Abstract
Recently, we have disclosed that human KIAA1199 (hKIAA1199) is a hyaluronan (HA) binding protein implicated in HA depolymerization. Although a murine homologue (mKiaa1199) was previously cloned, no information about the function of the molecule was available. Here, we show that cells transfected with mKiaa1199 cDNA selectively catabolized HA via the clathrin-coated pit pathway. A glycosaminoglycan-binding assay demonstrated the specific binding of mKiaa1199 to HA. These results were similar to our observations with hKIAA1199, although slight differences were found in the peak sizes of the minimum degradates of HA. We conclude that like hKIAA1199, mKiaa1199 is a hyaladherin, leading to HA depolymerization.
Keywords: KIAA1199, Hyaluronan, Catabolism, Extracellular matrix
Abbreviations: HA, hyaluronan; GAG, glycosaminoglycan; HYAL, hyaluronidase; ORF, open reading frame; HEK, human embryonic kidney; FA, fluoresceinamine; CS, chondroitin sulfate; DS, dermatan sulfate; Hep, heparin; HS, heparan sulfate; GlcNAc, N-acetylglucosamine; HPLC, high performance liquid chromatography; CHC, clathrin heavy chain; siRNA, short interfering RNA; FBS, fetal bovine serum; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PCR, polymerase chain reaction; LDS, lithium dodecyl sulfate; CPC, cetylpyridinium chloride; PBS, phosphate-buffered saline; PBS-T, PBS containing 0.05% Tween 20
Highlights
-
•
Cells expressing murine Kiaa1199 (mKiaa1199) selectively digest hyaluronan (HA).
-
•
mKiaa1199 has the capability of binding specifically to HA.
-
•
mKiaa1199-mediated HA depolymerization involves the clathrin-coated pit pathway.
-
•
Like human KIAA1199, mKiaa1199 is a hyaladherin that takes part in HA catabolism.
1. Introduction
Hyaluronan (HA), a major component of the extracellular matrix, is ubiquitously present in all vertebrate animals including humans. HA is a unique linear glycosaminoglycan (GAG) composed of only two sugars, i.e. β(1,3)-linked-d-glucuronic acid and β(1,4)-linked-N-acetyl-d-glucosamine, and it provides structural and functional integrity within the tissue microenvironment to both cells and organs. HA is rapidly depolymerized under physiological conditions from extra-large native molecules to intermediate-size fragments in the extracellular milieu [1]. HA degradation is enhanced under certain pathological conditions, and its lower-molecular-weight products are commonly detected in diseases such as arthritis and cancers [2,3]. Two hyaluronidases (HYAL1 and HYAL2) and a cell surface HA receptor (CD44) were believed to have the key roles in HA degradation [4,5]. Recently, however, we have demonstrated that human KIAA1199 (hKIAA1199), what first had been identified as a deafness gene of unknown function, is clearly capable of binding HA and takes part in HA catabolism independently of the CD44 and HYAL enzymes in the dermis of healthy skin and the synovium of arthritis patients [6].
The hKIAA1199 protein has no substantial homology to HYAL enzymes, HA-binding proteins, or other molecules [5], lacking both HA-link modules and B(X7)B HA-binding motifs [7,8], although it has two GG domains, one G8 domain expected to take part in extracellular ligand binding, and four PbH1 domains believed to be involved in polysaccharide hydrolysis [9–11]. hKIAA1199 is reportedly expressed in a wide range of normal human tissues including brain [12]. Our previous study showed that hKIAA1199 is essential for endogenous HA degradation in human skin fibroblasts, and that cells transfected with hKIAA1199 cDNA degrade HA through specific binding with HA [6]. We have also demonstrated that hKIAA1199 is expressed by dermal fibroblasts in normal skin and over-expressed by synovial fibroblasts and tissues from arthritic joints [6]. All these data suggest that KIAA1199 plays a role in HA catabolism under certain physiological and pathological conditions in humans, although direct evidence on functional analyses of the molecule within tissues is still required. A murine homologue (mKiaa1199) of hKIAA1199 has been cloned and it is expressed in mouse tissues such as the inner ear [13]. However, no data about functions of the murine molecule have been provided. In the current study, therefore, we tried to analyze functional activity of mKiaa1199 by expressing this gene in HEK293 cells, and demonstrated for the first time that mKiaa1199 is a hyaladherin that takes part in HA depolymerization in a manner similar to hKIAA1199.
