-24 respectively). The 26S proteasome functions to degrade ubiquitinated proteins and consists of a 20S core particle and a 19S regulatory complex positioned at one or both ends of the core. We identified 28 proteasome proteins in the MacSP (Fig. 7) including PSMA1–6, PSMA8, a testis-specific paralog of PSMA7, PSMB1–7, which comprises the entire 20S core particle, PSMC 3, 4, 5, 6 and PSMD1–2, which comprises most of the 19S base structure, and PSMD3, 7, and 11–14, which comprises most of the 19S lid structure. A previous study found that one component of the alpha proteasome complex, PSMA3, was reduced in patients with asthenozoospermia as compared with normozoospermic patients, suggesting that proteasome activity (or some unknown component of the proteasome) may play a role in sperm motility (43). We also identified PSMG2, which forms a heterodimer with PSMG1 (not identified) and promotes assembly of the 20S core subunit (44).
Fig. 7.
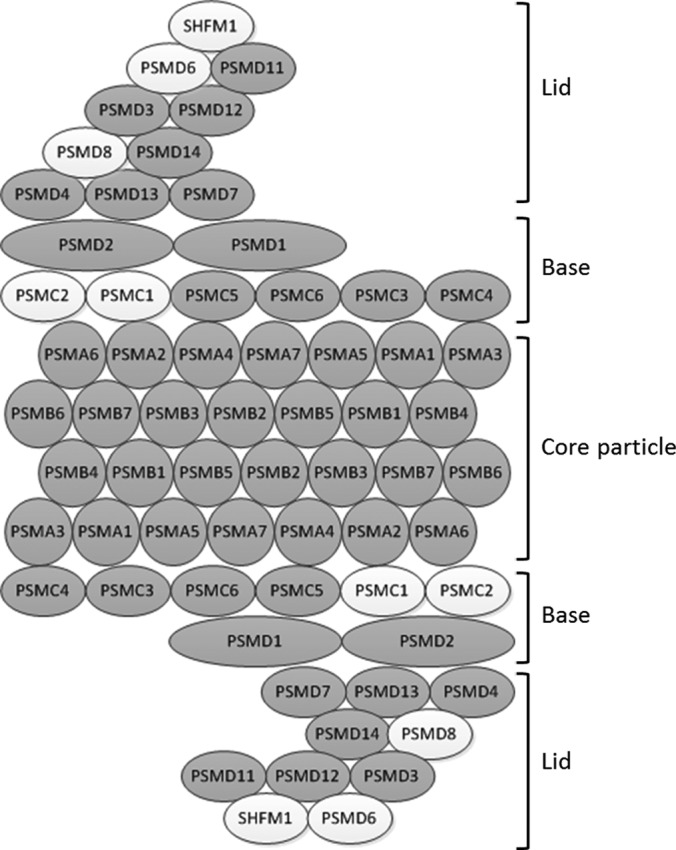
Graphical representation of the 26S proteasome. Subunits identified in the MacSP are shaded, whereas those which were not identified remain white. Every subunit comprising the 20S core particle (PSMA1–6, PSMA8 and PSMB1–7) were identified, as well as PSMG2 (not pictured), which functions to facilitate 20S assembly. The majority of the base and lid complex, which comprise the 19S regulatory particle, were also identified. Although not depicted, PSME4 (PA200) was also identified in the MacSP and functions to replace one or both of the 19S regulatory particles. Fig. adapted from KEGG (www.genome.jp/kegg/).
Finally, an alternate proteasome activator protein, PSME4 (PA200) was also identified in the MacSP. PSME4 has been proposed to replace the 19S regulatory complex at one or both ends of the 20S core particle (reviewed in (45)) and play a role in DNA double strand break repair (46), however mice lacking this gene show no developmental defects (47). Mice lacking PSME4, however, exhibited marked reduction in male, but not female, fertility as a result of pre- and postmeiotic defects in spermatogenesis resulting in a reduction of normal sperm (47). Interestingly, PSME4 exhibits broad tissue expression with the highest level of expression detected in the testis. Identification of a myriad of proteasome subunits in mature sperm suggests that mature proteasomes are present in sperm as they leave the epididymis and that various different proteasome complexes, such as those employing the 19S or PSME4 (PA200) regulatory complex may exist in mature sperm.
