Abstract
High abundance proteins like ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco) impose a consistent challenge for the whole proteome characterization using shot-gun proteomics. To address this challenge, we developed and evaluated Polyethyleneimine Assisted Rubisco Cleanup (PARC) as a new method by combining both abundant protein removal and fractionation. The new approach was applied to a plant insect interaction study to validate the platform and investigate mechanisms for plant defense against herbivorous insects. Our results indicated that PARC can effectively remove Rubisco, improve the protein identification, and discover almost three times more differentially regulated proteins. The significantly enhanced shot-gun proteomics performance was translated into in-depth proteomic and molecular mechanisms for plant insect interaction, where carbon re-distribution was used to play an essential role. Moreover, the transcriptomic validation also confirmed the reliability of PARC analysis. Finally, functional studies were carried out for two differentially regulated genes as revealed by PARC analysis. Insect resistance was induced by over-expressing either jacalin-like or cupin-like genes in rice. The results further highlighted that PARC can serve as an effective strategy for proteomics analysis and gene discovery.
One of the constant challenges for proteomics is inadequate protein identification because of the interference of high abundance proteins (1). The challenge is particularly critical for plant proteomics analysis because of the prevalence of Rubisco (Ribulose-1,5-bisphosphate carboxylase oxygenase) in green tissue. As a major enzyme involved in carbon fixation, Rubisco consists of 30 to 50% of total plant protein from green tissues and causes less sensitivity, dynamic range, and protein identification of plant proteomics (2–4). Influences of high abundance proteins like Rubisco affect both gel-based and shot-gun proteomics analysis. In one of the most popular shot-gun proteomics platforms with the data-dependent MS/MS acquisition, the peptides derived from the abundant proteins have more chance to be sampled by the MS instrument than the peptides from other functional proteins. Thus, the dynamic range and detection sensitivity will be sacrificed because of the prevalence of high abundance proteins (2). To address this challenge, we developed and evaluated a new method by combining PEI (Polyethyleneimine)1 precipitation and protein sample fractionation to improve the performance of multidimensional protein identification technology (MudPIT)-based proteomics analysis.
PEI is a positively charged polymer broadly used for removing nucleic acids from proteins (5). The compound can also be employed to remove acidic proteins like Rubisco from the total protein (6). PEI precipitation can be considered as a fractionation process to separate acidic proteins from the total protein, and thus can be used for both Rubisco removal as well as fractionation of plant proteins from green tissues. Despite the potential to be used for sample preparation, very few studies optimized the PEI precipitation for plant proteomics analysis and evaluated the effectiveness of the approach for enhancing proteomics performance. In this study, we demonstrated that PEI-Assisted Rubisco Cleanup (PARC) and fractionation could significantly improve protein identification. The method was also used to investigate the molecular mechanisms for plant insect interaction and discover two important regulators for rice defense against herbivorous insects.
Plant insect interaction represents an important traditional research field because of the relevance for crop growth and yield. Proteomics has emerged as a major approach to study plant physiological, pathological, and developmental processes at the systems level (7–9). Despite the progresses, only a few studies have been performed to comprehensively study plant defense against herbivorous insects using proteomics approach. Most of the previous research was carried out using gel-based platforms, resulting in the identification of limited numbers of differential proteins (10–13). Plants respond to insect attacks with systemic up-regulation of regulatory, metabolic and defense-related proteins, and the coverage of proteome is crucial for revealing in-depth mechanisms (14). Our previous transcriptomic analysis using rice as a model plant revealed molecular and genomic mechanisms for plant defenses, including the regulation of enzymes involved in secondary metabolisms and volatile production (15). Despite the transcriptomic analysis, the protein level regulation of rice defense against herbivorous insects is largely unknown. The comprehensive proteome profiling will thus help to reveal novel mechanisms for protein-level defense regulation and to identify the new regulators in plant defense that will guide the crop improvement essential for food security.
In this study, we developed and evaluated a novel plant protein isolation and fractionation method to improve the performance of MudPIT-based shot-gun proteomics. We further applied the method for a plant insect interaction study. The method increased protein identification as compared with the protein isolated by a commercial kit. The novel method also resulted in discovering almost three times more differential proteins. The significantly improved differential protein identification helped to reveal new proteome-level mechanisms for plant defense against herbivorous insects and to identify key genes involved in defense regulation. In particular, the proteomics results revealed dynamic re-allocation of carbon resources as a defense mechanism against herbivorous insects in plant. Furthermore, we carried out two levels of validation of proteomics data. The first was real-time PCR analysis to verify the gene expression of the up-regulated proteins. The second was the functional validation of two key regulatory proteins for plant defense. The first gene was a cupin-like protein specifically identified in PARC. The cupin-like protein was previously reported to be important for reactive oxygen species (ROS) production during the defense response. For example, in a study researched by Carrillo et al., a cupin-like protein Os03g48470 was demonstrated to be an oxalate oxidase to produce H2O2 (16). However, it was not clear which one of more than 100 cupin-like proteins in the protein family was actually involved in plant defense against herbivorous insects (17–19). The second gene is a jacalin-like protein with unknown molecular function. Transgene over-expression experiments validated both genes to be important regulators for rice defense against herbivorous insects. Overall, our results highlighted that the PARC method could significantly improve the performance of shot-gun proteomics to enable the in-depth analysis of plant defense mechanisms and the discovery of new regulators for plant defense against herbivorous insects.
EXPERIMENTAL PROCEDURES
Plant and Insect Growth
Rice (Oryza sativa ssp. Japonica cv. Nipponbare) seeds were germinated at 30 °C in the dark for 5 days. Then the seedlings were grown at 28 °C with 14 h of light for 2 weeks. Fall armyworm (FAW) (Spodoptera frugiperda) eggs were obtained from Benzon Research (Carlisle, PA, USA) and hatched at 28 °C. FAW larvae were raised on an artificial diet and second-instar FAW were used for herbivore treatment. Two larvae were placed on the leaves of a single two-week old rice seedling around nightfall at 10 pm. After 12 h, ∼5–10% of the leaf area was consumed. Insects were then removed and the rice plants were harvested to snap-freeze in liquid nitrogen. The samples were stored in ultralow freezer at −80 °C until further analysis.
Protein Sample Preparation Methods
The protein sample preparation integrates both PEI precipitation optimization (Fig. 1) and sample fractionation. The flow of the different fractionation and protein extraction approaches was as shown in Fig. 2. For the reference, the total plant protein was extracted with Plant Total Protein Extraction Kit (Sigma-Aldrich, St Louis, MO) according to the manufacture's instruction with minor modifications. The protein sample is referred to as TP (Total Protein) fraction, which represented plant total protein extracted by conventional method. Briefly, 100–250 mg of leaf tissue was ground into a fine powder in liquid nitrogen. The powder was washed with methanol and acetone, then pelleted and dried with a SpeedVac. The plant tissue pellet was dissolved in Reagent Type 4 Working Solution supplied by the kit. The protein extraction solution provided by the commercial Sigma kit contains 7 m urea, 2 m thiourea, 40 mm Triszma base, and 1% 3-(4-Heptyl)phenyl-3-hydroxypropyl dimethylammoniopropanesulfonate (C7BzO) as the detergent. The solution was adjusted to pH 10.4. The TP sample preparation using this commercial kit involves chaotropic reagents, particularly, with a zwitterionic detergent to dissolve most proteins including hydrophobic membrane proteins. After the extraction and removal of plant tissue debris, the TP samples were ready for proteomic analysis. However, the TP fraction cannot be used for PEI precipitation to remove Rubisco (Fig. 1B) potentially because of the changes in protein charges by extraction buffer for the high concentration of the zwitterionic detergent.
Fig. 1.