2. Materials and methods
2.1. Cell cultures
HEK293 cell line (DS Pharma Biomedical) was maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% (vol/vol) FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. The cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
2.2. Assay for cellular [3H]HA depolymerization
High-molecular-weight [3H]-labeled HA of >1000 kDa ([3H]HA) was prepared as described previously [6]. Cellular HA depolymerization was assayed by culturing confluent cells in medium containing [3H]HA (40,000 dpm/ml) and by applying the media to a Sepharose CL-2B (GE Healthcare) column (1 × 60 cm) equilibrated with 0.5 M NaCl in distilled water. The radioactivity of each fraction was measured by a scintillation counter (Aloka LSC-6100). The column was calibrated with fluoresceinamine-labeled HA (FA-HA): FA-HA H1 (1760 kDa), M1 (907 kDa), L1 (197 kDa), S1 (56 kDa), T1 (28 kDa) and U1 (9.8 kDa) (peak top kDa), all of which were purchased from PG Research. For detection of FA, an excitation wavelength of 490 nm and an emission wavelength of 525 nm were used.
2.3. Antibodies
Rat monoclonal antibody against hKIAA1199 was previously developed using a peptide of CA762RYSPHQDADPLKPRE777, which corresponds to amino acid residues Ala762 to Glu777 of hKIAA1199 (GenBank Accession No. NM_018689) [6]. Since mKiaa1199 has the same amino acid sequence in the molecule (GenBank Accession No. AB_103331), this antibody specifically recognized both hKIAA1199 and mKiaa1199. Antibodies against clathrin heavy chain (CHC), α-adaptin, caveolin-1 and GAPDH were purchased from Santa Cruz Biotechnology.
2.4. Immunoblotting
Cell homogenate supernatants were separated by electrophoresis on NuPAGE 4–12% Bis–Tris gels (Invitrogen) and proteins were transferred onto polyvinylidene difluoride membranes. The membranes were reacted with the antibodies specific to KIAA1199, CHC, α-adaptin, caveolin-1 or GAPDH. Then, they were incubated with horseradish peroxidase-conjugated secondary antibodies: donkey anti-rat IgG antibody for KIAA1199 (Jackson ImmunoResearch); goat anti-rabbit IgG antibody for caveolin-1 and GAPDH (DAKO); rabbit anti-mouse IgG antibody for α-adaptin (DAKO); and rabbit anti-goat IgG antibody for CHC (DAKO). Immunoreactive bands were detected by SuperSignal West Pico Chemiluminescent Substrate system (Thermo Scientific).
2.5. Preparations of plasmids and transfectants
The cDNA of mKiaa1199 was amplified by PCR using brain cDNA template (Takara Bio). The authenticity of the cDNA was verified by sequencing using Applied Biosystems 3730xl DNA Analyzer (Life Technologies). Plasmid was prepared by inserting the cDNA of mKiaa1199 into the expression vector pcDNA3.1(−) (Invitrogen) according to the manufacture's protocol. Transient transfection was done using Lipofectamine LTX (Invitrogen), and transfectants were used for the experiments at 48 h after transfection. Stable transfectants of mKiaa1199 in HEK293 cells (mKiaa1199/HEK293 cells) were prepared by transfection with the pcDNA3.1(−) vectors containing the mKiaa1199 cDNA and selection by culturing in medium containing 800 μg/ml of G418 (Sigma). The expression and activity of mKiaa1199 were monitored by immunoblotting and by the HA-degrading assay, respectively. The pcDNA3.1(−) vectors containing the hKIAA1199 cDNA and stable transfectants of hKIAA1199 in HEK293 cells (hKIAA1199/HEK293 cells), both of which were previously prepared [6], were used as positive controls.
2.6. Identification of non-reducing terminal sugar of depolymerized HA
mKiaa1199/HEK293 cells were cultured in medium containing [3H]HA (1,000,000 dpm/ml) for 72 h, and radiolabeled products in the medium were detected on a Sepharose CL-2B column. Depolymerized [3H]HA in the fractions was precipitated with ethanol, dissolved in distilled water, and then fractionated on a column (1 × 107 cm) of Sephadex G-25 (GE Healthcare) after digestion with β-N-acetylglucosaminidase (Sigma) or sequential digestion with β-glucuronidase (Sigma) and β-N-acetylglucosaminidase. The elution position of N-acetylglucosamine (GlcNAc) used as a standard was determined according to previous methods [14].
