Abstract
Patients with advanced head and neck squamous cell carcinoma (HNSCC) have a poor prognosis with the currently available therapy, and tumor recurrence is frequently observed. The discovery of specific membrane-associated cancer stem cell (CSC) markers is crucial for the development of novel therapeutic strategies to target these CSCs. To address this issue, we established sphere cultures to enrich CSCs and used them for plasma membrane proteomics to identify specific membrane signatures of the HNSCC spheres. Of a dataset that included a total of 376 identified proteins, 200 were bona fide membrane proteins. Among them, 123 proteins were at least 1.5-fold up- or down-regulated in the spheres relative to the adherent cultures. These proteins included cell adhesion molecules, receptors, and transporter proteins. Some of them play key roles in wnt, integrin, and TGFβ signaling pathways. When we compared our dataset with two published hESC membrane protein signatures, we found 18 proteins common to all three of the databases. CD166 and CD44 were two such proteins. Interestingly, the expression of CD166, rather than that of the well-established HNSCC CSC marker CD44, was significantly related to the malignant behavior of HNSCC. Relative to CD166low HNSCC cells, CD166high HNSCC cells had a greater sphere-formation ability in vitro and tumor formation ability in vivo. Patients whose tumors expressed high levels of CD166 had a significantly poorer clinical outcome than those whose tumors expressed low levels of CD166 (cohort 1: 96 cases, p = 0.040), whereas the level of CD44 expression had only a marginal influence on the clinical outcome of patients with HNSCC (p = 0.078). The level of CD166 expression in HNSCC tumors was also associated with the tumor recurrence rate (cohort 2: 104 cases, p = 0.016). This study demonstrates that CD166 is a valuable cell surface marker for the enrichment of HNSCC stem cells and that plasma membrane proteomics is a promising biological tool for investigating the membrane proteins of CSCs.
Head and neck squamous cell carcinoma (HNSCC)1 is the sixth most common cancer worldwide. Despite ongoing improvement in traditional treatments, the long-term survival rate of patients with HNSCC has not significantly improved over the past several decades. More than 60% of patients with advanced tumors or localized lymph node metastases die within five years of their diagnosis (1). Tumor recurrence and resistance to therapy are the major causes of death. Recently, newly recognized cancer stem cells (CSCs) or tumor-initiating cells have been associated in a cause-and-effect manner with tumor recurrence and resistance to therapy. The concept of CSCs was established because of the heterogeneous nature of cancer and suggests that CSCs are a subpopulation of cancer cells with stem-cell-like traits and the source of all cells in the cancer. Conventional cancer therapies such as chemotherapy and radiotherapy may destroy only those cells that form the bulk of the tumor, leaving the CSCs intact and able to give rise to tumor recurrence. Based on this theory, researchers are searching for therapies that would destroy CSCs in the hope of finally curing cancer (2). In order to develop strategies that target CSCs, experimental assays are required to determine how to distinguish CSCs from their progeny. Different methods have been used to isolate CSCs from a range of hematopoietic and solid tumors, and some CSC-specific cell surface markers have been found. These markers are primarily selected from the corresponding normal stem-cell markers based on their heterogeneous expression in the pertinent cancers. Despite some controversy, the CD34+CD38- marker signature was chosen to define the CSCs of leukemia (3), the CD44+CD24- signature was chosen to define breast cancer CSCs (4), and the CD44 marker was chosen to define the CSCs of HNSCC (5). Though membrane proteins represent only one-third of the proteins encoded by the human genome, they represent more than two-thirds of the known protein targets of drugs. These cell surface markers are not only useful for enriching CSCs from different tumors, but also of significant interest for drug discovery.
However, as more cell surface markers for different cancers have been identified, conflicting results have been reported regarding the usefulness of some of the markers and the reproducibility of some of the marker profiles (6). Quintana et al. examined the expression of 22 common CSC markers in melanoma and found that none of them were exclusively enriched in tumorigenic cells relative to non-tumorigenic cells derived from melanoma (7). CD133 is a widely accepted cell surface marker for glioblastoma CSCs, but Beier et al. found that some glioblastoma CSCs were CD133- (8). CD44 is a CSC marker that is commonly expressed by different malignancies of hematopoietic and epithelial origin, including HNSCC (5). However, increasing data have demonstrated a high level of expression of CD44 in the great majority of cells in head and neck tissues, including normal mucosa and carcinomas, and its subsequent expression could not be used to distinguish normal from benign or malignant epithelia of the head and neck. These observations suggest the need for a comprehensive investigation and greater understanding of the cell surface molecules of CSCs.
Many different “omic” technologies have shown promise as means to identify markers for cancer stem cells and tumors (9). Among them, membrane proteomics can directly detect changes in the cell surface content and provide insights into the post-translational regulation of cell surface functions. Therefore, in this study, we chose to use membrane proteomics both to investigate the cell surface molecules of CSCs that were enriched from the HNSCC cell populations based on their ability to form spheres and to relate their expression to that of stem cell traits. Our results may contribute to further clinical applications of CSCs by providing tools for purifying and identifying CSCs.
EXPERIMENTAL PROCEDURES
Cell Culture
Four HNSCC cell lines (i.e. CAL27, WSU-HN6, WSU-HN12, and WSU-HN13) were grown in DMEM supplemented with 10% fetal bovine serum or in tumor sphere medium consisting of serum-free DMEM/F12, N2 supplement, 10 ng/ml of human recombinant bFGF, and 10 ng/ml of EGF. The cells were plated at a density of 1 × 105 live cells/10-mm dish, and the medium was changed every other day until spheres were observed, ∼3 to 4 weeks later.
Confocal Microscope Analysis of the Expression of Stem Cell Markers
The adherent cells and the microspheres were placed onto glass coverslips, washed twice with PBS, and fixed with 4% paraformaldehyde on ice for 30 min. After blocking in 10% goat whole serum for 2 h, the cells were incubated with primary antibodies at 4°C overnight. The primary antibodies were mouse monoclonal anti-ABCG2 (Abcam, Cambridge, MA, 1:40), mouse monoclonal anti-Bmi-1 (Millipore, Billerica, MA, 1:100), mouse monoclonal anti-CD29 (Ebioscience, San Diego, CA, 1:50), mouse monoclonal anti-EpCAM (Abcam, Cambridge, MA, 1:100), mouse monoclonal anti-CD140 (Ebioscience, 1:50), rabbit polyclonal anti-CD166 (Epitomics, Burlingame, CA, 1:50), mouse monoclonal anti-CD44 (Epitomics, 1:50), purified mouse IgG1 (Ebioscience, 1:50), and rhodamine phalloidin (Invitrogen, Grand Island, NY, 1:200). After washing with PBS containing 0.02% Tween 20, Alexa Fluor 488 IgG F(ab′)2 fragment (Invitrogen, 1:200) was added to the coverslips for a 1-h incubation in a dark chamber. The coverslips were washed with PBS containing 0.02% Tween 20 and stained with 0.5 mg/ml of 4′,6-diamidino-2-phenylindole. Cells in three randomly selected fields were counted under 200-fold magnification using a Leica TCS SP2 confocal spectral microscope to calculate the percentage of positive cells.
In Vivo Tumorigenicity of Adherent Cells and Cells in Spheres in HNSCC Cell Cultures
The animal experiments were performed in accordance with the institutional guidelines for the use of laboratory animals. Four-week-old BALB/C nude mice were supplied by the Shanghai Experimental Animal Center, Chinese Academy of Sciences, Shanghai, China. To assess the in vivo tumorigenicity of the adherent cells and cells that formed spheres in cultures of HNSCC, CAL27, and WSU-HN13 cells, 1 × 105, 1 × 104, and 1 × 103 tumor cells, respectively, of the two types of cells of each line were bilaterally subcutaneously injected into 18 nude mice in 100 μl of medium/Matrigel (1:1), yielding three mice per group. Tumor development was monitored every other day until sacrifice. The volumes of the tumors were calculated using the following formula: volume = (length × width2)/2. At 16 weeks post-transplantation or whenever the tumor's volume was greater than 150 mm3, the mice were sacrificed. The tumors were excised and fixed with 10% neutral buffered formalin before paraffin embedding and sectioning.
Isolation and Preparation of Membrane Proteins
Membrane proteins were isolated using a Pierce cell surface protein isolation kit (Thermo). The cells were first labeled with EZ-Link Sulfo-NHS-SS-Biotin, a thiol-cleavable amine-reactive biotinylation reagent. Then, the cells were collected and precipitated via centrifugation at 500 × g for 3 min. The pellets were subsequently washed twice with TBS and lysed with lysis buffer, and the labeled proteins were isolated using NeutrAvidin Agarose. The bound proteins were released via incubation with SDS-PAGE sample buffer containing 50 mm DTT. The proteins were diluted and quantified using the Pierce BCA protein assay reagent (Thermo).