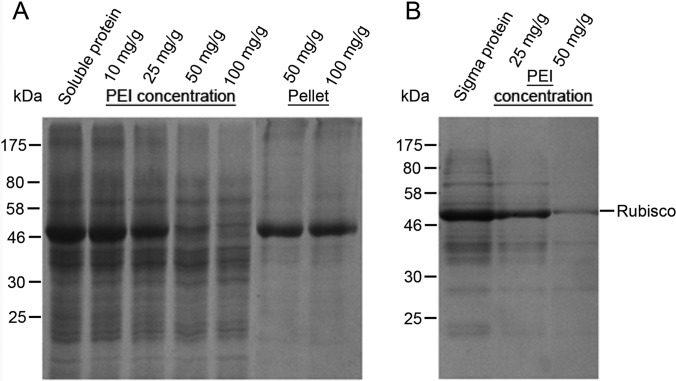
Efficiency of Rubisco removal by PEI (Polyethyleneimine) precipitation. A, PEI precipitation of total soluble protein isolated by soluble protein extraction buffer. From the left to the right, lane 1 shows the total soluble protein extracted by soluble protein extraction buffer. Lane 2 to 5 shows the supernatant of soluble protein after precipitation with 10, 25, 50, and 100 mg/g PEI (weight/total protein), respectively. Lane 6 and 7 are dissolved protein pellets after PEI precipitation. B, Incompatibility between PEI precipitation and total protein extracted by Sigma Aldrich Plant Total Protein Extraction kit. From the left to the right, lane 1 shows the total protein extracted by the kit. Lane 2 and 3 shows total protein supernatant after PEI precipitation at 25 and 50 mg/g, respectively. PEI concentration (mg/g) is as indicated at the top. Molecular weight markers in kDa are shown on the left.
Fig. 2.
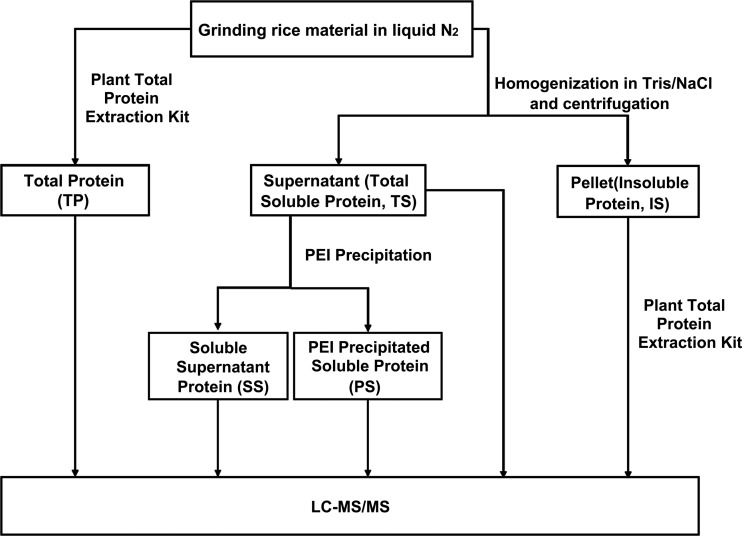
Flowchart of protein extraction, Rubisco removal and fractionation method. Rice protein samples were extracted and fractionized into five fractions. Aerial part of rice seedlings were ground into fine powder in liquid nitrogen. Total protein was extracted by Sigma Plant Total Protein Extraction Kit (TP). Total soluble protein TS and Insoluble protein (IS) were obtained by centrifugation after the powder was homogenized in extraction buffer. PEI precipitation was used to fractionize the TS fraction into Soluble Supernatant protein (SS) and PEI Precipitated Soluble protein (PS).
Besides the TP sample prepared by the commercial kit, a modified SDS (sodium dodecyl sulfate)-based total protein extraction method was used to validate the effectiveness of the commercial kit (20). The SDS solubilized total protein was subject to SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) analysis. The gel fragment containing proteins were washed, distained, dehydrated, and further processed for in-gel digestion according to previous studies (21).
To carry out PEI precipitation, a soluble protein extraction method was developed with a nonionic detergent. Approximately 200 mg rice leaves were ground into a fine powder in liquid nitrogen using mortar and pestle. 1 ml of extraction buffer (50 mm Tris-HCl pH 7.5, 10 mm MgCl2, 150 mm NaCl, 0.1% Nonidet P-40, 1 mm PMSF) was then added and incubated with the sample. The extract was then centrifuged at 13,000 rpm for 10 min at 4 °C, and the supernatant was then transferred to a new micro-centrifuge tube. The supernatant contained most of the soluble protein in the cell and was thus referred to as TS (total soluble protein). The pellet was used to continue to extract protein using the same Plant Total Protein Extraction Kit (Sigma-Aldrich) according to the manufacturer's instruction. The sample contained mostly the insoluble protein and was thus referred to as IS. In addition to PEI precipitation, we also evaluated ammonium sulfate for its capacity to selectively precipitate Rubisco. 40% ammonium sulfate was used to precipitate Rubisco according to previous publication (22). The ammonium sulfate precipitated pellets were then re-suspended and analyzed by SDS-PAGE gel in the same way as PEI precipitated proteins.
PEI precipitation was carried out to remove the Rubisco contamination in the TS fraction. A titration was performed to determine the effective concentration for PEI precipitation as shown in Fig. 1A. For LC-MS/MS experiments, the TS samples were first mixed with 100 mg/g of PEI, vortexed vigorously for 10 s, and then precipitated on ice for 5 min. Samples were then centrifuged for 15 min at 13,000 rpm. The supernatant was removed by pipetting and collected as SS (Supernatant Soluble) fraction. The SS fraction is the Rubisco removed soluble protein. The PEI precipitated pellets were then resuspended in resuspension buffer and centrifuged for 10 min at 13,000 rpm to re-pellet the debris. The supernatant was collected as PS (precipitated soluble protein). The fraction should be Rubisco enriched fraction containing many acidic proteins. The protein concentration for each fraction was measured by Bradford Protein Assay Kit (Thermo Scientific Pierce).
MudPIT and Shot-gun Proteomics
MudPIT-based shot-gun proteomics were carried out to analyze each aforementioned fraction as described by Washburn et al. (23). Approximately 100 μg protein was digested by Mass Spectrometry Grade Trypsin Gold (Promega, Madison, WI) with 1:40 w/w at 37 °C for 24 h. The digested peptides were desalted using a Sep-Pak C18 plus column (Waters Corporation, Milford, MA) and then loaded onto a biphasic (strong cation exchange/reversed phase) capillary column using a high pressure tank. The two-dimensional liquid chromatography separation and tandem mass spectrometry were carried out as previously described (24).
The separated peptides were analyzed using a linear ion trap mass spectrometer, Finnigan LTQ (Thermo Finnegan). The mass spectrometry was set to the data-dependent acquisition mode. Then the full mass spectra were recorded on the peptides over a 300–1700 m/z range, followed by five tandem mass (MS/MS) events for the most abundant ions from the first MS analysis. The Xcalibur data system (Thermo Fisher Scientific) was used to control the LC-LTQ system and collect the data. The experiments were carried out with duplicate biological samples for all fractions except the TP fraction.
Data Analysis
A data preprocessing pipeline based on Xcalibur Development Kit was used to generate DTA files in the same way as the ThermoFinnigan Bioworks (2.0) software. Tandem mass spectra were extracted from the raw files and converted into the MS2 file. The MS2 file was searched against the rice protein database containing 66,338 protein sequences from the MSU Rice Genome Annotation (Release 7), the same number of reverse sequence, and common contaminant proteins. The reverse sequences of the original data set were included calculate confidence levels and false-positive rates. The common containment proteins were included for data quality control. The ProLuCID (version 1.0) algorithm was used to assign peptide sequence to peptide fragmentation spectra using the Texas A&M Supercomputing Facility (25). The ProLuCID parameters were set as the follows: the precursor mass accuracy was set at 100 ppm; fragment ion mass accuracy was set at 600 ppm. No fixed or variable modifications were considered. At least two distinct peptides (semitryptic) were required to identify a protein with no sequence coverage assigned.
Protein quantification was achieved by spectral counting. The validity of peptide/spectrum matches were assessed in DTASelect v.2.0 using a 0.05 false discovery cutoff, with a cross-correlation score (XCorr) that's larger than 1, and normalized difference in cross-correlation scores (DeltaCN) larger than 0.08. Semitryptic peptide was included in the final calculation. DTASelect software listed the accession numbers together for those proteins combined into one group. Each individual accession numbers were then listed in the supplementary data. Number of peptides and spectra used for protein quantification was shown in supplemental Table S1. Detailed information was shown in supplemental Table S6. Proteins identified with more than two peptides were used for quantification. Protein identification was based on both specific and nonspecific peptides. Quantification is based on duplicate biological samples.