2.7. Analysis of HA-specific depolymerization
mKiaa1199/HEK239 cells were cultured in medium containing FA-GAGs: FA-HAs (H1, M1, L1, S1, T1 and U1), chondroitin sulfate A, C and D (FA-CSA, FA-CSC and FA-CSD), dermatan sulfate (FA-DS), heparin (FA-Hep), and heparan sulfate (FA-HS) (10 μg/ml each). All these FA-GAGs were purchased from PG Research. After a 72-h incubation period, the media were harvested and fractionated on a Sepharose CL-6B (GE Healthcare) column (1 × 35 cm) equilibrated with PBS. The amounts of FA-GAGs in each fraction were determined by fluorescence counting.
2.8. Determination of minimum HA size depolymerized from high-molecular-weight HA by mKiaa1199/HEK293 and hKIAA1199/HEK293 cells
mKiaa1199/HEK293 and hKIAA1199/HEK293 cells were cultured in the medium containing FA-HA H1 (10 μg/ml). After a 72-h culture, the media were applied to a TSK-GEL G5000PW (7.5 mm inner diameter × 30 cm, Tosoh Corporation) using a 0.05 M CAPS buffer, pH 10.0, containing 0.8 M NaNO3 at a flow rate of 0.5 ml/min at 30 °C, and FA-HA was detected by fluorescence counting. Size of the fragments was determined using FA-HAs including FA-HA H1, M1, L1, S1, T1, U1 and 3K (2.6 kDa) (PG Research) under the same chromatographic condition.
2.9. Co-precipitation of mKiaa1199 protein with GAGs
mKiaa1199/HEK293 cells were homogenized in 50 mM phosphate buffer, pH 6.0 containing a cocktail of proteinase inhibitors (Roche Diagnostics). Aliquots of the homogenate supernatant (50 μl) were mixed with 50 μl of 1 mg/ml aqueous solutions of non-labeled GAGs: CSA, CSC, CSD, DS, Hep, HS, HA-H2 (1452 kDa), HA-M2 (1039 kDa), HA-L2 (219 kDa), HA-S2 (52 kDa) and HA-T2 (28 kDa) (PG Research) or H2O (negative control). They were incubated for 1 h at 37 °C, and 1% cetylpyridinium chloride (CPC) (final concentration; wt/vol) was added to the GAG/lysate mixtures. The precipitates were electrophoresed on NuPAGE 4–12% Bis–Tris gels (Invitrogen) under reducing conditions and immunoblotted with anti-KIAA1199 monoclonal antibody.
2.10. RNA interference
Two different siRNAs for CHC, caveolin-1 and α-adaptin, and non-silencing control siRNAs were purchased from Invitrogen. The sequences of the siRNAs are described previously [6]. siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen).
2.11. Immunofluorescence microscopy for mKiaa1199 in mKiaa1199/HEK293 cells
For double immunostaining of mKiaa1199 and CHC, mKiaa1199/HEK293 cells were grown on glass multi-chamber slides (BD Biosciences) to 70–80% confluence, and fixed with 4% (wt/vol) paraformaldehyde in PBS. After washing in PBS containing 0.05% (vol/vol) Tween 20 (PBS-T), the cells were reacted with rat anti-KIAA1199 monoclonal antibody conjugated to Alexa-Flour 488 and goat anti-CHC antibody conjugated to Alexa-Flour 555. The samples were counterstained with TO-PRO-3 (Invitrogen) and mounted in vectashield (Vector). As for controls, samples were reacted with non-immune IgG conjugated to Alexa-Flours. These samples were observed using Zeiss LSM 510 confocal microscope (Carl Zeiss).