Divergence of Mating Systems and Proteome Diversification
Aminopeptidases
Although their precise function in sperm has yet to be elucidated, aminopeptidases appear to play an important role in sperm development and function and several members are found to be constituents of sperm across diverse taxa. In Drosophila the leucine aminopeptidase gene family has undergone significant expansion and functional diversification and represents the most abundant proteins in sperm, however, their function(s) have yet to be determined (48). Four aminopeptidases were identified in the MacSP including leucine aminopeptidase 3, LAP3; aminopeptidase-like 1, NPEPL1; an aspartyl aminopeptidase, DNPEP; and a puromycin-sensitive aminopeptidase NPEPPS. All four aminopeptidases identified in the MacSP have been shown in other mammals to play crucial roles in sperm development or function and are likely conserved constituents of sperm across a variety of taxa, however further studies are required to understand their roles in sperm biology.
LAP3 has also been identified in the mouse (9), rat (7), human (6) and Drosophila (2) sperm proteomes and thus appears to be a conserved sperm constituent across taxa. The MacSP also includes a paralog of LAP3, NEPEL1 (ensembl.org), also identified in the rat sperm proteome (7). A previous study identified both LAP3 and NEPEL1 as potential targets for protein S- nitrosylation (49), which has been proposed to occur in the female reproductive tract because oviduct cells produce biologically significant levels of NO (50). It is thus possible that this modification plays a key role in activating these proteins and that they play a role in capacitation, the acrosome reaction, or sperm-egg binding.
DNPEP has also been identified in the mouse (9), rat (7) and human (5) sperm proteomes and a recent MS study identified DNPEP in bovine epididymosomes from both the caput and cauda epididymis (51). Epididymosomes have been proposed to play a role in the transfer of proteins to sperm during the maturation process in the epididymis. However, only a few proteins are known to be acquired by this mechanism. Although it remains unclear whether DNPEP is added to sperm via epididymosomes during the maturation process, this protein has been suggested to play a role in remodeling epididymal sperm components (51). A previous study identified DNPEP in mouse cauda epididymal sperm and found that peptide intensity, as measured by MS, increased more than twofold in capacitated sperm suggesting significant post-translational modification (52). This study also predicted that DNPEP has a phosphorylation motif, and thus may undergo post-translational modification during capacitation.
NPEPPS, a puromycin sensitive aminopeptidase, was also identified in the rat and human sperm proteomes and is believed to play a role in male fertility and protection against protein aggregation (53). NPEPPS deficient mice exhibit a wide range of phenotypes including loss of behavioral mating activity, germ cell degeneration, lower testis weight, sperm number, and sperm motility (54).
ADAM Proteins
Hydrolases were one category of catalytic proteins found to be highly enriched in the MacSP (p = 4.83 e-12) and include peptidases and, in particular, metalloproteases. The MacSP contains several members of the ADAM (a-disintegrin and metalloprotease) family of proteins including ADAM1, 2, 10, 30, 32. These ADAMS, excluding ADAM10, are known to exhibit testis specific or testis-biased expression and have been previously identified in mature sperm.
ADAMs 1 and 2 are expressed as heterodimers on the sperm surface and are essential for sperm migration through the female reproductive tract, zona pellucida binding on the egg, and have also been suggested to play a role in the regulation and localization of sperm proteins (55). Although present in many nonhuman primates, ADAM1 is nonfunctional in humans as a result of a variety of mutations that disrupt the reading frame (56). Although many ADAMs involved in reproduction and expressed on the sperm surface have found to be under positive selection (9, 57), in the macaque the adhesion domain of ADAM2 has been found to be under accelerated evolution (57). The function of ADAMs 30 and 32 in sperm is unknown, however previous work (58, 59) determined that they were localized on the sperm surface, and ADAM32 has been suggested to play a role in sperm development or fertilization. Notably, ADAM30 is not found on the surface of testicular sperm (58) suggesting that it may be acquired during epididymal transit.