Ontology and Pathway Analysis
PatternLab (26) software (version 2.1.1.5) was used for data analysis to discover differentially expressed proteins. Isoforms of protein were combined in groups by PatternLab. The Row Sigma normalization was carried out to adjust systemic errors. The TFold test was applied to derive differentially expressed proteins. The cutoff of p value (BH q-value) and FDR were 0.05 for both. F-stringency was optimized by software automatically. Gene ontology analysis was performed using agriGO (27). Each protein was classified with respect to its biological process using GO annotation. The pathway analysis of differentially regulated proteins was analyzed using KEGG (28).
Alignments and Phylogenetic Analysis
A phylogenetic tree was generated using sequences download from NCBI reference proteins and UniProtKB/Swiss-Prot database. The threshold for blasted sequences of Os03g48770 was 1e-54. The amino acid sequences were initially aligned using ClustalX (29). The trees were created using the neighbor-joining method of MEGA version 5.05 with the Poisson model having uniform rates for all sites. The phylogenetic trees were tested by 1000 bootstrapping (30). In addition to phylogenetic tree, multiple sequence alignment was carried out for wheat oxalate oxidase GF-3.8 (P26759), rice germin-like protein 3–6 (Os03g48780) and the cupin domain containing protein Os03g48770 by Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Real-time Quantitative PCR
For the qRT-PCR analysis, wild type rice seedlings were infested with FAW larvae and the gene expression was analyzed at series of time points of 0 h, 12 h, 24 h, and 48 h. RNA isolation using TRI Reagent was performed following manufacture's instruction (31).
Reverse transcription of total RNA was performed using SuperScript III First-Strand Synthesis kit (Invitrogen, Carlsbad, CA). Triplicate quantitative assays were performed using the SYBR Green Master Mix (Applied Biosystems, Foster City, CA) with an ABI 7900HT sequence detection system. All data were normalized with 18S rRNA as an internal reference gene using the following primers: forward 5′-CGGCTACCACATCCAAGGAA-3′; reverse 5′-TGTCACTACCTCCCCGTGTCA-3′. Primers used for quantitative qRT-PCR analysis were listed in supplemental Table S2. The relative quantification method (modified ΔΔCT) was used to evaluate the differential gene expression. The ΔCTs were normalized against the lowest expressed samples within the group to derive ΔΔCT, where the smallest ΔΔCT for each group will be 0. The 2ΔΔCT will represent the ratio of gene expression over the lowest expressed sample in the group. Both the ratio of gene expression and 95% confidence intervals derived from triplicate assays were presented.
Rice Transformation
For overexpression of Os03g48770 and Os12g14440, full-length cDNA were subcloned into a binary vector pCXUN with ubiquitin promoter (32) and introduced into Nipponbare rice embryonic calli by Agrobacterium tumefaciens-mediated transformation (33). Transgenic plants were selected on selection plates with 50 mg/L hygromycin. T1 generation plants were used to perform insect feeding assay.
Leaf Area Damage Measurement
Transgenic and wild type rice leaves from T1 plants were collected and cut into small pieces. The pieces were inserted into 10 ml 0.7% agarose in a 30 ml plastic cup. One 3rd instar larva was put on the leaf and allowed it to feed for 48 h. The percentage of leaf area damage was calculated by ImageJ (34). ANOVA analysis was carried out to compare control groups versus transgenic lines with 95% confidence intervals. Three validated transgenic lines from each target genes were used. For each transgenic line, six leaf segments were used for insect treatment bioassay and leaf area measurement.
RESULTS
The study integrated method development with functional proteomics analysis. First, a new strategy for plant shot-gun proteomics, PARC (PEI-Assisted Rubisco Cleanup) was developed to improve the protein identification and differential expression analysis. Second, the method was used to investigate the proteomic and molecular mechanisms for plant defense against herbivorous insects, which further validated PARC as an effective method to reveal in-depth mechanisms and novel regulators for plant insect interaction.
PARC as an Efficient Approach to Remove Rubisco
The method development started with optimization of PEI precipitation for removing Rubisco from extracted protein samples (Fig. 1). PEI precipitation was carried out using two types of input protein, the total soluble protein (TS) extracted by soluble protein extraction buffer and the total protein (TP) extracted by the urea/thiourea based buffer in the Plant Total Protein Extraction Kit (See Experimental Procedures for details). As shown in Fig. 1A, PEI precipitation could effectively remove Rubisco at the concentration of 50 to 100 mg/g from the total soluble proteins TS. However, PEI precipitation could not selectively precipitate Rubisco from the total protein (TP) extracted by the commercial Sigma Aldrich kit (Fig. 1B). As shown in Fig. 1B, when 25 and 50 mg/g concentrations of PEI were used, both Rubisco and other proteins were precipitated from the total protein sample TP. The difference in selectivity might result from the change of protein charges in the TP sample caused by the urea and thiourea buffer.
Based on the results, we developed the strategy as described in materials and supplies to remove Rubisco and to fractionate protein (Fig. 2). In this strategy, the total soluble protein TS was first extracted and then fractionated into supernatant Rubisco reduced fraction (SS) and pelletized Rubisco enriched fraction (PS) using 100 mg/g PEI. In the meantime, the insoluble protein (IS) can be extracted by the aforementioned the Sigma Aldrich Plant Total Protein Extraction Kit, and the IS, PS and SS constituted the three fractions of the total protein, which can be compared with TP. Shot-gun proteomics analysis was then carried out for each fraction (SS, PS, IS, TS, and TP).
Two additional control experiments were also carried out. First, the effectiveness and suitability of the commercial kit as reference was further evaluated by comparison of the protein identification using SDS-based protein sample preparation with that of TP. As shown in supplemental Fig. S1, more nonredundant proteins were identified in the TP fraction than those from the SDS-based method. Second, different chemical compounds including PEI and ammonium sulfate can be used to “selectively” precipitate Rubisco. We briefly compared the effectiveness of PEI versus ammonium sulfate. As shown in supplemental Fig. S2, the addition of ammonium sulfate resulted in the precipitation more non-Rubisco proteins as compared with PEI. Our PARC method development thus is majorly based on PEI because of its higher specificity. In particular, the efficacy of PARC for protein identification and differential protein expression can be evaluated by comparing (1) SS and PS versus TS for soluble proteins as well as (2) SS, PS, and IS versus TP for total proteins.
Effective Removal of Rubisco, Improved Protein Identification and Enhanced Differential Analysis of Shot-gun Proteomics
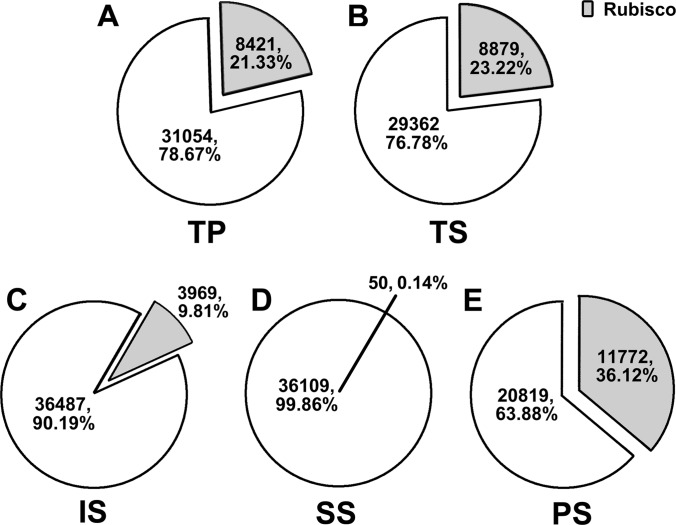
The aforementioned protein fractions were subject to the MudPIT-based proteomics analysis. The effectiveness of PARC was evaluated from three aspects. First, we analyzed the percentage of spectral counts from Rubisco as part of the total spectral count for each fraction (TS, SS, PS, IS, TP). As shown in Fig. 3, PARC could efficiently remove Rubisco from soluble proteins. The Rubisco spectra counts were 21.33% of the total protein spectral counts TP. Similar to TP sample, the total soluble protein TS fraction contained 23.22% of Rubisco spectral counts. The insoluble fraction (IS) contained a surprisingly low percentage of Rubisco peptide counts at 9.81% of total spectral counts. SS and PS fractions were derived from TS when the sample was subject to PEI precipitation. As expected, the supernatant Rubisco-reduced fraction (SS) had very little Rubisco contamination with the Rubisco at 0.14% of total spectral counts. Meanwhile, the precipitated Rubisco-enriched fraction (PS) had a significantly higher percentage of Rubisco spectral counts at 36.12%.