2.12. Cellular distribution of exogenously added HA in mKiaa1199/HEK293 cells
The cells on glass multi-chamber slides were incubated in the presence or absence of 0.1 mg/ml biotin-labeled high-molecular-weight HA of 1410 kDa (PG research) at 37 °C for 1 h, and then fixed with 4% (wt/vol) paraformaldehyde in PBS. After washing in PBS-T, incubation with streptavidin conjugated to Alexa-Fluor 488 (Invitrogen), and nuclear counterstaining with TO-PRO-3, they were observed using Zeiss LSM 510 confocal microscope. As for a control, the cells were incubated with biotin-labeled high-molecular-weight HA digested with Streptomyces hyaluronidase (Seikagaku Corporation), followed by the immunostaining described above.
3. Results and discussion
3.1. Cells transfected with mKiaa1199 cDNA acquire HA-degrading capability
The overall homology of the coding regions between mKiaa1199 and hKIAA1199 proteins was 91% identical, and four PbH1 domains, which may be involved in polysaccharide hydrolysis [10,11], are completely conserved (Supplementary Fig. 1). To examine the implication of mKiaa1199 in HA depolymerization, we transfected HEK293 cells, a cell line with no HA depolymerizing activity, with a full-length mKiaa1199 cDNA. As shown in Fig. 1A, mKiaa1199 protein was observed at exactly the same position as hKIAA1199 of 150 kDa by immunoblotting. The mKiaa1199 transfectants depolymerized exogenous [3H]HA to intermediate-size fragments, and released the catabolites into the medium, whereas mock transfectants showed negligible HA depolymerization (Fig. 1A). The mKiaa1199-mediated depolymerization of HA seems to be independent of HYAL1 and HYAL2, as both molecules were expressed at negligible levels in HEK293 cells [6]. The non-reducing terminal sugar of the fragments was determined to be glucuronic acid by incubation of the depolymerized [3H]HA with β-N-acetylglucosaminidase or β-glucuronidase followed by β-N-acetylglucosaminidase (Fig. 1B). This finding indicates that HA is cleaved at the β-endo-N-acetylglucosamine bonds. Taken together, the evidence suggests that cells which overexpress mKiaa1199 depolymerize HA to intermediate-size fragments in an endo-β-N-acetylglucosaminidase-dependent manner and accumulate the catabolites extracellularly. These observations are consistent with the results previously reported with hKIAA1199 [6].
Fig. 1.
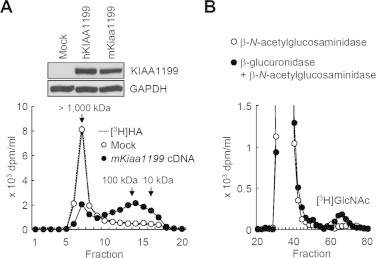
HA depolymerization by mKiaa1199 transfectants and determination of HA cleavage sites. (A) HEK293 cells were transiently transfected with empty vector (Mock) or vector containing hKIAA1199 or mKiaa1199 cDNA. The expression of hKIAA1199 and mKiaa1199 proteins in each transfectant was assessed by immunoblotting using anti-KIAA1199 monoclonal antibody (inset). Mock and mKiaa1199 transfectants were incubated with [3H]HA for 48 h, and HA depolymerization was then examined by size exclusion chromatography. GAPDH, a loading control. (B) Determination of the non-reducing terminal sugar of depolymerized HA. [3H]HA fragments obtained from the culture media were incubated with β-N-acetylglucosaminidase (open circles) or β-glucuronidase followed by incubation with β-N-acetylglucosaminidase (closed circles), and then applied to Sephadex G-25 column chromatogram.