One-dimensional SDS-PAGE and Standard In-gel Digestion
For this assay, we mixed equal amounts of the total membrane proteins isolated from the spheres or the adherent cells derived from the four HNSCC cell lines to create a sphere group and an adherent group. For each group, 50 μg of total membrane protein was dissolved in SDS-PAGE loading buffer, boiled for 10 min, and loaded onto an 8% SDS-PAGE (1.5-mm-thick) gel. After silver staining, 25 equally spaced pieces of gel were excised from each lane, spanning 40 to 300 kDa. The proteins in each gel piece were reduced by treatment with 50 mm DTT in 100 NH4HCO3 for 45 min at 56°C and were alkylated by treatment with a solution of 55 mm iodoacetamide in 100 mm NH4HCO3 in the dark for 1 h at room temperature. After washing, the gel pieces were dehydrated with acetonitrile and dried using a SpeedVac instrument. Trypsin (12.5 ng/μl in 25 mm NH4HCO3, pH 8.0) was added to each gel piece, and the pieces were allowed to swell at 4°C for 30 min. The excess trypsin was removed and replaced with 25 mm NH4HCO3, 10% acetonitrile, pH 8.0. The mixture was incubated overnight at 37°C. The digest was collected, and the peptides were extracted twice with 50% acetonitrile/0.5% formic acid. The combined extracts were then dried in a SpeedVac instrument for subsequent LC-MS/MS analysis.
LC-MS/MS
The peptides were separated via reverse-phase nano-liquid chromatography using an Agilent 1200 Series nano-LC system equipped with an orthogonal nanospray ion source HPLC-Chip Cube MS interface coupled to a 1200 Series LC/MSD Trap XCT Ultra ion trap mass spectrometer (all from Agilent Technologies, Santa Clara, CA). The system was equipped with an HPLC-Chip (Agilent Technologies) that incorporated a 40-nl enrichment column and a 75 mm × 150 μm analytical column that was packed with 5-μm Zorbax 300SB-C18 particles. The peptides were loaded with 98% solvent A (0.1% formic acid in water) and 2% solvent B (0.1% formic acid in 90% acetonitrile) at a flow rate of 4 μl/min. A stepwise gradient of 5%–15% solvent B for 10 min, 15%–30% solvent B for 60 min, 30%–90% solvent B for 20 min, 90% solvent B for 10 min, and 95%–5% B for 5 min was used to separate the peptides. The total run time, including the time required for column reconditioning, was 120 min.
The LC/MSD Trap XCT Ultra system was operated in the standard enhanced-scan mode with positive ionization. The drying gas flowed at 4 l/min at a temperature of 325°C. The capillary was set to 1.8 kV with the skimmer at 40 V, and the capillary exit was set to 158.5 V. The trap drive was set to 82.9 V. The ion charge control was on, with a maximum accumulation time of 200 ms; the smart target was 500,000, and the MS scan range was 400–1800 m/z, with an average of 2. Automatic MS/MS was performed in the ultrascan mode, with the number of precursor ions set at 3, the fragmentation amplitude set at 1.00 V, the active exclusion on (after 2 spectra for 0.5 min), the exclusion of singly charged ions on, an MS/MS scan range of 100–2200 m/z, and ultrascan on.
Protein Identification
MS/MS spectra were used to search the SwissProt human database using the Spectrum Mill MS Proteomics Workbench software (Agilent Technologies, Rev. A.03.03.084). Peak lists were created using the Spectrum Mill Data Extractor program with the following attributes: scans with the same precursor ± 1.4 m/z were merged within a time frame of ±15 s. Precursor ions had a minimum signal-to-noise value of 25. Charges up to a maximum of 7 were assigned to the precursor ion, and the 12C peak was determined by Data Extractor. The SwissProt human protein database (version 56.9, 20,402 sequences) was searched for tryptic peptides with a mass tolerance of ±2.5 Da for the precursor ions and a mass tolerance of ±0.7 Da for the fragmentations. The search parameters included the fixed/static modification of carbamidomethylation of cysteines, trypsin digestion with two missed cleavages, and variable modifications including oxidized methionines and pyroglutamic acid (supplemental Table S1). The false positive rate was estimated by repeating the process using a database in which all of the protein sequences were reversed. All of the protein hits found in a distinct database search using Spectrum Mill software were nonredundant. To eliminate redundancy, the protein summary mode groups all of the proteins that have at least one common peptide, but only the highest scoring member of each protein group is shown and counted in the protein list. After the Spectrum Mill auto-validation tools (e.g. “Auto-validate after Search”) and their default settings had been accepted, spectra and protein identities that fulfilled the workbench scoring threshold criteria peptide modes were selected (supplemental Table S1). The number of proteins that passed these thresholds was used to calculate the false positive rate using the formula 100 × (2 × number of decoy database IDs)/total IDs. No proteins were identified in the reversed database, so the estimated overall false positive identification rates were 0% among the six LC-MS/MS runs. The annotated spectra for all those single peptide identifications are listed in supplemental Table S2 and supplemental Fig. S1. In addition, all of the experiments described above were repeated thrice, and those proteins that appeared more than twice were used in the follow-up analysis.
In order to compare the proteomes of cells in spheres and adherent cells, we calculated the MS/MS spectral counts of proteins and normalized them to the total spectral count of each raw dataset. The ratios of the normalized protein spectral counts of cells in spheres to those of adherent cells were calculated. Proteins with a ratio of greater than 1.5 were considered to be up-regulated in the spheres, and proteins with a ratio of less than 0.667 were considered to be down-regulated in the spheres.
Protein Annotation
The subcellular localization information was predicted using five different databases. Ingenuity Pathway Analysis Knowledge Base (build version: 131235; content version: 11904312; release date: December 15, 2011) was used to derive extracted proteins located on the “plasma membrane.” Transmembrane hidden Markov model protein sequences were downloaded using their UNIPROT accession number, and transmembrane helices were predicted by comparing the protein sequences against the transmembrane hidden Markov model sequences (Server v.2.0). (iii) PSORT II was used to predict proteins with subcellular locations. The final two information sources were the GO:0005886: plasma membrane database in Gene Ontology and the subcellular location information in Uniprot. The proteins that were located on the “plasma membrane” (Ingenuity Pathway Analysis, PSORT, Gene Ontology databases) or that had a transmembrane helix received a score of 1; otherwise, the protein received a score of 0. The sum of each database-predicted score was the total membrane protein prediction score. Molecular function annotations, KEGG pathway analysis, and information linked to the functional network of all identified proteins were analyzed using the Ingenuity Pathway Analysis Knowledge Base, the online DAVID Bioinformatics Resource 6.7, and GeneGo MetaCore (version: 6.10), respectively.
In addition, we compared our results with two datasets for the hESC membrane protein signature that were published by Prokhorova et al. (10) and Gu et al. (11) to investigate whether hESC and HNSCC CSCs share membrane proteins. International Protein Index protein IDs were converted into gene IDs. A Venn diagram was drawn to identify the overlapping proteins, which were further annotated using MetaCore software.
Western Blot Analysis
Cells or clinical samples were lysed with lysis buffer (Sigma), and the proteins were separated via SDS-PAGE. Western blotting was performed according to a standard protocol using rabbit polyclonal antibodies directed against CD49f (Epitomics, 1:1000), CD71 (Epitomics, dilution 1:1000), CD98 (Epitomics, 1:1000), CD104 (Epitomics, 1:1000), CD166 (Epitomics, 1:1000), CD44 (Epitomics, 1:1000), NOMO1 (Epitomics, 1:1000), and anti-β-actin mAb (Sigma-Aldrich, St. Louis, MO, 1:10,000) at the optimized dilutions. Labeled bands were detected using an IRDye800-conjugated affinity purified anti-mouse IgM or anti-rabbit secondary antibody (Rockland, Gilbertsville, PA). The membrane was then washed several times, scanned using the Odyssey infrared imaging system (LI-COR, Lincoln, NE) at a channel wavelength of 700 nm/800 nm, and analyzed using Odyssey software.
Immunohistochemical Analysis of CD44 and CD166 Expression in HNSCC Samples
The study was approved by the Institutional Review Board of Shanghai Ninth People's Hospital. Two independent cohorts were obtained. The first cohort included 96 samples of primary HNSCC obtained from patients who underwent surgery with curative intent and had negative resection margins and 12 samples of normal oral mucosa obtained from patients who underwent surgery for benign oral or maxillofacial conditions in the Ninth People's Hospital, Shanghai Jiao Tong University, between 1989 and 1993. Tissue microarrays were constructed by Shanghai Biochip Co Ltd, Shanghai, China. Duplicates of 1-mm-diameter cylinders from two representative areas were included for each case, along with various control samples (12). All of the patients in the first cohort had complete clinicopathologic documentation and records of their follow-up data. Follow-up was completed on December 31, 1999. The median follow-up time was 73 months (range: 1 to 120 months). The clinical feature of the tumors was determined according to the TNM classification system of the International Union Against Cancer (Edition 6). Overall survival (OS) was defined as the interval between surgery and death or the last observation taken. The data were censored at the last follow-up for living patients. The second cohort included samples obtained from 104 patients who underwent curative surgery in the Department of Oral and Maxillofacial Surgery, Ninth People's Hospital, Shanghai Jiao Tong University, between 1999 and 2005, including 68 cases of primary tumors and 36 cases of recurring tumors. The patients in this group had complete clinicopathologic documentation, but no follow-up data.