Additionally, we identified ADAM10 in the MacSP, which to our knowledge is the first time this ADAM has been identified as a constituent of mature sperm. ADAM10 is expressed in a variety of tissues and is highly expressed in the central nervous system (60). Previous studies have attempted to generate ADAM10 knockout mice, and whereas heterozygous males have normal phenotypes and are fertile, homozygous mice are not viable and die early in embryogenesis (61). Although fertile, this study did not assay whether heterozygous males exhibited a reduction in fertility or abnormal sperm physiology or number, which remains to be elucidated. ADAM10 did not exhibit signs of positive selection in the mammalian lineage (57), which suggests that ADAM10 may play a role in a conserved cellular process unrelated to sperm-egg binding. Although the role of ADAM10 in mature sperm has yet to be investigated, it may play a role in DNA damage induced apoptosis as ADAM10, along with ADAM17, increase in abundance and cell surface localization in rat spermatocytes during DNA damage induced apoptosis (62). ADAM10 has previously been implicated in the NOTCH or EGFR signaling pathways. Given the known role of the EGFR signaling pathway in both capacitation and the acrosome reaction (reviewed in (63)), ADAM10 may play a role in one or both of these processes.
The MacSP also contains 3 ADAM-like proteins annotated as ADAM18, 20, and 21-like. Protein BLAST queries of these proteins identified higher similarity to other ADAMs, and further examination of the ADAM family phylogenies using all available ADAM sequences (obtained from NCBI) for the human, chimpanzee, mouse, and rat clearly places ADAM18, 20 and 21 in their own monophyletic group (Fig. 6). This analysis allowed us to assign ADAM18-like to the ADAM3-, ADAM20-like to the ADAM4- and ADAM21-like to the ADAM6-groups. Interestingly, human ADAMs 3, 4, and 6 are annotated as pseudogenes (64, 65) and thus are not present in the human protein database. This most likely explains the mis-annotation of the macaque ADAMs 3, 4 and 6 given that annotation of the macaque proteome was likely largely based on the human (12). Although the human genome contains two copies of ADAM3 (CYRN1 and CYRN2), a previous study demonstrated that 27% of fertile men carry a large deletion in CYRN1 and CYRN2 contains stop codons in all three reading frames (66). Although CYRN1 and CYRN2 are transcribed, they are not translated and are thus nonfunctional (66). Most pseudogenes exist as disabled copies of functional parent genes, however a recent study identified ADAM3 as a unitary pseudogene in humans. A unitary pseudogene has no correlated functional parental gene, and loss of ADAM3 function is thought to have occurred in the human lineage after human-chimpanzee divergence (65). Likewise, the human ADAM4 genes contain a number of stop codons, whereas the ADAM6 gene has a frameshift mutation, resulting in nonfunctional pseudogenes (64). The identification of orthologs of ADAM3, 4 and 6 in the MacSP provides empirical evidence of sperm genes functioning in primate taxa, which have been subsequently lost in the human lineage. In species for which ADAMs 3, 4 and 6 have been studied, they exhibit testis-biased expression and have been identified in sperm (reviewed in (59)).
Despite the widespread use of rodents in studies of male infertility, nonhuman primates (especially Old World Monkeys such as the Rhesus macaque) are indispensable models for the study of a wide range of physiological, hormonal and molecular reproductive processes in humans. Characterization of the macaque sperm proteome, using highly sensitive and robust proteomic and statistical methodologies, has identified a wealth of proteins with identified or predicted functions in human or mouse sperm and therefore provides a power foundation for future molecular genetic studies of human spermatogenesis and, importantly, studies that link population-level genetic variation to sperm function and competitive ability. Previous studies using similar proteomic approaches in humans have been limited by the availability of “pure” sperm isolated form the epididymis, further emphasizing the relevance of the macaque as a model for the study of male fertility. The macaque sperm proteome has also highlighted specific cases where the molecular basis of sperm is divergent between closely related primate species and thus provides novel empirical support for the rapid evolution of genetic systems which differentiate humans from the closest relatives.
Supplementary Material
Footnotes
*
 This article contains supplemental Fig. S1 and Tables S1 to S4.
This article contains supplemental Fig. S1 and Tables S1 to S4.
1 The abbreviations used are:
- MacSP
- Macaque Sperm Proteome
- BLAST
- Basic Local Alignment Search Tool
- CCD
- Charge-coupled Device
- DAPI
- 4′,6-diamidino-2-phenylindole
- DIC
- Differential Interference Contrast
- FDR
- False Discovery Rate
- GO
- Gene Ontology
- LTQ
- Linear Trap Quadrapole.