Fig. 3.
Rubisco removal efficiency evaluated by proteomics. The pie charts showed the percentage of spectral counts for Rubisco in total spectral counts identified in each fraction. A, TP. B, TS. C, IS. D, SS. E, PS.
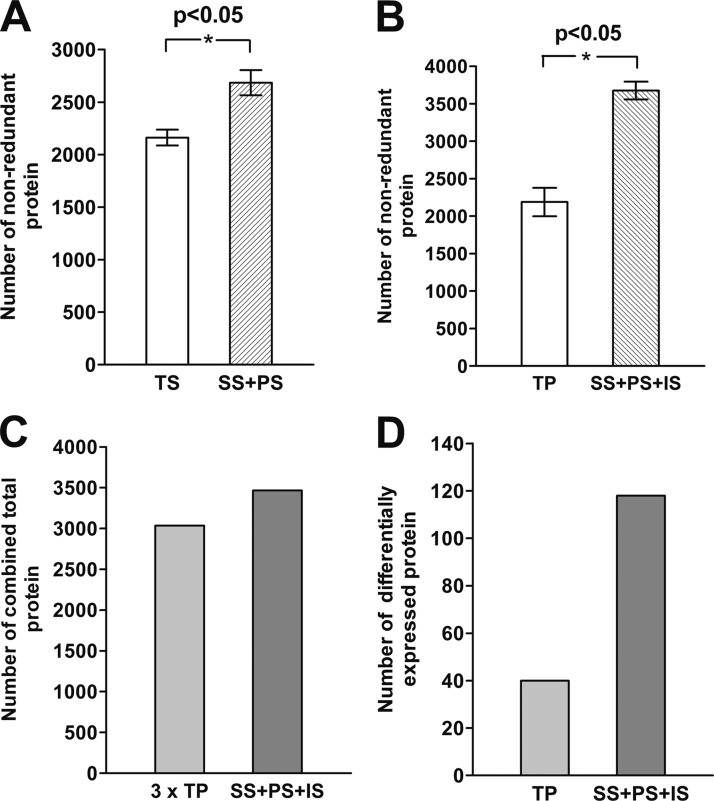
Second, the protein identification was improved by fractionation and Rubisco removal as shown in Fig. 4. Total soluble protein TS was fractionated into SS and PS fractions through Rubisco removal and PEI precipitation. The fractionation process led to the identification of 523 more proteins and a total of 24.2% increase in the number of protein identified (Fig. 4A). Furthermore, the Rubisco removed SS, Rubisco enriched PS, and the total insoluble proteins (IS) should represent the total proteins, which can be compared with the total protein (TP) extracted by the commercial kit. The three fractions together identified 1488 more proteins than TP. In other words, PARC increased the protein identification by 68.0% (Fig. 4B). Further validation was carried out by comparison of the number of proteins identified by three biological TP samples versus the one set of SS, PS, and IS fractions (Fig. 4C). This comparison is justified by the same number of LC gradient to remove the bias caused by more experimental LC-MS/MS runs for combination of the three fractions. As shown in Fig. 4C, the combined SS, PS and IS fractions still identify 431 more proteins than the three times of TP sampling. The result highlighted that the fractionation can increase protein identification, even though the exact level of improvement is hard to define.
Fig. 4.
Improvement of protein identification and differential protein expression analysis. A, Comparison of the number of protein identified by TS and the combination of SS and PS. B, Comparison of the number of protein identification for TP and the combination of homogenizations, SS, PS, and IS. C, Comparison of total number of combined proteins between 3 independent TP biological samples and 1 biological samples of SS, PS, and IS. D, Comparison of the number of differentially expressed proteins identified in FAW larvae treated and untreated rice samples with the same fractionations of B. The standard errors were calculated based on duplicates of the biological samples from different factions.
Third and most importantly, the improved protein identification led to the better evaluation of differential protein expression. To further validate the effectiveness of PARC and to investigate mechanisms for plant insect interaction at the systems level, differential protein expression analysis was carried out between insect treated rice and untreated reference. The study revealed superior performance of the PARC, as shown in Fig. 4D. The comparison of TP samples between insect treated and control only identified 40 differentially expressed proteins. PARC enabled significantly improved differential protein discovery. One hundred and eighteen differentially expressed proteins were identified when SS, PS, and IS fractions were analyzed. PARC therefore increased the identification of differentially expressed proteins by almost three times.
Overview of Differential Protein Expression for Plant Insect Interaction as Revealed by the Proteomics
The systems-level overview of proteomics results included both Gene Ontology (GO) and pathway analysis. Considering that PARC significantly improved the performance of proteomics analysis, the functional proteomics analysis of rice-FAW (Fall Armyworm) interaction was focused on the PARC-based study (i.e. SS, PS, and IS fractions) rather than the traditional approach (i.e. TP fraction). In fact, the PARC analysis identified much more relevant pathways than those of the traditional methods.
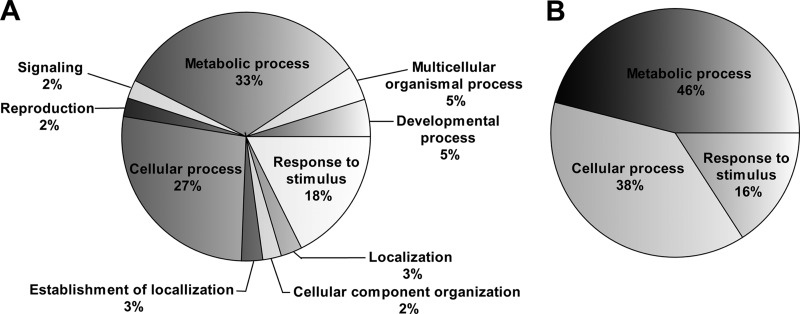
Gene Ontology analyses revealed that proteomics analysis based on PARC can identify much more GO terms than the reference sample (TP), assumingly because of the greater amount of proteins identified in the PARC-based analysis (Fig. 5). Even though both methods identified metabolic processes and responses to stimuli as major GO groups, the PARC-based analyses enabled the identification of new GO terms such as signaling and cellular component organization.
Fig. 5.
Gene Ontology analysis of differentially expressed protein. Pie charts shows GO distribution of (A) combination of SS, PS, and IS as well as (B) TP according to their biological processes.
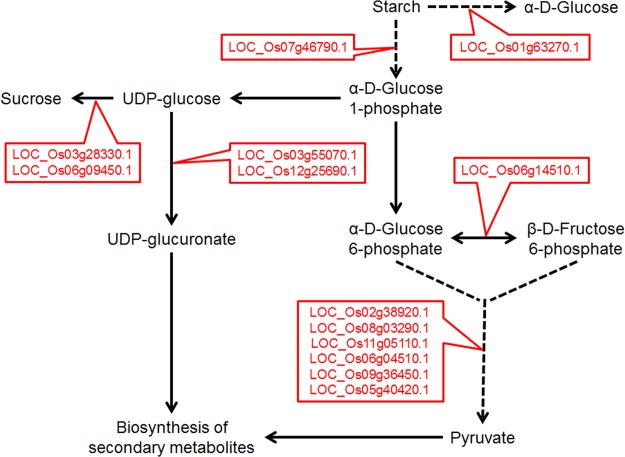
A group of differentially expressed metabolic proteins were shown in Table I and detailed functional classification of all differentially expressed proteins was shown in supplemental Table S3. The analyses revealed that plant responded to herbivore insect damage by up-regulating proteins involved in a broad range of processes including primary metabolism, secondary metabolism, upstream jasmonic acid biosynthesis like lipoxygenase, defense relevant proteins and others. An important aspect of metabolism changes was the carbohydrate biosynthesis and degradation. As shown in Fig. 6, both the starch and carbohydrate degradation pathway and sucrose biosynthesis were up-regulated, leading to the net carbon fluxes toward both sucrose and secondary metabolite biosynthesis.