Immunohistochemical staining was performed using the standard SP method. The primary antibodies were rabbit polyclonal CD166 antibody (Epitomics, 1:150) and mouse monoclonal CD44 antibody (Changdao Biotech, Shanghai, China, dilution 1:50). Formalin-fixed paraffin tissue sections were treated with 10 mmol/l citrate buffer to retrieve antigenicity. Hydrogen peroxide treatment (0.3%) was performed to quench the endogenous peroxidase activity. The slides were blocked with 5% normal goat serum for 30 min and subsequently incubated with primary antibody at 4°C overnight. The next day, the sections were incubated with goat anti-rabbit second antibody for 1 h at room temperature in a humidified chamber, washed three times with PBS, and incubated in a development solution containing 0.06% diaminobenzidine and 0.1% hydrogen peroxide. The sections were finally counterstained with hematoxylin and mounted. Sections that were not treated with primary antibodies and those that were treated with non-immune rabbit serum served as negative controls. The sections were examined microscopically and scored at 200× magnification. Because the staining intensity was evenly distributed in the overwhelming majority of the samples, sections were scored according to the staining percentage as low or high, with cut-off values of 50% for CD166 and 90% for CD44, which were the median staining values of CD166 and CD44. All of the immunostained slides were evaluated by three independent observers.
The associations among the CD166 expression values, the CD44 expression values, and the clinicopathologic parameters of the patients were analyzed using the chi-squared test and Kruskal–Wallis H analysis. Estimated survival curves were calculated according to the Kaplan–Meier product limit method, and the survival times of the patients were compared by means of the log rank test. Clinical and pathologic characteristics were analyzed for their association with OS using the Cox proportional hazards regression analysis.
Flow Cytometric Analysis and Cell Sorting
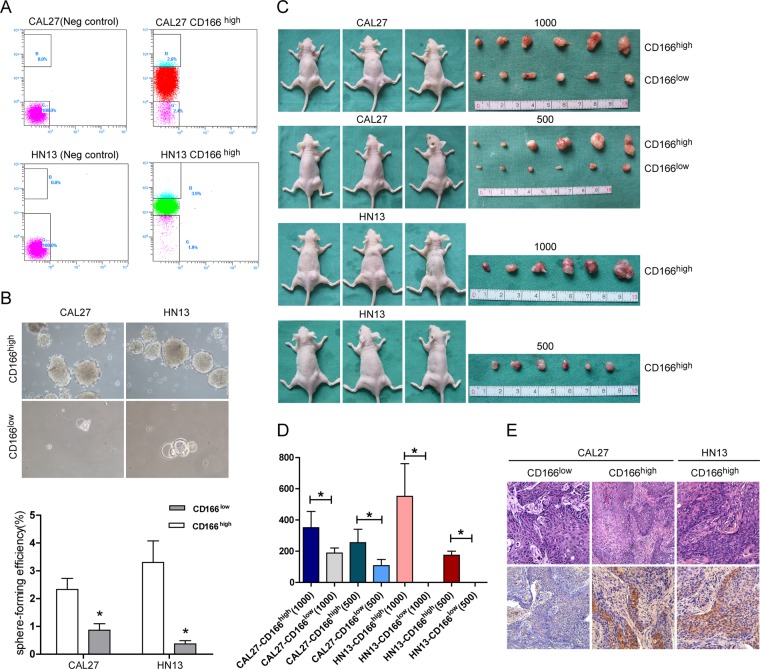
The cells from the microspheres and the adherent cells from CAL27 and HN13 cultures were washed with PBS and dissociated into single cells with trypsin. The cells were harvested, suspended in eFluor NC Flow cytometry staining buffer to a final concentration of 2 × 107/ml, pre-incubated with 20 μl of affinity-purified human FcγR-binding inhibitor per 100 μl for 20 min on ice to block nonspecific Fc-mediated interactions, incubated with CD44 (eBioscience, 1:100) or CD166 (eBioscience, 1:100) primary antibodies on ice for 60 min, washed twice, incubated with FITC-labeled secondary antibody (Invitrogen) for 30 min, and then analyzed and sorted using a Beckman Coulter EPICS ALTRA flow cytometric system. For negative controls, the cells were incubated with an isotype control. Both the CD166high and the CD166low cells in the CAL27 and HN13 populations were collected for further study.
Tumorigenicity of CD166high and CD166Low Cells in Nude Mice
To assess in vivo tumorigenicity, 5 × 103, 1 × 103, or 5 × 102 freshly sorted CD166high or CD166low CAL27 and HN13 cells were subcutaneously injected into both sides of 18 nude mice in 100 μl of medium/Matrigel (1:1). Each treatment group included three mice (six injections) that were injected with adherent cultured cells on the left side and sphere cultured cells on the right side. Tumor development was monitored from the sixth week after inoculation. The mice were sacrificed at the 16th week after inoculation or whenever the tumor's volume was greater than 150 mm3. The harvested tumors were fixed in 10% formaldehyde, and paraffin sections were prepared for H&E staining and CD166 immunohistochemistry.
RESULTS
Overexpression of Stem-cell-related Molecules in HNSCC Spheres
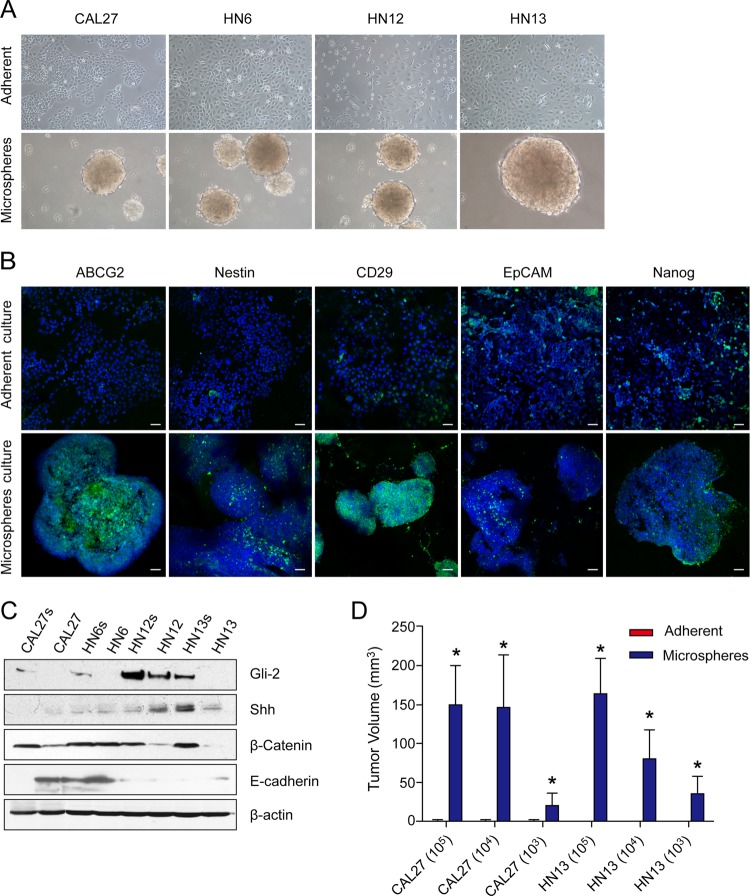
Four HNSCC cell lines—CAL27, HN6, HN12, and HN13—were digested into single cell suspension and cultured in low-attachment plates in defined serum-free DMEM/F12 containing bFGF and EGF. Two weeks later, small sphere-like bodies were observed in each culture (Fig. 1A). These spheres grew gradually, and some reached a diameter of 0.5 to 1 mm in 3 to 4 weeks. To investigate whether these spheres had enriched stem cell features, we examined their expression of various stem-cell-related molecules. Compared with the adherent cells, these spheres had a markedly increased expression of multiple stem-cell-related molecules, including ABCG2, nestin, CD29, EpCAM, and nanog (Fig. 1B). Gli2, one of the three key transcription factors in the hedgehog signaling pathway, was up-regulated in all sphere-forming cells, although the expression of shh, the main ligand of the hedgehog signaling pathway, was not obviously increased. In addition, the expression of β-catenin, the key regulator of the wnt signaling pathway, was increased in the sphere-forming cells, and the expression of E-cadherin was down-regulated (Fig. 1C). These data indicate that the cells in HNSCC spheres exhibit more stem-cell-like features than do their adherent counterparts.
Fig. 1.
Cells that formed spheres in HNSCC cultures enhanced cancer stem-cell-like behavior and tumorigenicity. A, sphere-like bodies formed in cultures of four HNSCC cell lines (CAL27, WSU-HN6, WSU-HN12, and WSU-HN13) grown for 2 weeks in defined serum-free DMEM/F12 supplemented with bFGF and EGF. B, HNSCC sphere cells exhibited higher expression of ABCG2, nestin, CD29, EpCAM, and nanog than did the adherent cultured cells. Scale bar, 50 μm. C, the expression of shh, Gli-2, β-catenin, and E-cadherin in the HNSCC spheres and adherent cells. D, different quantities (105, 104, and 103 cells) of adherent cells or sphere cells obtained from CAL27 and HN13 cell cultures were subcutaneously inoculated into nude mice to assess their in vivo tumorigenicity. The volume of the tumors was determined using the following formula: volume = (length × width2)/2. *p < 0.05.