REFERENCES
- 1. Dorus S., Busby S. A., Gerike U., Shabanowitz J., Hunt D. F., Karr T. L. (2006) Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38, 1440–1445 [DOI] [PubMed] [Google Scholar]
- 2. Wasbrough E. R., Dorus S., Hester S., Howard-Murkin J., Lilley K., Wilkin E., Polpitiya A., Petritis K., Karr T. L. (2010) The Drosophila melanogaster sperm proteome-II (DmSP-II). J. Proteomics 73, 2171–2185 [DOI] [PubMed] [Google Scholar]
- 3. Martínez-Heredia J., Estanyol J. M., Ballescà J. L., Oliva R. (2006) Proteomic identification of human sperm proteins. Proteomics 6, 4356–4369 [DOI] [PubMed] [Google Scholar]
- 4. Oliva R., de Mateo S., Estanyol J. M. (2009) Sperm cell proteomics. Proteomics 9, 1004–1017 [DOI] [PubMed] [Google Scholar]
- 5. de Mateo S., Castillo J., Estanyol J. M., Ballesca J. L., Oliva R. (2011) Proteomic characterization of the human sperm nucleus. Proteomics 11, 2714–2726 [DOI] [PubMed] [Google Scholar]
- 6. Baker M. A., Reeves G., Hetherington L., Muller J., Baur I., Aitken R. J. (2007) Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin. Appl. 1, 524–532 [DOI] [PubMed] [Google Scholar]
- 7. Baker M. A., Hetherington L., Reeves G., Muller J., Aitken R. J. (2008) The rat sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics 8, 2312–2321 [DOI] [PubMed] [Google Scholar]
- 8. Baker M. a., Hetherington L., Reeves G. M., Aitken R. J., Müller J. (2008) The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics 8, 1720–1730 [DOI] [PubMed] [Google Scholar]
- 9. Dorus S., Wasbrough E. R., Busby J., Wilkins E. C., Karr T. L. (2010) Sperm Proteomics Reveals Intensified Selection on Mouse Sperm Membrane and Acrosome Genes. Mol. Biol. Evolution 27, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnston D. S., Wooters J., Kopf G. S., Qiu Y., Roberts K. P. (2005) Analysis of the human sperm proteome. Annals of the New York Academy of Sciences 1061, 190–202 [DOI] [PubMed] [Google Scholar]
- 11. Peddinti D., Nanduri B., Kaya A., Feugang J. M., Burgess S. C., Memili E. (2008) Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst. Biol. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibbs R. A., Rogers J., Katze M. G., Bumgarner R., Weinstock G. M., Mardis E. R., Remington K. A., Strausberg R. L., Venter J. C., Wilson R. K., Batzer M. A., Bustamante C. D., Eichler E. E., Hahn M. W., Hardison R. C., Makova K. D., Miller W., Milosavljevic A., Palermo R. E., Siepel A., Sikela J. M., Attaway T., Bell S., Bernard K. E., Buhay C. J., Chandrabose M. N., Dao M., Davis C., Delehaunty K. D., Ding Y., Dinh H. H., Dugan-Rocha S., Fulton L. A., Gabisi R. A., Garner T. T., Godfrey J., Hawes A. C., Hernandez J., Hines S., Holder M., Hume J., Jhangiani S. N., Joshi V., Khan Z. M., Kirkness E. F., Cree A., Fowler R. G., Lee S., Lewis L. R., Li Z., Liu Y. S., Moore S. M., Muzny D., Nazareth L. V., Ngo D. N., Okwuonu G. O., Pai G., Parker D., Paul H. A., Pfannkoch C., Pohl C. S., Rogers Y. H., Ruiz S. J., Sabo A., Santibanez J., Schneider B. W., Smith S. M., Sodergren E., Svatek A. F., Utterback T. R., Vattathil S., Warren W., White C. S., Chinwalla A. T., Feng Y., Halpern A. L., Hillier L. W., Huang X., Minx P., Nelson J. O., Pepin K. H., Qin X., Sutton G. G., Venter E., Walenz B. P., Wallis J. W., Worley K. C., Yang S. P., Jones S. M., Marra M. A., Rocchi M., Schein J. E., Baertsch R., Clarke L., Csuros M., Glasscock J., Harris R. A., Havlak P., Jackson A. R., Jiang H., Liu Y., Messina D. N., Shen Y., Song H. X., Wylie T., Zhang L., Birney E., Han K., Konkel M. K., Lee J., Smit A. F., Ullmer B., Wang H., Xing J., Burhans R., Cheng Z., Karro J. E., Ma J., Raney B., She X., Cox M. J., Demuth J. P., Dumas L. J., Han S. G., Hopkins J., Karimpour-Fard A., Kim Y. H., Pollack