3.2. Stable transfectants of mKiaa1199 in HEK293 cells selectively bind and degrade HA species of different molecular weights
We first prepared stable transfectants of mKiaa1199 in HEK293 cells (mKiaa1199/HEK293 cells), and examined the activity of mKiaa1199 for binding to and depolymerization of HA. When mKiaa1199 protein isolated from the transfectants were incubated with HA or other GAGs (CSA, CSC, CSD, DS, Hep, and HS), and then precipitated with CPC, mKiaa1199 was co-precipitated with HA, whereas the other GAGs showed negligible precipitates (Fig. 2A). In addition, we observed co-precipitation of mKiaa1199 with various HA species with different molecular weights (HA-H2, 1452 kDa; HA-M2, 1039 kDa; HA-L2, 219 kDa; HA-S2, 52 kDa; HA-T2, 28 kDa) (Fig. 2B). Then, we further studied degradation of FA-labeled HA (FA-HA) added to mKiaa1199/HEK293 cells. As shown in Fig. 3A, the cells selectively digested several FA-HA species with different molecular weights, i.e. FA-HA H1 (1760 kDa), M1 (907 kDa), L1 (197 kDa), S1 (56 kDa), T1 (28 kDa) and U1 (9.8 kDa) (peak top kDa), into fragments of a constant size, whereas they showed no digestion of other FA-GAGs (CSA, CSC, CSD, DS, Hep, and HS). Meanwhile, no HA-degrading activity was detected in cell lysates from mKiaa1199/HEK293 cells (data not shown). All these data on HA specific binding and depolymerization of mKiaa1199 match our previous findings with hKIAA1199 cells [6]. Interestingly, however, the peak size of minimum degradates depolymerized from FA-HA H1 by mKiaa1199/HEK293 cells (about 4.1 kDa, corresponding to ten disaccharide units) differed slightly from the peak size obtained by hKIAA1199/HEK293 cells under the same condition (about 3.3 kDa, corresponding to eight disaccharide units) (Fig. 3B and C). In particular, a low-molecular-weight shoulder (<2 kDa) was observed in the elution profile of the fragments depolymerized by mKiaa1199 (Fig. 3B). To examine how the mKiaa1199 expression levels affected the sizes of the HA fragments, we performed a time course digestion of HA by stable transfectants of mKiaa1199 with maximal mKiaa1199 expression (mKiaa1199/HEK293) or low mKiaa1199 expression (mKiaa1199/HEK293-L) (Supplementary Fig. 2A) and then measured the fragment sizes. As shown from Supplementary Fig. 2B and C, the peak size of the HA degradates from the mKiaa1199/HEK293-L cells (about 17.9 kDa) was larger than the peak size of the HA degradates from the mKiaa1199/HEK293 cells (about 4.1 kDa). It thus appears that mKiaa1199 expression levels may affect the sizes of the degradates. We were unable, however, to rule out the possibility that the degradates from the mKiaa1199/HEK293-L cells were not end products. This possibility must be expressly allowed, given that mKiaa1199/HEK293-L cells failed to completely degrade the high-molecular-weight HA after the 72 h incubation in our time course experiment. Although the reason why the expression of mKiaa1199 results in the HA degradation product with slightly different molecular size compared to that by hKIAA1199 is not clear, it may be interesting to speculate that this is due to less sequence homology of mouse G8 domain (82% to that of hKIAA1199), which may be involved in extracellular ligand binding [10].
Fig. 2.
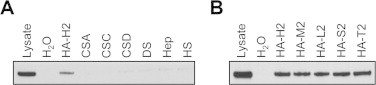
HA-specific binding of recombinant mKiaa1199 protein expressed in mKiaa1199/HEK293 cells. (A) Binding assay of mKiaa1199 protein to different GAGs. Lysates of mKiaa1199/HEK293 cells were incubated with H2O (negative control) or unlabeled GAGs including HA (HA-H2), CSA, CSC, CSD, DS, Hep and HS. The samples were precipitated with CPC, and analyzed by immunoblotting with anti-KIAA1199 antibody. (B) Binding of mKiaa1199 to HA species with different molecular sizes. The lysates were incubated with H2O (negative control) or HA with different sizes including HA-H2, HA-M2, HA-L2, HA-S2 and HA-T2, and then subjected to immunoblotting with anti-KIAA1199 antibody.
Fig. 3.