Improved in Vivo Tumorigenic Ability of Cells in HNSCC Spheres
Although some controversy remains about its use, the tumorigenic ability of nude mice is still the gold standard for identifying stem-cell-like cells in solid cancers; therefore, in this study, we compared the tumorigenic capacities of HNSCC cells that formed spheres or were adherent in cultures using nude mice. We subcutaneously injected 1 × 105, 1 × 104, or 1 × 103 cells derived from spheres or that were adherent in cultures in bilateral sites (both the fore and hind legs) of three mice, with six inoculations per group. Cell-number-dependent tumor development was observed, and the tumor latency period decreased as the number of injected cells increased. Three months later, tumors had developed in all six of the mice injected with 1 × 105 CAL 27 sphere cells, all six of the mice injected with 1 × 104 CAL27 sphere cells, five of the six mice injected with 1 × 103 CAL27 sphere cells, all six of the mice injected with 1 × 105 WSU-HN13 sphere cells, four of the six mice injected with 1 × 104 WSU-HN13 sphere cells, and two of the six mice injected with 1 × 103 WSU-HN13 sphere cells. Of the mice in the adherent cell group, only one of the six injected with 1 × 105 CAL 27 adherent cells, one of the six injected with 1 × 104 CAL27 adherent cells, one of the six injected with 1 × 103 CAL27 adherent cells, and one of the six injected with 1 × 105 WSU-HN13 adherent cells developed tumors, with an average latency period of 12 to 14 weeks, compared with 4 to 9 weeks for mice injected with sphere cells; none of the mice injected with 1 × 104 WSU-HN13 adherent cells or 1 × 103 WSU-HN13 adherent cells developed tumors (supplemental Figs. S2A and S2C, supplemental Table S3). Pathology of HE-stained sections of the xenografted tumors confirmed that the tumors were moderately differentiated squamous cell carcinomas, and more blood vessels were observed in the tumors derived from cells that formed spheres in culture than in tumors derived from their adherent counterparts (supplemental Fig. S2B). Furthermore, the average volumes of tumors derived from the CAL 27 cultured sphere group were 149.74 ± 50.89 mm3 in the 105 cells subgroup, 146.38 ± 67.78 mm3 in the 104 cells subgroup, and 19.92 ± 15.98 mm3 in the 103 cells subgroup, compared with volumes of 0.024 ± 0.024 mm3 in the 105 cells subgroup, 0.004 ± 0.004 mm3 in the 104 cells subgroup, and 0.192 ± 0.129 mm3 in the 103 cells subgroup for the tumors derived from the CAL27 adherent cultured cell group. The average volumes of the tumors from the sphere-forming WSU-HN13 group were 163.39 ± 45.72 mm3 in the 105 cells subgroup, 80.67 ± 37.03 mm3 in the 104 cells subgroup, and 35.45 ± 22.85 mm3 in the 103 cells subgroup, compared with volumes of 0.018 ± 0.018 mm3 in the 105 cells subgroup and 0 mm3 in the 104 and 103 cells subgroups for the tumors derived from the adherent WSU-HN13 cell culture group (Fig. 1D). These data indicate that the HNSCC spheres were enriched with stem-like cells.
Analysis of the Membrane Proteins of the HNSCC Sphere-forming Cells (Spectrum Count)
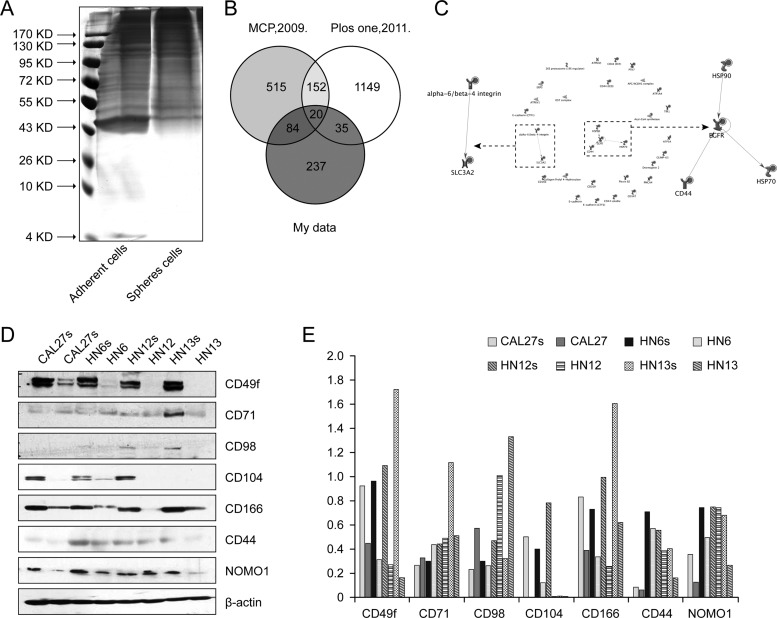
To identify the specific membrane proteins that may contribute to the more stem-cell-like traits of the sphere-forming cells, we prepared membrane preparations from both spheres and adherent cells from cultures of four different HNSCC cell lines. After separating the proteins using one-dimensional SDS-PAGE (Fig. 2A), we used nano-LC-MS/MS to identify their membrane protein expression profiles (supplemental Fig. S3A). All of the LC-MS/MS data were searched against the Uniprot human protein database using the Spectrum Mill software. A total of 376 proteins were identified based on one or more matching mass spectra (supplemental Table S4). Of these proteins, 200 were predicted to be membrane proteins based on the subcellular location prediction from five databases (score ≥ 1), and others were predicted to be located in extracellular spaces, cellular projections, or endoplasmic membranes (supplemental Table S5). The spectral counts of each protein were normalized to the total spectral counts of each sample and then compared with the data for the different cell lines. A total of 123 membrane proteins were expressed differentially (fold change ≥ 1.5-fold) in sphere-forming and adherent cells, among which 65 proteins were up-regulated and 58 were down-regulated in the sphere-forming cells. The proteins that were overexpressed in the sphere-forming cells included enzymes (e.g. CNTN1 and GPNMB), kinases (e.g. EGFR and PTK7), transmembrane receptors (ITGB4), transporters (e.g. BSG, ATP4A, and ITGB4), and some well-recognized stem-cell-related membrane proteins (e.g. CD44 and CD49f) that the DAVID bioinformatics database identifies as being involved in “pathways in cancer,” “regulation of the actin cytoskeleton,” “adherent junctions,” “focal adhesion,” and “cell adhesion molecules (CAMs)” (supplemental Fig. S3C).
Fig. 2.
Membrane proteomic analysis of the sphere-forming cells and the adherent cells in HNSCC cell cultures. A, membrane proteins extracted from adherent cultured cells and spheres (the gel was stained with silver nitrate). B, Venn diagrams of the identified HNSCC membrane proteins and those found in two datasets of hESC membrane protein signatures published by Prokhorova et al. (10) and Gu et al. (11). C, functional networks of the proteins common to the datasets were analyzed using GeneGo MetaCore software. Two networks were identified: EGFR/CD44/HSP70/HSP90 and integrin alpha-6/beta-4/SLC3A2. D, the expression of seven membrane proteins identified via MS was evaluated with Western blotting, and five of them were consistently up-regulated in the sphere cells. E, densitometric analysis of the Western blot staining levels.
By comparing the HNSCC membrane signature with two published hESC signatures, we found that 20 proteins (the “3/3 signature”) were consistently detected in all three signatures (Fig. 2B). Among them, the fold changes of 18 proteins were greater than 1.5, including CD166 (gene symbol: ALCAM), CD44, CD98 (gene symbol: SLC3A2), and CD49f (gene symbol: ITGA6) (Table I). MetaCore analysis disclosed two types of networks among these 18 proteins: the EGFR/CD44/HSP70/HSP90 and integrin alpha-6/integrin beta-4/SLC3A2 networks (Fig. 2C).