J. R., Vinar T., Addo-Quaye C., Degenhardt J., Denby A., Hubisz M. J., Indap A., Kosiol C., Lahn B. T., Lawson H. A., Marklein A., Nielsen R., Vallender E. J., Clark A. G., Ferguson B., Hernandez R. D., Hirani K., Kehrer-Sawatzki H., Kolb J., Patil S., Pu L. L., Ren Y., Smith D. G., Wheeler D. A., Schenck I., Ball E. V., Chen R., Cooper D. N., Giardine B., Hsu F., Kent W. J., Lesk A., Nelson D. L., O'brien W. E., Prufer K., Stenson P. D., Wallace J. C., Ke H., Liu X. M., Wang P., Xiang A. P., Yang F., Barber G. P., Haussler D., Karolchik D., Kern A. D., Kuhn R. M., Smith K. E., Zwieg A. S. (2007) Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234 [DOI] [PubMed] [Google Scholar]
- 13. Hewitson L., Schatten G. (2002) The Use of Primates as Models for Assisted Reproduction. Reproductive Biomedicine Online 5, 50–55 [DOI] [PubMed] [Google Scholar]
- 14. Gatti J.-L., Castella S., Dacheux F., Ecroyd H., Métayer S., Thimon V., Dacheux J.-L. (2004) Post-testicular sperm environment and fertility. Animal Reproduction Sci. 82–83, 321–339 [DOI] [PubMed] [Google Scholar]
- 15. Jones R. C. (1998) Evolution of the vertebrate epididymis. J. Reproduction Fertility. Supplement 53, 163. [PubMed] [Google Scholar]
- 16. Pilch B., Mann M. (2006) Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 7, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 18. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 19. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry abilities that proteins are present in a sample on the basis. Society 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 20. Reimand J., Kull M., Peterson H., Hansen J., Vilo J. (2007) g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193-W200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z., Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones D. T., Taylor W. R., Thornton J. M. (1992) The Rapid Generation of Mutation Data Matrices From Protein Sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 23. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 783–791 [DOI] [PubMed] [Google Scholar]
- 25. Duan F., Spindel E. R., Li Y.-H., Norgren R. B. (2007) Intercenter reliability and validity of the rhesus macaque GeneChip. BMC Genomics 8, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lundgren D. H., Hwang S. I., Wu L., Han D. K. (2010) Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 7, 39–53 [DOI] [PubMed] [Google Scholar]
- 27. Brown P. R., Miki K., Harper D. B., Eddy E. M. (2003) A-kinase anchoring protein 4 binding proteins in the fibrous sheath of the sperm flagellum. Biol. Reproduction 68, 2241–2248 [DOI] [PubMed] [Google Scholar]
- 28. Mochida K., Rivkin E., Gil M., Kierszenbaum a. L. (2000) Keratin 9 is a component of the perinuclear ring of the manchette of rat spermatids. Developmental Biol. 227, 510–519 [DOI] [PubMed] [Google Scholar]
- 29. Moreno R. D., Urriola-Munoz P., Lagos-Cabre R. (2011) The emerging role of matrix metalloproteases of the ADAM family in male germ cell apoptosis. Spermatogenesis 1, 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Goor H., Melenhorst W. B., Turner A. J., Holgate S. T. (2009) Adamalysins in biology and disease. J. Pathol. 219, 277–286 [DOI] [PubMed] [Google Scholar]
- 31. Vibranovski M. D., Lopes H. F., Karr T. L., Long M. (2009) Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5, e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zamudio N. M., Chong S., O'Bryan M. K. (2008) Epigenetic regulation in male germ cells. Reproduction 136, 131–146 [DOI] [PubMed] [Google Scholar]