Table I. Selected significantly up-regulated metabolic proteins after FAW herbivory as identified by PARC.
| Gene function | Gene ID | pValue | Ratio |
|---|---|---|---|
| Carbohydrates biosynthesis and degradation | |||
| Sucrose synthase | Os03g28330.1 | 0.000177 | 22.30 |
| Sucrose synthase | Os06g09450.1 | 0.004775 | 8.20 |
| Glucose-6-phosphate isomerase | Os06g14510.1 | 0.046986 | 3.94 |
| Triosephosphate isomerase | Os09g36450.1 | 0.029399 | 2.92 |
| 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | Os05g40420.1 | 0.032074 | 4.22 |
| Enolase | Os06g04510.1 | 0.022889 | 3.57 |
| Pyruvate kinase | Os11g05110.1 | 0.006019 | 1.98 |
| Glyceraldehyde-3-phosphate dehydrogenase | Os02g38920.1 | 0.015524 | 2.47 |
| Glyceraldehyde-3-phosphate dehydrogenase | Os08g03290.1 | 0.003714 | 1.79 |
| Ribulose bisphosphate carboxylase small chain | Os12g17600.1 | 0.006673 | 5.30 |
| Triosephosphate isomerase | Os09g36540.1 | 0.029399 | 2.92 |
| UDP-glucose 6-dehydrogenase | Os12g25690.1 | 0.036053 | 4.38 |
| Alpha-glucan phosphorylast isozyme | Os01g63270.1 | 0.003241 | 2.56 |
| 4-alpha-glucanotransferase | Os07g46790.1 | 0.031757 | 2.82 |
| Amino acids biosynthesis | |||
| 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | Os12g42876.1 | 0.008758 | 7.21 |
| 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | Os12g42884.1 | 0.011412 | 7.47 |
| Aminotransferase | Os02g55420.1 | 0.035815 | 3.94 |
| Glutamine synthetase | Os03g12290.1 | 0.036183 | 3.36 |
| Secondary metabolites biosynthesis | |||
| Caffeoyl-CoA O-methyltransferase | Os08g38900.1 | 0.011654 | 3.71 |
| Isoflavone reductase | Os01g01660.1 | 0.039708 | 3.20 |
| Lipoxygenase 2.1 | Os12g37260.1 | 0.000453 | 3.36 |
Fig. 6.
Pathway analysis of up-regulated proteins by FAW herbivory as revealed by PARC. The figure focused on the carbohydrate related metabolisms.
The correlation of protein and mRNA changes induced by insect damage was evaluated at both global and gene-specific levels. The comparison of this proteomics study with previous transcriptomics analysis of FAW treated rice revealed correlated mRNA and protein expression for some key secondary metabolism genes. These genes include Os10g28200, an NAD dependent epimerase and dehydratase family protein, and Os12g37260, a lipoxygenase chloroplast precursor (15). In addition, Os12g14440 (jacalin-like lectin domain containing protein) was also shown to be up-regulated by the microarray study. This gene was chosen for the downstream functional analysis. It should be noted that previous study used an early version of long-oligo microarray which contains only 22,000 features and a relatively high error rate (15). A more comprehensive comparison of protein and mRNA expression can be achieved by comparing the proteomics data with the MPSS data for insect treatment (35). As shown in supplemental Table S4, among the 88 up-regulated proteins caused by FAW treatment, 32 were up-regulated at mRNA level when treated with beet armyworm (BAW). For reference, all proteins identified from proteomics data were included in supplemental Table S5.
Validation of Proteomics Results by qRT-PCR
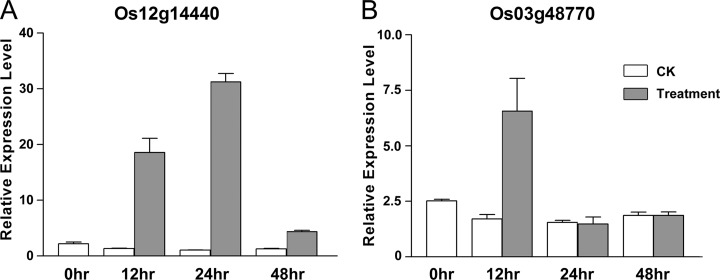
The gene-specific validation was focused on the up-regulated proteins as identified by the PARC-based proteomics analysis. Eleven up-regulated proteins were chosen for gene expression validation using real-time PCR. Most of these proteins were chosen because they are related to insect defense according to previous studies. As shown in Fig. 7 and supplemental Fig. S3 eight out of 11 genes showed correlated mRNA and protein expression level. In other words, the triplicate qRT-PCR assays revealed that eight genes have significantly increased mRNA expression between insect treatment and control plants at 12 h after treatments.
Fig. 7.
qRT-PCR validation of mRNA expression for selected differentially expressed proteins. qRT-PCR validation of (A) Os12g14440 and (B) Os03g48770 gene expression after 0 h, 12 h, 24 h, and 48 h treatments. The relative expression levels as compared with the lowest expressing sample in the group were derived from modified ΔΔCT method. The average ratio and 95% confidence interval were derived from triplicate assays.
Among the 8 genes verified by qRT-PCR, Os03g48770 (cupin domain containing protein) and Os12g14440 (jacalin-like lectin domain containing protein, JRL), were chosen for functional study using transgenic analysis. The cupin protein was exclusively identified as an up-regulated protein by the PARC approach. Both genes were up-regulated at 12 h after insect treatment according to the qRT-PCR results (Fig. 7).
Biological Validation of Proteomics Results by Transgenic Analysis
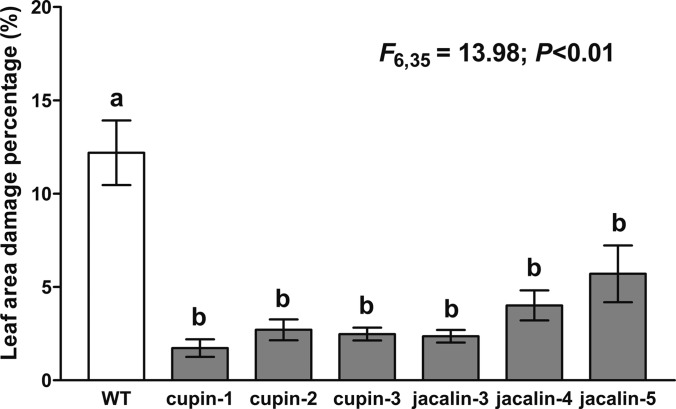
To verify the gene function of Os03g48770 and Os12g14440, transgenic lines have been generated to over-express the two target genes for insect treatment bioassay. The transgene overexpression was validated for T1 rice (supplemental Fig. S4). Six leaf segments from each transgenic line and the wild-type rice were subject to insect treatment bioassay by feeding the leaves with the FAW larvae (Fig. 8 and supplemental Fig. S5). After 2 days of larvae feeding, the leaf area damage was measured and calculated. The average percentages of leaf area damage were 1.73%, 2.71%, and 2.48% for the Os03g48770 over-expressing lines and 2.36%, 4.01%, and 5.71% for the Os12g14440 over-expressing lines, respectively. In contrast, the wild type rice underwent 12.19% leaf damage.
Fig. 8.
Comparison of leaf area damage caused by FAW larvae feeding on wild type and transgenic rice. The figure shows ANOVA analysis of three transgenic lines for both Os03g48770 (Cupin) and Os12g14440 (Jacalin) after FAW consumption. Wild type rice (group a) showed significant difference with transgenic lines (group b) (F6,35 = 13.98; p < 0.01). The measurement is based on bioassay of 6 leave segments of each independent line. The average percentage of leaf area damage and 95% confidence intervals were presented.