Table I. The membrane proteins expressed on the surface of HNSCC microspheres and hESCs as indicated by two proteomics datasets published by Prokhorova et al. (10) and Gu et al. (11).
| Protein ID | Symbol | Fold change (spheres/adherent) | Type(s) | Membrane prediction total score |
|---|---|---|---|---|
| P35613 | BSG | a | Transporter | 5 |
| Q13308 | PTK7 | a | Kinase | 3 |
| Q14126 | DSG2 | a | Other | 5 |
| P00533 | EGFR | 3.0042337 | Kinase | 5 |
| Q13740 | ALCAM | 3.0042337 | Other | 4 |
| P08195 | SLC3A2 | 2.253175275 | Transporter | 4 |
| P23229 | ITGA6 | 2.253175275 | Other | 5 |
| P05023 | ATP1A1 | 2.002822467 | Transporter | 4 |
| O15031 | PLXNB2 | 1.50211685 | Other | 3 |
| P16070 | CD44 | 1.50211685 | Other | 5 |
| Q07065 | CKAP4 | 1.50211685 | Other | 3 |
| P14625 | HSP90B1 | 0.751058425 | Other | 0 |
| P23634 | ATP2B4 | 0.751058425 | Transporter | 4 |
| P11021 | HSPA5 | 0.584156553 | Enzyme | 0 |
| Q15084 | PDIA6 | 0.450635055 | Enzyme | 2 |
| O60488 | ACSL4 | 0 | Enzyme | 2 |
| P04843 | RPN1 | 0 | Enzyme | 2 |
| P07237 | P4HB | 0 | Enzyme | 2 |
| P12830 | CDH1 | 0 | Other | 5 |
| P38606 | ATP6V1A | 0 | Transporter | 1 |
a The protein has been identified in spheres only.
A fold change of 0 indicates that the protein has been identified only in adherent cultured cells.
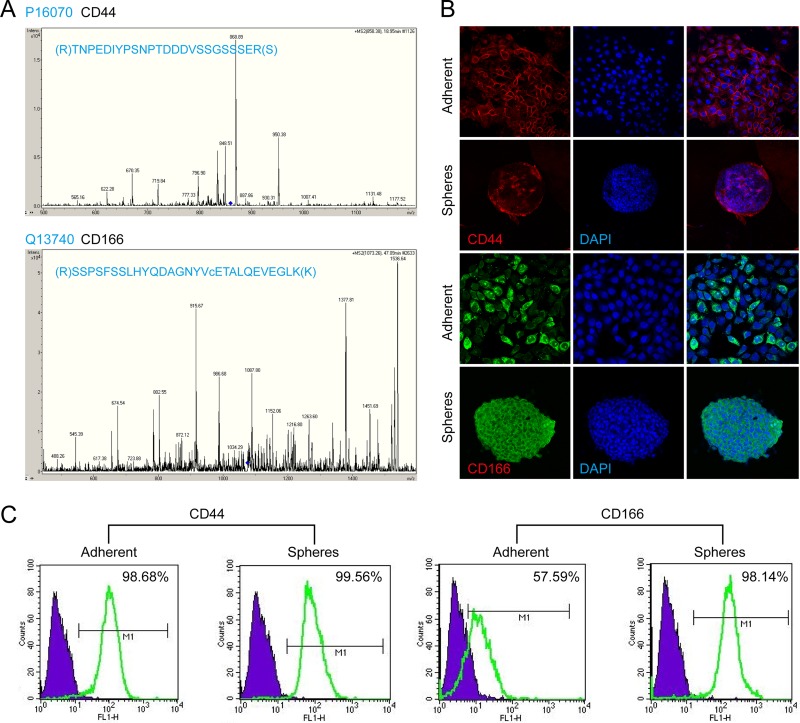
To validate the significance of the membrane proteins identified via mass spectrometry, we selected 7 membrane proteins (CD49f, CD98, CD166, CD44, CD104, CD71, and NOMO1) from the 18 candidates shared by the HNSCC CSCs and hESCs to investigate their expression by the sphere and adherent cells derived from the four different HNSCC cells (CAL27, HN6, HN12, and HN13). As shown in Fig. 2D, Western blots showed that five of these proteins were more highly expressed in sphere cells than in adherent cells. Fig. 2E shows the densitometric analysis of the Western blot results, showing that the expression levels of CD49f, CD104, CD166, CD44, and NOMO1 were all significantly higher in the sphere cells than in the adherent cells and that CD166 was the membrane protein most significantly increased in sphere cells. Fig. 3A shows the identified type spectra of CD44 and CD166 in MS/MS. We then further investigated the distribution of CD44+ and CD166+ cells in populations of sphere and adherent cells using immunofluorescence staining and flow cytometry. As shown in Fig. 3B, more than 90% of the adherent cells were CD44+; this percentage was very close to but still lower than the percentage of CD44+ sphere cells, whereas the percentage of CD166+ cells was much higher in sphere cells than in adherent cells. Flow cytometry (Fig. 3C) confirmed that the CD44-positive rates of the adherent and sphere cells derived from the CAL27 line were 98.68% and 99.56%, respectively, whereas the CD166-positive rates of the adherent and sphere cells were 57.59% and 98.14%, respectively.
Fig. 3.
Expression of CD44 and CD166 by adherent and sphere-forming cultured cells. A, MS/MS spectrum of peptide TNPEDIYPSNPTDDDVSSGSSSER from CD44 and peptide SSPSFSSLHYQDAGNYVcETALQEVEGLK from CD166 were identified via LC-MS/MS. B, immunofluorescence staining of CD44 and CD166 in adherent and sphere-forming cultured HNSCC cells. The expression of CD166 was significantly increased in the sphere-forming cells. C, the percentage of CD44- and CD166-positive cells in the adherent and sphere-forming populations from the cell cultures was determined using flow cytometry.
The Expression of CD166 Is an Independent Predictor of HNSCC Patients' Poor Prognosis
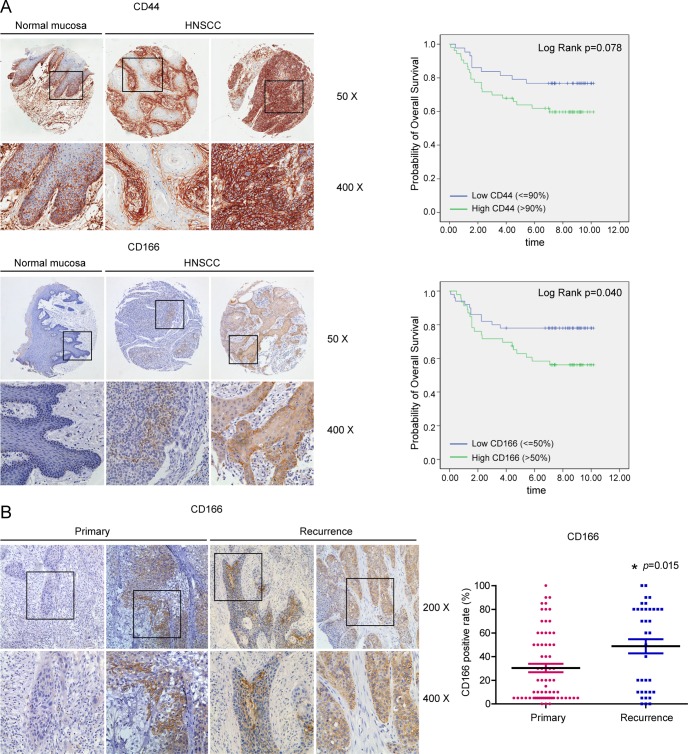
To investigate the clinical value of the membrane proteins overexpressed by HNSCC sphere cells that were identified via mass spectrometry, we used tissue arrays to examine the expression of CD44 and CD166 in HNSCC clinical samples and analyzed the relationship between their expression levels and the patients' clinicopathologic parameters and prognosis. In total, samples from 96 cases of cancer and 12 cases of normal mucosa were included. For negative controls, consecutive sections were stained with murine preimmune serum, and these sections were used to test the specificity of the detection system. As shown in Fig. 4A, CD44 was strongly expressed in both normal mucosa and the HNSCC samples. Two-thirds of the normal mucosa samples were CD44-positive, with variable intensity ranging from + to +++. The CD44-positive cells were localized mainly in the stratum basale and stratum spinosum. The average positive rate for CD44 expression in the carcinomas was 88.20% ± 13.88%; in more than half of the samples (53/96), over 90% of the cancer cells were CD4-positive. A high level of expression of CD44 was frequently observed in poorly differentiated tumors (p = 0.008). There was no significant association between the CD44 expression level and age (p = 0.763), gender (p = 0.150), anatomic site of tumor (p = 0.949), smoking history (p = 0.155), alcohol use history (p = 0.068), lymph node metastasis (p = 0.282), TNM stage (p = 0.305), or the patient's prognosis (p = 0.088, Mann–Whitney U test) (Table II, Fig. 4A).
Fig. 4.
Expression of CD166 and CD44 in clinical tissues. A, the expression of CD44 and CD166 in tumor samples was first analyzed using HNSCC tissue arrays that included samples from 96 patients with cancer and 12 patients with normal mucosa. For each case, the level of CD44 and CD166 staining was classified as low or high based on the median of the labeling index. CD44 was abundantly expressed in both normal oral mucosa and HNSCC. The cutoff values were 90% for CD44 staining and 50% for CD166 staining. The Kaplan–Meier survival curve prepared using the CD44 scores showed that the CD44 expression level was marginally associated with a patient's prognosis (p = 0.078). A significant association between the level of expression of CD166 and a patient's prognosis was observed (Kaplan–Meier, p = 0.040). B, the expression of CD166 in primary HNSCC and recurrent HNSCC samples was examined in another cohort that included 68 primary tumors and 36 recurrent tumors. The CD166-positive rate in the primary tumor group was 30.37% ± 29.32%, whereas it was 48.75% ± 35.64% in the recurrent tumor group. A chi-squared test disclosed a significant increase of CD166 expression in recurrent tumor group (*p = 0.015).