- 33. Wang J., Long M., Vibranovski M. D. (2012) Retrogenes moved out of the z chromosome in the silkworm. J. Mol. Evol. 74, 113–126 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y. E., Vibranovski M. D., Landback P., Marais G. A., Long M. (2010) Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dorus S., Skerget S., Karr T. L. (2012) Proteomic discovery of diverse immunity molecules in mammalian spermatozoa. Syst. Biol. Reprod. Med. 58, 218–228 [DOI] [PubMed] [Google Scholar]
- 36. Inoue N., Ikawa M., Nakanishi T., Matsumoto M., Nomura M., Seya T., Okabe M. (2003) Disruption of mouse CD46 causes an accelerated spontaneous acrosome reaction in sperm. Mol. Cell Biol. 23, 2614–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Naaby-Hansen S., Herr J. C. (2010) Heat shock proteins on the human sperm surface. J. Reproductive Immunol. 84, 32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Lamirande E. (2007) Semenogelin, the main protein of the human semen coagulum, regulates sperm function. Semin. Thromb. Hemost. 33, 60–68 [DOI] [PubMed] [Google Scholar]
- 39. Milardi D., Grande G., Vincenzoni F., Messana I., Pontecorvi A., De Marinis L., Castagnola M., Marana R. (2012) Proteomic approach in the identification of fertility pattern in seminal plasma of fertile men. Fertil. Steril. 97, 67–73.e1 [DOI] [PubMed] [Google Scholar]
- 40. Ulvsback M., Lazure C., Lilja H., Spurr N. K., Rao V. V., Loffler C., Hansmann I., Lundwall A. (1992) Gene structure of semenogelin I and II. The predominant proteins in human semen are encoded by two homologous genes on chromosome 20. J. Biol. Chem. 267, 18080–18084 [PubMed] [Google Scholar]
- 41. Zimmerman S., Sutovsky P. (2009) The sperm proteasome during sperm capacitation and fertilization. J. Reprod. Immunol. 83, 19–25 [DOI] [PubMed] [Google Scholar]
- 42. Zimmerman S. W., Manandhar G., Yi Y. J., Gupta S. K., Sutovsky M., Odhiambo J. F., Powell M. D., Miller D. J., Sutovsky P. (2011) Sperm proteasomes degrade sperm receptor on the egg zona pellucida during mammalian fertilization. PLoS One 6, e17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siva A. B., Kameshwari D. B., Singh V., Pavani K., Sundaram C. S., Rangaraj N., Deenadayal M., Shivaji S. (2010) Proteomics-based study on asthenozoospermia: differential expression of proteasome alpha complex. Mol. Hum. Reprod. 16, 452–462 [DOI] [PubMed] [Google Scholar]
- 44. Hirano Y., Hendil K. B., Yashiroda H., Iemura S., Nagane R., Hioki Y., Natsume T., Tanaka K., Murata S. (2005) A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437, 1381–1385 [DOI] [PubMed] [Google Scholar]
- 45. Rechsteiner M., Hill C. P. (2005) Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 15, 27–33 [DOI] [PubMed] [Google Scholar]
- 46. Ustrell V., Hoffman L., Pratt G., Rechsteiner M. (2002) PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21, 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khor B., Bredemeyer A. L., Huang C. Y., Turnbull I. R., Evans R., Maggi L. B. J., White J. M., Walker L. M., Carnes K., Hess R. A., Sleckman B. P. (2006) Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell Biol. 26, 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dorus S., Wilkin E. C., Karr T. L. (2011) Expansion and functional diversification of a leucyl aminopeptidase family that encodes the major protein constituents of Drosophila sperm. BMC Genomics 12, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lefievre L., Chen Y., Conner S. J., Scott J. L., Publicover S. J., Ford W. C., Barratt C. L. (2007) Human spermatozoa contain multiple targets for protein S-nitrosylation: an alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics 7, 3066–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosselli M., Keller P. J., Dubey R. K. (1998) Role of nitric oxide in the biology, physiology and pathophysiology of reproduction. Hum. Reprod. Update 4, 3–24 [DOI] [PubMed] [Google Scholar]