Statistical analysis of leaf area damage after FAW larvae feeding showed significant difference between the six transgenic lines and wild type rice with p < 0.01 (Fig. 8). The statistical analysis were also carried out for each individual line, which separated the samples into two groups (a and b). The samples in group b (all transgenic lines) have significantly decreased leaf area damage as compared with group a (wild type). The result indicated that the overexpression of the Os03g48770 and Os12g14440 in rice contributed to the resistance to the FAW larvae. More reliable functional studies need to be carried out in T3 transgenic lines in the future and is beyond the scope of PARC method development. The functional validation suggested that the differentially regulated proteins identified by PARC played important roles in insect defense.
DISCUSSION
PARC as an Effective Platform for Shot-gun Proteomics
Even though proteomics has emerged as a powerful platform to study systems and molecular mechanisms for biological processes like plant defense, the application of the technology is still limited by some inherent challenges. One example is that high abundance proteins will reduce sensitivity and hinder protein identification. Different strategies have been developed to address these issues caused by high abundance proteins and to improve the detection limit and performance of shot-gun proteomics. These strategies included target proteomics, fractionation, antibody-based protein removal, improved algorithm for protein identification, and others. Recently, we developed an organelle-enriched method to enhance the identification and differential expression analysis of mitochondrial proteins (24). The method involved both organelle enrichment for sample preparation and bioinformatics classification. Even though the method improved the identification of mitochondrial proteins, the impact of Rubisco was still not properly mitigated. We hereby presented another approach to improve the sensitivity, detection, protein identification and differential expression analysis for MudPIT-based shot-gun proteomics. Moreover, the method was used to dissect the proteomic and molecular mechanisms for plant defense against herbivorous insects, leading to the identification of two key regulators in plant defense.
Several sample preparation methods were reported to deplete Rubisco from plant protein samples. These methods include sucrose density gradient centrifugation, Ca2+/phytate precipitation, and Rubisco antibody column (4, 36–39). Despite the progresses, these procedures were either labor intensive, time consuming, costly, or leading to limited improvement in protein identification. We therefore developed PARC as a new strategy to combine the Rubisco removal and protein fractionation. In PARC, the total protein was first separated into two fractions with a nonionic detergent. The two fractions were a total soluble protein (TS) and an insoluble protein (IS). The TS fraction was then treated with PEI to derive two other fractions as Rubisco-removed soluble protein (SS) and PEI precipitated fraction (PS). PEI is a polymer with repeating units that can contain primary, secondary and tertiary amino groups. The negative charge of the molecule can selectively precipitate positively charged proteins like Rubisco. Our results indicated that PEI could be more selective for Rubisco removal than ammonium sulfate, which is another common compound for Rubisco precipitation.
In the PS fraction, proteins other than Rubisco were also precipitated. In Fig. 3E, the spectra count from Rubisco composes 36.12% of the total spectra count in PS fraction, which suggested coprecipitation of other positively charged proteins. However, because the PEI precipitated fraction will also be subjected to the shotgun proteomics analysis, other co-precipitated proteins will still be included in the identification process, so that no extra information will be lost because of Rubisco removal. Our studies eventually led us to the finding that the integration of fractionation and Rubisco removal by PEI led to the detection of more than 3500 proteins in the rice proteome. This translated into 68% increase of the number of proteins identified using a commercial kit without any fractionation. The number of identified proteins was also significantly higher than those previously published (40, 41). More importantly, the PARC method essentially tripled the number of differentially regulated proteins identified in the plant insect interaction study.
The identification of more plant proteins may also benefit from other novel approaches such as combining proteomic and genomic forces together (42). Briggs' lab has identified novel proteins by using the unique peptide sequence information from a comprehensive proteomic survey of the Arabidopsis proteome. The new proteo-genomics and bioinformatics approach has led to discovery of more than 10,000 novel peptides. This suggests the current state-of-the-art Arabidopsis genome is still incomplete. The informatics-intensive proteo-genomics research can be further pursued in future researches.
Nevertheless, considering the limitation of current proteomics platforms, fractionation is still one of the viable alternatives to expand the dynamic range of proteomics detection. The disadvantage of the fractionation approach is prolonged analysis time and increased reagent cost. However, the gain of more protein identification and in-depth differential protein expression analysis in this study could compensate the tripled instrumentation time and the increased data analysis time, because the proteomic setup and data analysis are heavily based on computerized and instrumentation-based platform. The principle of abundant protein precipitation and fractionation can be adapted and applied to studies beyond the plant species as PEI can be used to precipitate acidic proteins in general. More importantly, the improved protein identification led to significantly improved differential protein expression analysis, enabling much more in-depth systems and molecular mechanism analysis in the rice-FAW interaction study. The increased number of differential expressed proteins led to many more GO terms being discovered, more comprehensive pathway annotation, and therefore a more in-depth understanding of mechanisms for plant defense.
Systems and Molecular Mechanisms of Plant Defense against Herbivorous Insects as Revealed by PARC
As aforementioned, the PARC-based analysis enabled a greater comprehensive proteome profiling than the traditional approach, delivering in-depth mechanisms from the systems levels to the molecular levels. The GO and protein classification indicated the up-regulation of pathways involved in secondary metabolism, primary metabolism, defense, and other processes. The data correlates with the fact that plants respond to herbivore attack through many dimensions, such as direct and indirect, physical and chemical, constitutive and inducible defenses (14, 43–46). The proteomics data suggested a complex, yet surprising protein-level regulation of carbon utilization. As shown in Fig. 6, the starch degradation enzymes were up-regulated, which could potentially lead to increased production of α-d-Glucose-1P. Furthermore, the α-d-Glucose-1P could be used for either glycolysis and secondary metabolite production or sucrose biosynthesis. Some enzymes leading to the channeling of carbon flux to secondary metabolites were up-regulated. In addition, enzymes involved in sucrose biosynthesis were also up-regulated. Previous studies suggested that sucrose biosynthesis was up-regulated during plant defense against pathogen and insects in a wide range of crop and model plant species, including rice, cotton, tomato, and Arabidopsis (47–54). The study correlated with previous studies indicating the important roles for sucrose in plant defense against herbivorous insects. In addition, as compared with our previous transcriptomic analysis, the proteomic analysis successfully identified new up-regulated pathways in carbohydrate metabolism during defense (15). Overall, active changes in carbon metabolism seemed to be a major feature of plant defense against herbivorous insects. Further validation of the hypothesis will rely on metabolite analysis, which is a major focus in plant defense field.
The study also revealed the correlations of transcriptomic and proteomic regulations. Even though the comparison of previous microarray data and this proteomics analysis only revealed a few over-lapping genes, the comparison of the more comprehensive MPSS analysis with this study revealed almost 36% correlation between transcriptomic and proteomic up-regulated genes. The correlation is significant as several factors contributed to the differences between transcriptomics and proteomics dynamics. First, the insect species, treatment conditions, and plant growth stages were all different between the previous transcriptomics and current proteomics studies. Second, as shown in the qRT-PCR experiments (supplemental Fig. S3), the mRNA level changes are often transient and dynamic, which might not correlate with protein level changes at a given point. Third, several biological processes including RNA degradation, protein translation, and protein stabilization could lead to the differences between protein and gene expression level. Despite these key issues, we still found that 32 out of 88 up-regulated proteins were also up-regulated at mRNA levels. The study indicated that transcriptional regulation of plant defense is crucial in regulating the defense-related protein level. Despite the significant correlation of up-regulated genes and proteins, the down-regulated proteins are not well correlated with mRNA level down-regulation. This could be because of the dynamic protein degradation during defense process.