Table II. The expression of CD166 and CD44 in clinical samples.
| CD166 |
P | CD44 |
P | |||
|---|---|---|---|---|---|---|
| ≤50% | >50% | ≤90% | >90% | |||
| Age | ||||||
| >60 | 12 | 13 | 14 | 11 | ||
| >45, ≤60 | 18 | 18 | 12 | 24 | ||
| ≤45 | 20 | 15 | 0.463 | 17 | 18 | 0.763 |
| Sex | ||||||
| Male | 24 | 27 | 19 | 24 | ||
| Female | 26 | 19 | 0.297 | 32 | 21 | 0.150 |
| Site | ||||||
| Tongue | 39 | 34 | 33 | 40 | ||
| Buccal | 5 | 7 | 4 | 8 | ||
| Gingival | 6 | 3 | 6 | 3 | ||
| Lip | 0 | 1 | 0 | 1 | ||
| Oral floor | 0 | 1 | 0.662 | 0 | 1 | 0.949 |
| Smoking | ||||||
| No | 29 | 23 | 27 | 25 | ||
| Yes | 13 | 15 | 10 | 18 | ||
| Unknown | 8 | 8 | 0.494 | 6 | 10 | 0.155 |
| Drinking | ||||||
| No | 30 | 30 | 32 | 28 | ||
| Yes | 11 | 8 | 4 | 15 | ||
| Unknown | 9 | 8 | 0.658 | 7 | 10 | 0.068 |
| Lymph | ||||||
| Metastasis | 14 | 11 | 9 | 16 | ||
| No metastasis | 35 | 33 | 0.698 | 33 | 35 | 0.282 |
| Differentiation | ||||||
| I | 35 | 26 | 33 | 28 | ||
| I–II,II | 10 | 15 | 9 | 16 | ||
| II–III,III | 5 | 5 | 0.220 | 1 | 9 | 0.008 |
| TNM | ||||||
| I | 5 | 6 | 6 | 5 | ||
| II | 18 | 16 | 17 | 17 | ||
| III | 16 | 15 | 12 | 19 | ||
| IV | 10 | 7 | 0.594 | 7 | 10 | 0.305 |
| Prognosis | ||||||
| Die | 11 | 20 | 10 | 21 | ||
| Survive | 39 | 26 | 0.025 | 33 | 32 | 0.088 |
Compared with the expression of CD44, the expression of CD166 in the HNSCC samples was highly variable. CD166 either was not detected or was scattered in the basal layer of the normal oral mucosa (Fig. 4A). In the HNSCC samples, CD166-positive cells were more likely to be localized at the front of invasive cancer cell nests and clusters, and the CD166-positive rate ranged from 0% to 95% (mean value ± S.E.: 42.66% ± 29.03%). Among the HNSCC samples, 52.08% (50/96) exhibited low or moderate expression of CD166 (staining rate ≤ 50%), and 47.92% (46/90) of the samples displayed strong expression of CD166 (>50%). CD166 was localized mainly on the membrane of the cancer cells, and occasionally in the cytoplasm. There was no significant association between CD166 positivity and age (p = 0.463), gender (p = 0.297), anatomic site (p = 0.662), smoking history (p = 0.494), alcohol use history (p = 0.658), lymph node metastasis (p = 0.698), differentiation status (p = 0.220), or TNM stage (p = 0.594); however, a significant association was observed between the level of CD166 expression and the patient's prognosis (p = 0.025, Mann–Whitney U test) (Table II, Fig. 4A).
The univariate analysis of OS revealed that CD166 expression (≤50% versus >50%) (p = 0.046), differentiation (p = 0.02), TNM stage (p = 0.036), and lymph node metastasis (p = 0.003) were significantly associated with the OS of the HNSCC patients, whereas CD44 expression (≤90% versus >90%) was not significantly associated with the OS (p = 0.085) (supplemental Table S6). In the multivariate analysis, only CD166 expression (p = 0.044), differentiation (p = 0.012), and lymph node status (p = 0.007) were found to be independent prognostic factors for overall survival (supplemental Table S6). To further validate the results of immunohistochemistry, the expression of CD44 and CD166 in three pairs of normal mucosa and HNSCC was evaluated using Western blots. As shown in supplemental Fig. S4, CD44 was highly expressed in both the normal mucosa and the HNSCC tissues; however, the level of expression was slightly higher in tumors, whereas the level of expression of CD166 was much higher in the HNSCC tissues than in the normal mucosa.
We then investigated the expression of CD166 via immunohistochemistry in samples of primary HNSCC and recurrent HNSCC from the second cohort, which included 68 patients with primary tumors and 36 patients with recurrent tumors (Fig. 4B). The CD166-positive rate in the primary tumor group was 30.37% ± 29.32%, whereas it was 48.75% ± 35.64% in the recurrence group (Fig. 4B). The chi-squared test disclosed a significant increase in CD166 expression in the recurrence group (p = 0.015) (supplemental Table S7).
Enhanced Sphere-formation Ability and in Vivo Tumorigenicity of the CD166high HNSCC Cells
To determine whether CD166 might be a novel marker for enriching cancer stem-like cells from HNSCC, we examined the sphere-formation ability and the in vivo tumorigenic ability of CD166high and CD166low cells. We sorted CD166high cells and CD166low cells from CAL27 and HN13 cell cultures (Fig. 5A) and seeded 1000 sorted cells per group in low-attachment 10-cm plates in serum-free DMEM/F12 (with 10 ng/ml of bFGF and 10 ng/ml of EGF). Two weeks later, the number of spheres was counted, and the sphere-forming efficiency was calculated. As shown in Fig. 5B, the CD166high cells from both CAL27 and HN13 cell cultures demonstrated a greater sphere-forming ability than did their CD166low counterparts (p = 0.033 and p = 0.02, respectively).
Fig. 5.
CD166high cells displayed increased sphere-formation ability and tumorigenicity. A, CD166high and CD166low cells were sorted from CAL27 and WSU-HN13 cell populations using flow cytometry. B, sphere-formation by CD166high and CD166low cells was analyzed, revealing that CD166high cells from both HNSCC cell lines had significantly greater sphere-forming ability than did CD166low cells (*p < 0.05). C, the tumorigenicity of CD166high and CD166low cells in nude mice. All of the animals that were subcutaneously injected with CD166high cells developed tumors ∼10 to 13 weeks after injection, whereas only very small tumors had formed in the CAL27 CD166low groups by ∼16 weeks after inoculation, and no tumors formed in the HN13 CD166low groups. D, the volume of the tumors derived from the CD166high and CD166low cells (*p < 0.05). E, H&E staining and CD166 expression in the xenografted tumors. Compared with the tumors derived from CD166low cells, the level of CD166-positivity was much higher in the tumors derived from CD166high tumor cells.
To assess the in vivo tumorigenicity of the CD166high and CD166low cells, 1 × 103 or 5 × 102 CD166high or CD166low cells sorted from CAL27 and HN13 cell populations (Fig. 5A) were subcutaneously injected bilaterally into 12 nude mice in 100 μl of medium/Matrigel (1:1); each group consisted of three mice. All of the animals injected with CD166high cells developed tumors ∼10 to 13 weeks after injection. Very small tumors were found in the CAL27 CD166low groups ∼16 weeks after injection, and no tumors were found in the HN13 CD166low groups (Fig. 5C). Four months later, all of the animals were sacrificed. The tumors derived from 1 × 103 and 5 × 102 CAL27 CD166high cells were 351.2 ± 103.8 mm3 and 255.0 ± 85.91 mm3 in size, respectively, and the tumors derived from 1 × 103 and 5 × 102 CAL27 CD166low cells were 188 ± 32.58 mm3 and 107.6 ± 40.41 mm3 in size, respectively. The sizes of the tumors derived from 1 × 103 and 5 × 102 HN13 CD166high cells were 551.7 ± 209.1 mm3 and 174.3 ± 26.29 mm3, respectively (Fig. 5D). All of the growths were confirmed to be tumors via histology of H&E-stained sections. CD166 expression in the xenografted tumors was evaluated by means of immunohistochemical staining. As shown in Fig. 5E, the tumors derived from CD166high cells contained both CD166+ and CD166− cells, whereas very few CD166+ cells were observed in the tumors derived from CD166low cells. Relative to the tumors derived from CD166low cells, the percentage of CD166-positive cells was much higher in the tumors derived from CD166high tumor cells.
DISCUSSION
In this study, we used membrane proteomics approaches to study the specific membrane proteins associated with the CSCs of HNSCC based on their sphere-formation ability. Cultures that allow sphere formation are often used to enrich CSCs, which are believed to partly recapitulate stem cell features, including withstanding anoikis, proliferating under anchorage-independent conditions, and possessing the ability to form clonal spheroids. To validate the stem-cell-like features of the HNSCC spheres, we found the increased expression of a series of stem-cell-related molecules in the HNSCC spheres. More important, compared with adherent culture cells, the HNSCC spheres exhibited a significantly enhanced ability to form tumors in nude mice. Therefore, the comparative membrane proteomics analysis of the HNSCC spheres may lead to an effective method for identifying the cell surface markers of HNSCC CSCs (13, 14).