- 51. Girouard J., Frenette G., Sullivan R. (2011) Comparative proteome and lipid profiles of bovine epididymosomes collected in the intraluminal compartment of the caput and cauda epididymidis. Int. J. Androly. 34, e475–e486 [DOI] [PubMed] [Google Scholar]
- 52. Baker M. A., Reeves G., Hetherington L., Aitken R. J. (2010) Analysis of proteomic changes associated with sperm capacitation through the combined use of IPG-strip pre-fractionation followed by RP chromatography LC-MS/MS analysis. Proteomics 10, 482–495 [DOI] [PubMed] [Google Scholar]
- 53. Menzies F. M., Hourez R., Imarisio S., Raspe M., Sadiq O., Chandraratna D., O'Kane C., Rock K. L., Reits E., Goldberg A. L., Rubinsztein D. C. (2010) Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum. Mol. Genet. 19, 4573–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Osada T., Watanabe G., Kondo S., Toyoda M., Sakaki Y., Takeuchi T. (2001) Male reproductive defects caused by puromycin-sensitive aminopeptidase deficiency in mice. Mol. Endocrinol. 15, 960–971 [DOI] [PubMed] [Google Scholar]
- 55. Cho C., Bunch D. O., Faure J. E., Goulding E. H., Eddy E. M., Primakoff P., Myles D. G. (1998) Fertilization defects in sperm from mice lacking fertilin beta. Science 281, 1857–1859 [DOI] [PubMed] [Google Scholar]
- 56. Jury J. A., Frayne J., Hall L. (1998) Sequence analysis of a variety of primate fertilin alpha genes: evidence for non-functional genes in the gorilla and man. Mol. Reprod. Dev. 51, 92–97 [DOI] [PubMed] [Google Scholar]
- 57. Finn S., Civetta A. (2010) Sexual Selection and the Molecular Evolution of ADAM Proteins. J. Mol. Evol. 71, 231–240 [DOI] [PubMed] [Google Scholar]
- 58. Han C., Choi E., Park I., Lee B., Jin S., Kim D. H., Nishimura H., Cho C. (2009) Comprehensive analysis of reproductive ADAMs: relationship of ADAM4 and ADAM6 with an ADAM complex required for fertilization in mice. Biol. Reproduction 80, 1001–1008 [DOI] [PubMed] [Google Scholar]
- 59. Kim T., Oh J., Woo J.-M., Choi E., Im S. H., Yoo Y. J., Kim D. H., Nishimura H., Cho C. (2006) Expression and relationship of male reproductive ADAMs in mouse. Biol. Reproduction 74, 744–750 [DOI] [PubMed] [Google Scholar]
- 60. Karkkainen I., Rybnikova E., Pelto-Huikko M., Huovila A. P. (2000) Metalloprotease-disintegrin (ADAM) genes are widely and differentially expressed in the adult CNS. Mol. Cell Neurosci. 15, 547–560 [DOI] [PubMed] [Google Scholar]
- 61. Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lubke T., Lena Illert A., von Figura K., Saftig P. (2002) The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 62. Lizama C., Rojas-Benitez D., Antonelli M., Ludwig A., Moreno R. D. (2012) Involvement of TACE/ADAM17 and ADAM10 in etoposide-induced apoptosis of germ cells in rat spermatogenesis. J. Cell Physiol. 227, 829–838 [DOI] [PubMed] [Google Scholar]
- 63. Breitbart H., Etkovitz N. (2011) Role and regulation of EGFR in actin remodeling in sperm capacitation and the acrosome reaction. Asian J. Androl. 13, 106–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Choi I., Oh J., Cho B. N., Ahnn J., Jung Y. K., Han Kim D., Cho C. (2004) Characterization and comparative genomic analysis of intronless Adams with testicular gene expression. Genomics 83, 636–646 [DOI] [PubMed] [Google Scholar]
- 65. Zhang Z. D., Frankish A., Hunt T., Harrow J., Gerstein M. (2010) Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 11, R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grzmil P., Kim Y., Shamsadin R., Neesen J., Adham I. M., Heinlein U. A., Schwarzer U. J., Engel W. (2001) Human cyritestin genes (CYRN1 and CYRN2) are non-functional. Biochem. J. 357, 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.