Functional Validation of Plant Defense Genes as Revealed by PARC
Besides the systems and pathway level analysis, further functional verification was carried out by transgenic functional study of two genes identified from the proteomics study. The Os03g48770 (cupin-like protein) and Os12g14440 (jacalin-like lectin protein) over-expressing lines showed improved resistance against rice herbivore FAW. These two genes represented new regulators for rice defense. Jacalin-related lectins (JRLs) can bind specifically with different carbohydrates to regulate biological processes (55). Previous studies showed that several JRLs were up-regulated significantly during insect defense and it was suggested that binding with foreign glycans might be important for the defense process (56). Other research has indicated that JRLs might regulate the signal transduction by modulating plant protein-carbohydrate interactions (57). For example, JLRs can regulate the size of Arabidopsis beta-glucosidase PYK10 complex antagonistically to retain the activity against pathogens instead of losing the enzyme activity (58). For the phytophagous insects, JRLs can bind with sugars on the gut epithelial cells and therefore inhibit nutrient uptake of insect (59). We hereby showed that a specific JRL induced by insect treatment can promote the rice defense against FAW larvae. However, the detailed mechanisms for this JRL to regulate plant defense need further investigation.
Os03g48770 belonged to the cupin super gene family and was identified by the PARC method only. The members of this superfamily have very diverse functions and their roles in plant disease defense have been studied in several plants species (19, 60, 61). In particular, one of the cupin superfamily members was proven to be an oxalate oxidase which produces H2O2 and can serve as an important signal in plant disease defense (62). In this study, we showed that insect treatment led to the overexpression of a member of rice cupin superfamily genes and the overexpression of this gene resulted in significantly improved defense against FAW. The further phylogenic analysis indicated that this particular cupin gene is a homology of a wheat oxalate oxidase, GER3 (63, 64) (supplemental Fig. S6A and B). Os03g48770 also shares 97% similarity with Os03g48780, which encodes an oxalate oxidase involved in ROS signaling (16). It will be interesting to further study if this gene is involved in rice defense against both insect and pathogen, and how the up-stream and down-stream regulation is involved. The Os03g48770 gene has the potential to be directly used in crop improvement. Even though the F1 transgenic lines have shown promising insect resistance for both genes, extensive research in homozygous T3 transgenic lines need to be carried out to validate if these genes can be used in engineering crop resistance to herbivorous insects.
Overall, PARC represented an effective method to improve the sensitivity, protein identification and differential expression characterization in shot-gun proteomics studies. The method was implemented to identify both novel mechanisms and important regulators for plant defense against herbivorous insects. The technique can be broadly applied to proteomics studies for new gene discovery and mechanism elucidation.
Supplementary Material
Footnotes
* This work is supported by Texas A&M University AgriLife.
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S6.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S6.
1 The abbreviations used are:
- MudPIT
- Multidimensional Protein Identification Technology
- PARC
- Polyethyleneimine-Assisted Ribulose-1,5-bisphosphate carboxylase oxygenase Cleanup
- ROS
- Reactive oxygen species
- FAW
- fall armyworm
- GO
- Gene Ontology
- OEM
- organelle enrichment method
- OXO
- oxalate oxidase
- JRLs
- jacalin-related lectins.
REFERENCES
- 1. Wagner K., Miliotis T., Marko-Varga G., Bischoff R., Unger K. K. (2002) An automated on-line multidimensional HPLC system for protein and peptide mapping with integrated sample preparation. Anal. Chem. 74, 809–820 [DOI] [PubMed] [Google Scholar]
- 2. Rose J. K., Bashir S., Giovannoni J. J., Jahn M. M., Saravanan R. S. (2004) Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J. 39, 715–733 [DOI] [PubMed] [Google Scholar]
- 3. Widjaja I., Naumann K., Roth U., Wolf N., Mackey D., Dangl J. L., Scheel D., Lee J. (2009) Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics 9, 138–147 [DOI] [PubMed] [Google Scholar]
- 4. Krishnan H. B., Natarajan S. S. (2009) A rapid method for depletion of Rubisco from soybean (Glycine max) leaf for proteomic analysis of lower abundance proteins. Phytochemistry 70, 1958–1964 [DOI] [PubMed] [Google Scholar]
- 5. Atkinson A., Jack G. W. (1973) Precipitation of nucleic acids with polyethyleneimine and the chromatography of nucleic acids and proteins on immobilised polyethyleneimine. Biochim. Biophys. Acta 308, 41–52 [DOI] [PubMed] [Google Scholar]
- 6. Holler C., Zhang C. M. (2008) Purification of an acidic recombinant protein from transgenic tobacco. Biotechnol. Bioeng. 99, 902–909 [DOI] [PubMed] [Google Scholar]
- 7. Jorrín-Novo J. V., Maldonado A. M., Echevarria-Zomeño S., Valledor L., Castillejo M. A., Curto M., Valero J., Sghaier B., Donoso G., Redondo I. (2009) Plant proteomics update (2007–2008): Second-generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J. Proteomics 72, 285–314 [DOI] [PubMed] [Google Scholar]
- 8. Yuan J. S., Galbraith D. W., Dai S. Y., Griffin P., Stewart C. N., Jr. (2008) Plant systems biology comes of age. Trends Plant Sci. 13, 165–171 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Gao P., Yuan J. S. (2010) Plant protein-protein interaction network and interactome. Curr. Genomics 11, 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lippert D., Chowrira S., Ralph S. G., Zhuang J., Aeschliman D., Ritland C., Ritland K., Bohlmann J. (2007) Conifer defense against insects: Proteome analysis of Sitka spruce (Picea sitchensis) bark induced by mechanical wounding or feeding by white pine weevils (Pissodes strobi). Proteomics 7, 248–270 [DOI] [PubMed] [Google Scholar]
- 11. Zhang J. H., Sun L. W., Liu L. L., Lian J., An S. L., Wang X., Zhang J., Jin J. L., Li S. Y., Xi J. H. (2010) Proteomic Analysis of Interactions Between the Generalist Herbivore Spodoptera exigua (Lepidoptera: Noctuidae) and Arabidopsis thaliana. Plant Mol. Biol. Rep. 28, 324–333 [Google Scholar]
- 12. George D., Babalola O. O., Gatehouse A. M. R. (2011) Differential protein expression in maize (Zea mays) in response to insect attack. Afr. J .Biotechnol. 10, 7700–7709 [Google Scholar]
- 13. Ferry N., Stavroulakis S., Guan W., Davison G. M., Bell H. A., Weaver R. J., Down R. E., Gatehouse J. A., Gatehouse A. M. (2011) Molecular interactions between wheat and cereal aphid (Sitobion avenae): Analysis of changes to the wheat proteome. Proteomics 11, 1985–2002 [DOI] [PubMed] [Google Scholar]
- 14. Howe G. A., Jander G. (2008) Plant immunity to insect herbivores. Annu. Rev. Plant Biol . 59, 41–66 [DOI] [PubMed] [Google Scholar]
- 15. Yuan J. S., Köllner T. G., Wiggins G., Grant J., Degenhardt J., Chen F. (2008) Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. Plant J. 55, 491–503 [DOI] [PubMed] [Google Scholar]
- 16. Carrillo M. G. C., Goodwin P. H., Leach J. E., Leung H., Cruz C. M. V. (2009) Phylogenomic relationships of rice oxalate oxidases to the cupin superfamily and their association with disease resistance QTL. Rice 2, 67–79 [Google Scholar]
- 17. Banerjee J., Maiti M. K. (2010) Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem. Bioph. Res. Co. 394, 178–183 [DOI] [PubMed] [Google Scholar]
- 18. Davidson R. M., Reeves P. A., Manosalva P. M., Leach J. E. (2009) Germins: A diverse protein family important for crop improvement. Plant Sci. 177, 499–510 [Google Scholar]
- 19. Lou Y., Baldwin I. T. (2006) Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol. 140, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Méchin V., Damerval C., Zivy M. (2007) Total protein extraction with TCA-acetone. Methods Mol. Biol. 355, 1–8 [DOI] [PubMed] [Google Scholar]