Comparing the membrane proteins of HNSCC spheres and adherent cells revealed 123 candidate membrane proteins that were differentially expressed in cultured HNSCC sphere cells and adherent cells, with 65 proteins being overexpressed and 58 proteins being underexpressed. These proteins are involved in cell differentiation (CAMD1 and CD44), cell adhesion (CD44, CD342, DSG2, NCAM, and CD166), protein transport, and embryonic development (CD49f and alpha E-cadherin). Some of the molecules are involved in key signaling pathways of development, such as the Wnt (CTNND1, CNTN1, and PTK7), integrin (integrin alpha2, integrin alpha6, and integrin beta4) and TGFβ (CD109) signaling pathways. Some of the molecules have been shown to play a role in stem cell functions, such as CD4 and CD49f. Next, we compared our results with two published membrane protein datasets derived from embryonic stem cells, given that the hallmark traits of stem cells (i.e. self-renewal and differentiation capacity) were mirrored by the high proliferative capacity and phenotypic plasticity of tumors (9, 15, 16). The comparison showed that of the 20 common proteins, 18 candidate proteins were differentially expressed in sphere-forming and adherent cells, including the well-known HNSCC CSC marker, CD44, and many new candidates, including CD166, BSG, and PTK7. Further investigation showed that the expression of CD166, but not CD44, was remarkably up-regulated in the sphere-forming cells relative to the adherent cells. CD166, but not CD44, was significantly associated with a patient's prognosis and recurrent tumors. More important, both the in vitro and in vivo studies demonstrated that CD166high HNSCC cells had a greater sphere-formation ability and greatly enhanced tumorigenic ability. Therefore, CD166 might be a novel surface marker for cancer stem-like cells in HNSCC.
CD44 is one of the predominant CSC markers and is also a well-known HNSCC CSC marker (4). In 2007, Prince et al. demonstrated that CD44 could be used to enrich the CSCs from HNSCC. However, their data showed that although CD44+ HNSCC cells exhibited a significantly enhanced tumor-formation ability in nude mice relative to CD44− HNSCC cells (5), a relatively high number of the CD44+ HNSCC cells (>5000 cells) were needed to generate tumors in immune-deficient mice, indicating either a low frequency of CSC among the CD44+ HNSCC cells or that CD44 was not a specific marker for HNSCC CSCs. The other disadvantages of CD44 as a specific CSC marker in HNSCC are that CD44 is abundantly expressed in most HNSCC samples (60% to 100% positive cells, averaging 88% in our clinical samples) and in normal mucosa (strongly positive in the stratum basale and stratum spinosum of two out of three samples) and that CD44 expression does not distinguish normal epithelia from benign or malignant epithelia of the head and neck (17, 18). Although the CD44 expression level was 1.5-fold higher in the spheres than in the adherent cultured cells, the clinical data showed that CD44 expression was not an independent predictor of a patient's prognosis and that its expression level was significantly associated only with the differentiation status of the clinical samples.
Compared with CD44, the level of CD166 expression in the spheres was much higher (3-fold) than in the adherent cultured cells. In the clinical samples, the CD166-positive cells were more often found at the periphery of tumor nests or in the invasive frontier. Both univariate and multivariate analyses showed that CD166 expression was significantly associated with the prognosis of HNSCC patients and that the level of CD166 expression was much higher in the samples of recurrent tumors than in the primary tumor samples. CD166, also called activated leukocyte cell adhesion molecule (ALCAM), is a transmembrane glycoprotein and a member of the Ig superfamily that mediates cell–cell adhesion through homophilic (ALCAM–ALCAM) and heterophilic (ALCAM–CD6) interactions. CD166 is involved in hematopoiesis (19, 20), neurite extension (21), osteogenesis (22), and embryonic implantation in the uterus (23). The level of CD166 expression correlates with the aggregation and metastatic potential of a few human tumors (24), including melanoma (25, 26) and prostate (27, 28), ovarian (29), breast (30–32), colorectal (33, 34), esophageal (35), and pancreatic cancer (36, 37). Furthermore, ALCAM was also identified in medium conditioned by breast cancer cells and may be a potential serum breast cancer biomarker (38, 39). In recent years, CD166 has been proposed as a cell surface marker for mesenchymal progenitor cells and colon cancer stem cells (34). The expression of ALCAM at the invasive front of HNSCC tumors may be related to their potential for tumor invasion and lymph node metastasis (40–42). In this study, we did not observe a significant correlation between CD166 expression and lymph node metastasis, but we found that the CD166 expression level was an independent predictor for a patient's prognosis and that it was significantly associated with recurrent tumors. More important, we found that compared with CD166low HNSCC cells, CD166high cells had a remarkably greater sphere-formation ability and tumorigenicity. As few as 500 CD166high WSU-HN13 cells initiated a tumor in nude mice at 100% efficiency, compared with the minimum of 5000 CD44+ HNSCC cells needed to generate a tumor in immune-deficient mice (5), whereas WSU-HN13 CD166low cells did not generate tumors. These data demonstrate that CD166high WSU-HN13 cells have CSC features and suggest that CD166 is a novel marker for stem-cell-like components of HNSCC. Moreover, considering the high rate (>90%) of CD44+ cells in the CAL27 and WSU-HN13 populations, we did not try to investigate the behavior of CD44+CD166+ cells in this study.
Notably, both CD166high and CD166low CAL27 cells drove tumor formation in nude mice injected with 1000 or 500 cells, but the tumors derived from the CD166high cells were much larger and grew much more quickly than those derived from the CD166low cells. Most of the CD166low tumors had formed by ∼4 months post-injection. Immunohistochemical staining demonstrated the existence of some CD166+ cells in these tumors. One possible reason for this finding is that because the sorting efficiency is never 100% accurate, a few CD166-positive cells may be sorted into the CD166low fraction. Another possibility is that some more primitive CSC cells reside in the CD166low subpopulation. Further study of the therapeutic value of targeting the CD166high HNSCC subpopulation in vivo might help to define the molecular characteristics of CSCs.
In conclusion, we used mass spectroscopy to compare the plasma membrane proteomics of HNSCC CSC-like spheres and their adherent counterparts in this study. We discovered 123 membrane proteins, 65 of which were overexpressed and 58 of which were underexpressed. These membrane proteins are potentially valuable markers for HNSCC CSCs and included both well-known CSC markers and many new candidates. Compared with the well-established HNSCC CSC marker CD44, the level of CD166 expression serves as a cell surface marker to enrich CSCs from HNSCC cell populations, as demonstrated in both in vitro and in vivo studies. Our data demonstrate that plasma membrane proteomics is a promising biological tool for determining CSC-specific membrane proteins.