- 21. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protocols 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 22. Loewen P. C., Didychuk A. L., Switala J., Perez-Luque R., Fita I., Loewen M. C. (2013) Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate. Acta Crystallogr. 69, 10–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., Liu S., Dai S., Yuan J. (2012) Integration of shot-gun proteomics and bioinformatics analysis to explore plant hormone responses. Bmc Bioinformatics 13, S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu T., Venable J., Park S., Cociorva D., Lu B., Liao L., Wohlschlegel J., Hewel J., Yates III J. (2006) ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell. Proteomics 5, S174 [Google Scholar]
- 26. Carvalho P. C., Fischer J. S., Chen E. I., Yates J. R., 3rd, Barbosa V. C. (2008) PatternLab for proteomics: a tool for differential shotgun proteomics. Bmc Bioinformatics 9, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M. (2002) The KEGG database. Novart. Fdn. Symp. 247, 91–103 [PubMed] [Google Scholar]
- 29. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 30. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Portillo M., Fenoll C., Escobar C. (2006) Evaluation of different RNA extraction methods for small quantities of plant tissue: Combined effects of reagent type and homogenization procedure on RNA quality-integrity and yield. Physiol. Plantarum 128, 1–7 [Google Scholar]
- 32. Chen S., Songkumarn P., Liu J., Wang G. L. (2009) A Versatile Zero Background T-Vector System for Gene Cloning and Functional Genomics. Plant Physiol. 150, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toki S., Hara N., Ono K., Onodera H., Tagiri A., Oka S., Tanaka H. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976 [DOI] [PubMed] [Google Scholar]
- 34. Baviskar S. N. (2011) A Quick & Automated Method for Measuring Cell Area Using ImageJ to measure microbes. Am. Biol. Teach. 73, 554–556 [Google Scholar]
- 35. Venu R., Madhav M. S., Sreerekha M., Nobuta K., Zhang Y., Carswell P., Boehm M. J., Meyers B. C., Korth K. L., Wang G.-L. (2010) Deep and comparative transcriptome analysis of rice plants infested by the beet armyworm (Spodoptera exigua) and water weevil (Lissorhoptrus oryzophilus). Rice 3, 22–35 [Google Scholar]
- 36. Xi J., Wang X., Li S., Zhou X., Yue L., Fan J., Hao D. (2006) Polyethylene glycol fractionation improved detection of low-abundant proteins by two-dimensional electrophoresis analysis of plant proteome. Phytochemistry 67, 2341–2348 [DOI] [PubMed] [Google Scholar]
- 37. Peltier J. B., Ripoll D. R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K. J. (2004) Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279, 4768–4781 [DOI] [PubMed] [Google Scholar]
- 38. Hashimoto M., Komatsu S. (2007) Proteomic analysis of rice seedlings during cold stress. Proteomics 7, 1293–1302 [DOI] [PubMed] [Google Scholar]
- 39. Alam I., Sharmin S., Kim K. H., Kim Y. G., Lee J., Lee B. H. (2013) An improved plant leaf protein extraction method for high resolution two-dimensional polyacrylamide gel electrophoresis and comparative proteomics. Biotech. Histochem. 88, 61–75 [DOI] [PubMed] [Google Scholar]
- 40. Gammulla C. G., Pascovici D., Atwell B. J., Haynes P. A. (2011) Differential proteomic response of rice (Oryza sativa) leaves exposed to high- and low-temperature stress. Proteomics 11, 2839–2850 [DOI] [PubMed] [Google Scholar]
- 41. Mirzaei M., Soltani N., Sarhadi E., Pascovici D., Keighley T., Salekdeh G. H., Haynes P. A., Atwell B. J. (2012) Shotgun Proteomic Analysis of Long-distance Drought Signaling in Rice Roots. J. Proteome Res. 11, 348–358 [DOI] [PubMed] [Google Scholar]
- 42. Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci .U.S.A. 105, 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mithöfer A., Boland W. (2012) Plant Defense Against Herbivores: Chemical Aspects. Annual Review of Plant Biology 63, 431–450 [DOI] [PubMed] [Google Scholar]
- 44. Singh V., Louis J., Ayre B. G., Reese J. C., Pegadaraju V., Shah J. (2011) TREHALOSE PHOSPHATE SYNTHASE11-dependent trehalose metabolism promotes Arabidopsis thaliana defense against the phloem-feeding insect Myzus persicae. Plant J. 67, 94–104 [DOI] [PubMed] [Google Scholar]
- 45. Steinbrenner A. D., Gómez S., Osorio S., Fernie A. R., Orians C. M. (2011) Herbivore-Induced Changes in Tomato (Solanum lycopersicum) Primary Metabolism: A Whole Plant Perspective. J. Chem. Ecol. 37, 1294–1303 [DOI] [PubMed] [Google Scholar]
- 46. Uefune M., Kugimiya S., Sano K., Takabayashi J. (2012) Herbivore-induced plant volatiles enhance the ability of parasitic wasps to find hosts on a plant. J. Appl. Entomol. 136, 133–138 [Google Scholar]
- 47. Clancy K. M. (1992) The role of sugars in Western spruce budworm nutritional ecology. Ecol. Entomol. 17, 189–197 [Google Scholar]
- 48. Awmack C. S., Leather S. R. (2002) Host plant quality and fecundity in herbivorous insects. Annu. Rev. Entomol. 47, 817–844 [DOI] [PubMed] [Google Scholar]
- 49. Moghaddam M. R., Van den Ende W. (2012) Sugars and plant innate immunity. J. Exp. Bot. 63, 3989–3998 [DOI] [PubMed] [Google Scholar]
- 50. Gómez-Ariza J., Campo S., Rufat M., Estopa M., Messeguer J., San Segundo B., Coca M. (2007) Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Mol. Plant Microbe Interact 20, 832–842 [DOI] [PubMed] [Google Scholar]
- 51. Singh V., Shah J. (2012) Tomato responds to green peach aphid infestation with the activation of trehalose metabolism and starch accumulation. Plant Signal. Behav. 7, 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hao P., Liu C., Wang Y., Chen R., Tang M., Du B., Zhu L., He G. (2008) Herbivore-induced callose deposition on the sieve plates of rice: An important mechanism for host resistance. Plant Physiol. 146, 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmidt L., Schurr U., Röse U. S. R. (2009) Local and systemic effects of two herbivores with different feeding mechanisms on primary metabolism of cotton leaves. Plant Cell Environ. 32, 893–903 [DOI] [PubMed] [Google Scholar]
- 54. Heil M., Ibarra-Laclette E., Adame-Álvarez R. M., Martinez O., Ramirez-Chávez E., Molina-Torres J., Herrera-Estrella L. (2012) How Plants Sense Wounds: Damaged-Self Recognition Is Based on Plant-Derived Elicitors and Induces Octadecanoid Signaling. Plos One 7, e30537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sharon N., Lis H. (2004) History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology 14, 53R–62R [DOI] [PubMed] [Google Scholar]
- 56. Jiang S. Y., Ma Z., Ramachandran S. (2010) Evolutionary history and stress regulation of the lectin superfamily in higher plants. Bmc Evol. Biol. 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Van Damme E. J., Barre A., Rougé P., Peumans W. J. (2004) Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci. 9, 484–489 [DOI] [PubMed] [Google Scholar]
- 58. Nagano A. J., Fukao Y., Fujiwara M., Nishimura M., Hara-Nishimura I. (2008) Antagonistic jacalin-related lectins regulate the size of ER body-type beta-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol. 49, 969–980 [DOI] [PubMed] [Google Scholar]
- 59. Vasconcelos I. M., Oliveira J. T. A. (2004) Antinutritional properties of plant lectins. Toxicon 44, 385–403 [DOI] [PubMed] [Google Scholar]
- 60. Dunwell J. M., Khuri S., Gane P. J. (2000) Microbial relatives of the seed storage proteins of higher plants: Conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dunwell J. M., Gibbings J. G., Mahmood T., Naqvi S. M. S. (2008) Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 27, 342–375 [Google Scholar]
- 62. Berna A., Bernier F. (1999) Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol. Biol. 39, 539–549 [DOI] [PubMed] [Google Scholar]
- 63. Lane B. G., Dunwell J. M., Ray J. A., Schmitt M. R., Cuming A. C. (1993) Germin, a Protein Marker of Early Plant Development, Is an Oxalate Oxidase. J. Biol. Chem. 268, 12239–12242 [PubMed] [Google Scholar]
- 64. Ramputh A. I., Arnason J. T., Cass L., Simmonds J. A. (2002) Reduced herbivory of the European corn borer (Ostrinia nubilalis) on corn transformed with germin, a wheat oxalate oxidase gene. Plant Sci. 162, 431–440 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.