Supplementary Material
Footnotes
* This study was supported by the National Natural Science Foundation of China (Grant No. 81272978 and Grant No. 81102050), the Shanghai Municipal Health Bureau (Grant No. 2010Y146), and the program for professor of special appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (No. 1220000243). The authors thank the Bioinformatics Center of the Shanghai Institutes for Biological Sciences and CAS for help with the bioinformatics analysis.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ALCAM
- activated leukocyte cell adhesion molecule
- CSC
- cancer stem cell
- hESC
- human embryonic stem cell
- HNSCC
- head and neck squamous cell carcinoma
- OS
- overall survival.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Jordan C. T., Guzman M. L., Noble M. (2006) Cancer stem cells. N. Engl. J. Med. 355, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 3. Bonnet D., Dick J. E. (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 [DOI] [PubMed] [Google Scholar]
- 4. Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003) Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prince M. E., Sivanandan R., Kaczorowski A., Wolf G. T., Kaplan M. J., Dalerba P., Weissman I. L., Clarke M. F., Ailles L. E. (2007) Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 104, 973–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clevers H. (2011) The cancer stem cell: premises, promises and challenges. Nat. Med. 17, 313–319 [DOI] [PubMed] [Google Scholar]
- 7. Quintana E., Shackleton M., Foster H. R., Fullen D. R., Sabel M. S., Johnson T. M., Morrison S. J. (2010) Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell 18, 510–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beier D., Hau P., Proescholdt M., Lohmeier A., Wischhusen J., Oefner P. J., Aigner L., Brawanski A., Bogdahn U., Beier C. P. (2007) CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 67, 4010–4015 [DOI] [PubMed] [Google Scholar]
- 9. Pece S., Tosoni D., Confalonieri S., Mazzarol G., Vecchi M., Ronzoni S., Bernard L., Viale G., Pelicci P. G., Di Fiore P. P. (2010) Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 140, 62–73 [DOI] [PubMed] [Google Scholar]
- 10. Prokhorova T. A., Rigbolt K. T., Johansen P. T., Henningsen J., Kratchmarova I., Kassem M., Blagoev B. (2009) Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol. Cell. Proteomics 8, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu B., Zhang J., Wu Y., Zhang X., Tan Z., Lin Y., Huang X., Chen L., Yao K., Zhang M. (2011) Proteomic analyses reveal common promiscuous patterns of cell surface proteins on human embryonic stem cells and sperms. PLoS One 6, e19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao Q., Qiu S. J., Fan J., Zhou J., Wang X. Y., Xiao Y. S., Xu Y., Li Y. W., Tang Z. Y. (2007) Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J. Clin. Oncol. 25, 2586–2593 [DOI] [PubMed] [Google Scholar]
- 13. Leth-Larsen R., Lund R., Hansen H. V., Laenkholm A. V., Tarin D., Jensen O. N., Ditzel H. J. (2009) Metastasis-related plasma membrane proteins of human breast cancer cells identified by comparative quantitative mass spectrometry. Mol. Cell. Proteomics 8, 1436–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Houdt W. J., Emmink B. L., Pham T. V., Piersma S. R., Verheem A., Vries R., Fratantoni S. A., Pronk A., Clevers H., Borel Rinkes I. H., Jimenez C. R., Kranenburg O. (2011) Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies BIRC6 as a potential therapeutic target. Mol. Cell. Proteomics 10, M111.011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ben-Porath I., Thomson M. W., Carey V. J., Ge R., Bell G. W., Regev A., Weinberg R. A. (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barker N., Ridgway R. A., van Es J. H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A. R., Sansom O. J., Clevers H. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 [DOI] [PubMed] [Google Scholar]
- 17. Mack B., Gires O. (2008) CD44s and CD44v6 expression in head and neck epithelia. PLoS One 3, e3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen C., Wei Y., Hummel M., Hoffmann T. K., Gross M., Kaufmann A. M., Albers A. E. (2011) Evidence for epithelial-mesenchymal transition in cancer stem cells of head and neck squamous cell carcinoma. PLoS One 6, e16466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arai K., Shibahara T., Yamamoto N., Noma H. (2002) The presence of candidate tumor suppressor gene loci at chromosome 3p for oral squamous cell carcinomas. Oral Oncol. 38, 763–771 [DOI] [PubMed] [Google Scholar]
- 20. Chen N., Hudson J. E., Walczak P., Misiuta I., Garbuzova-Davis S., Jiang L., Sanchez-Ramos J., Sanberg P. R., Zigova T., Willing A. E. (2005) Human umbilical cord blood progenitors: the potential of these hematopoietic cells to become neural. Stem Cells 23, 1560–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato C., Matsuda T., Kitajima K. (2002) Neuronal differentiation-dependent expression of the disialic acid epitope on CD166 and its involvement in neurite formation in Neuro2A cells. J. Biol. Chem. 277, 45299–45305 [DOI] [PubMed] [Google Scholar]
- 22. Alsalameh S., Amin R., Gemba T., Lotz M. (2004) Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum. 50, 1522–1532 [DOI] [PubMed] [Google Scholar]
- 23. Fujiwara H., Tatsumi K., Kosaka K., Sato Y., Higuchi T., Yoshioka S., Maeda M., Ueda M., Fujii S. (2003) Human blastocysts and endometrial epithelial cells express activated leukocyte cell adhesion molecule (ALCAM/CD166). J. Clin. Endocrinol. Metab. 88, 3437–3443 [DOI] [PubMed] [Google Scholar]
- 24. Wiiger M. T., Gehrken H. B., Fodstad O., Maelandsmo G. M., Andersson Y. (2010) A novel human recombinant single-chain antibody targeting CD166/ALCAM inhibits cancer cell invasion in vitro and in vivo tumour growth. Cancer Immunol. Immunother. 59, 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swart G. W., Lunter P. C., Kilsdonk J. W., Kempen L. C. (2005) Activated leukocyte cell adhesion molecule (ALCAM/CD166): signaling at the divide of melanoma cell clustering and cell migration? Cancer Metastasis Rev. 24, 223–236 [DOI] [PubMed] [Google Scholar]
- 26. Klein W. M., Wu B. P., Zhao S., Wu H., Klein-Szanto A. J., Tahan S. R. (2007) Increased expression of stem cell markers in malignant melanoma. Mod. Pathol. 20, 102–107 [DOI] [PubMed] [Google Scholar]
- 27. Kristiansen G., Pilarsky C., Wissmann C., Stephan C., Weissbach L., Loy V., Loening S., Dietel M., Rosenthal A. (2003) ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate 54, 34–43 [DOI] [PubMed] [Google Scholar]
- 28. Kristiansen G., Pilarsky C., Wissmann C., Kaiser S., Bruemmendorf T., Roepcke S., Dahl E., Hinzmann B., Specht T., Pervan J., Stephan C., Loening S., Dietel M., Rosenthal A. (2005) Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. J. Pathol. 205, 359–376 [DOI] [PubMed] [Google Scholar]
- 29. Mezzanzanica D., Fabbi M., Bagnoli M., Staurengo S., Losa M., Balladore E., Alberti P., Lusa L., Ditto A., Ferrini S., Pierotti M. A., Barbareschi M., Pilotti S., Canevari S. (2008) Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin. Cancer Res. 14, 1726–1733 [DOI] [PubMed] [Google Scholar]
- 30. Ihnen M., Kilic E., Kohler N., Loning T., Witzel I., Hagel C., Holler S., Kersten J. F., Muller V., Janicke F., Milde-Langosch K. (2011) Protein expression analysis of ALCAM and CEACAM6 in breast cancer metastases reveals significantly increased ALCAM expression in metastases of the skin. J. Clin. Pathol. 64, 146–152 [DOI] [PubMed] [Google Scholar]
- 31. Davies S., Jiang W. G. (2010) ALCAM, activated leukocyte cell adhesion molecule, influences the aggressive nature of breast cancer cells, a potential connection to bone metastasis. Anticancer Res. 30, 1163–1168 [PubMed] [Google Scholar]
- 32. King J. A., Ofori-Acquah S. F., Stevens T., Al-Mehdi A. B., Fodstad O., Jiang W. G. (2004) Activated leukocyte cell adhesion molecule in breast cancer: prognostic indicator. Breast Cancer Res. 6, R478–R487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levin T. G., Powell A. E., Davies P. S., Silk A. D., Dismuke A. D., Anderson E. C., Swain J. R., Wong M. H. (2010) Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 139, 2072–2082, e2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dalerba P., Dylla S. J., Park I. K., Liu R., Wang X., Cho R. W., Hoey T., Gurney A., Huang E. H., Simeone D. M., Shelton A. A., Parmiani G., Castelli C., Clarke M. F. (2007) Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 10158–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tachezy M., Effenberger K., Zander H., Minner S., Gebauer F., Vashist Y. K., Sauter G., Pantel K., Izbicki J. R., Bockhorn M. (2011) ALCAM (CD166) expression and serum levels are markers for poor survival of esophageal cancer patients. Int. J. Cancer 131, 396–405 [DOI] [PubMed] [Google Scholar]
- 36. Kahlert C., Weber H., Mogler C., Bergmann F., Schirmacher P., Kenngott H. G., Matterne U., Mollberg N., Rahbari N. N., Hinz U., Koch M., Aigner M., Weitz J. (2009) Increased expression of ALCAM/CD166 in pancreatic cancer is an independent prognostic marker for poor survival and early tumour relapse. Br. J. Cancer 101, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hong X., Michalski C. W., Kong B., Zhang W., Raggi M. C., Sauliunaite D., De Oliveira T., Friess H., Kleeff J. (2010) ALCAM is associated with chemoresistance and tumor cell adhesion in pancreatic cancer. J. Surg. Oncol. 101, 564–569 [DOI] [PubMed] [Google Scholar]
- 38. Kulasingam V., Diamandis E. P. (2007) Proteomics analysis of conditioned media from three breast cancer cell lines: a mine for biomarkers and therapeutic targets. Mol. Cell. Proteomics 6, 1997–2011 [DOI] [PubMed] [Google Scholar]
- 39. Kulasingam V., Zheng Y., Soosaipillai A., Leon A. E., Gion M., Diamandis E. P. (2009) Activated leukocyte cell adhesion molecule: a novel biomarker for breast cancer. Int. J. Cancer 125, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van den Brand M., Takes R. P., Blokpoel-deRuyter M., Slootweg P. J., van Kempen L. C. (2010) Activated leukocyte cell adhesion molecule expression predicts lymph node metastasis in oral squamous cell carcinoma. Oral Oncol. 46, 393–398 [DOI] [PubMed] [Google Scholar]
- 41. Verma A., Shukla N. K., Deo S. V., Gupta S. D., Ralhan R. (2005) MEMD/ALCAM: a potential marker for tumor invasion and nodal metastasis in esophageal squamous cell carcinoma. Oncology 68, 462–470 [DOI] [PubMed] [Google Scholar]
- 42. Sawhney M., Matta A., Macha M. A., Kaur J., DattaGupta S., Shukla N. K., Ralhan R. (2009) Cytoplasmic accumulation of activated leukocyte cell adhesion molecule is a predictor of disease progression and reduced survival in oral cancer patients. Int. J. Cancer 124, 2098–2105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.