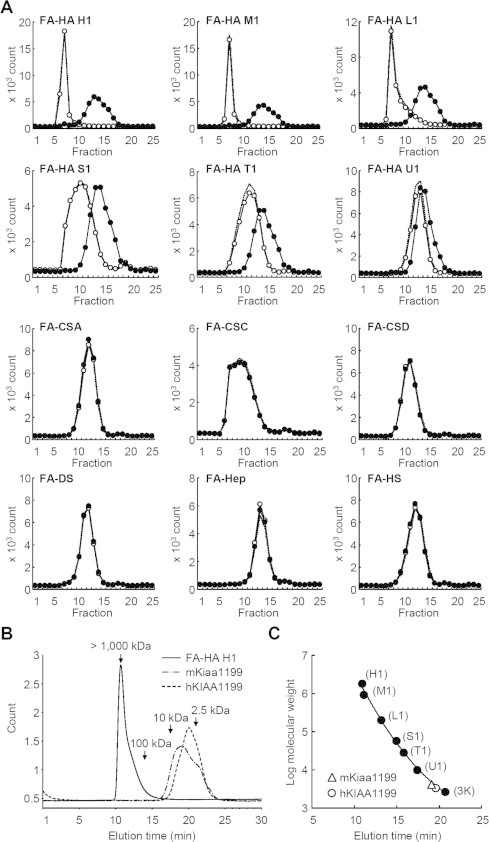
HA-specific depolymerization by mKiaa1199/HEK293 cells, and peak size of the minimum degradates. (A) Depolymerization of HA with different molecular sizes by mKiaa1199/HEK293 cells. Control HEK293 (open circles) and mKiaa1199/HEK293 cells (closed circles) were cultured for 72 h with HA species with different molecular weights (FA-HA H1, M1, L1, S1, T1 or U1) or FA-GAGs other than HA (FA-CSA, FA-CSC, FA-CSD, FA-DS, FA-Hep and FA-HS). Depolymerization of these GAGs in the media was determined by chromatography on a Sepharose CL-6B column. Dotted lines indicate GAG without incubation. (B) HPLC pattern of minimum degradates depolymerized from FA-HA H1 by mKiaa1199/HEK293 (chain line) and hKIAA1199/HEK293 (dotted line) cells. The solid line indicates FA-HA H1 without incubation. (C) Peak sizes of minimum fragments from FA-HA H1 depolymerized by mKiaa1199/HEK293 and hKIAA1199/HEK293 cells. Molecular weight of the fragments was determined using the following FA-HA species as a standard: FA-HA H1, M1, L1, S1, T1, U1 and 3K.
3.3. HA is catabolized by mKiaa1199 via the clathrin-coated pit pathway
Since our earlier study suggested the implication of the clathrin-coated vesicles or early endosomes in hKIAA1199-mediated HA depolymerization [6], we assessed the possible involvement of the clathrin-coated pit pathway in mKiaa1199-mediated HA degradation. As shown in Fig. 4A and B, HA degradation was reduced in mKiaa1199/HEK293 cells when the expression of CHC and α-adaptin subunit of AP-2, an adaptor protein complex functioning as a major organizer of clathrin coats, was knocked down by siRNAs. In contrast, the knockdown of caveolin-1, a protein involved in the caveolae pathway, caused no changes in mKiaa1199-mediated HA degradation (Fig. 4C). Immunohistochemistry revealed the localization of mKiaa1199 in vesicles in peripheral regions occasionally close to CHC-positive vesicles (Fig. 4D), but no fluorescence was observed by non-immune IgG (data not shown). High-molecular-weight HA added to the mKiaa1199/HEK293 cells was shown in some vesicles in the cell periphery (Fig. E), but no fluorescence was detected by incubation with Streptomyces hyaluronidase-digested HA (data not shown). These results suggest that HA is endocytosed via clathrin-coated pits and degraded in vesicles in the periphery of mKiaa1199/HEK293 cells. We also confirmed that the levels of CHC and α-adaptin expression were essentially identical between mKiaa1199/HEK293 and hKIAA1199/HEK293 cells (Supplementary Fig. 3). This matching of expression patterns suggests that CHC and α-adaptin are unlikely to be involved in the size determination of the end products.
Fig. 4.
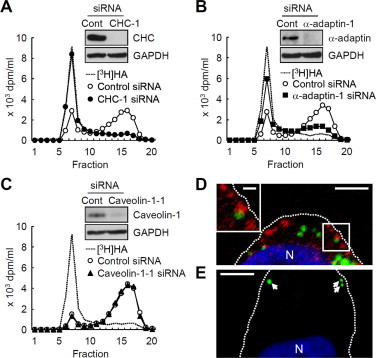
Clathrin-specific HA depolymerization and cellular localization of mKiaa1199 in mKiaa1199/HEK293 cells. (A–C) Changes in HA depolymerization in mKiaa1199/HEK293 cells by knockdown of CHC (A), α-adaptin (B) or caveolin-1 (C) with siRNAs. As for controls, cells were transfected with control non-silencing siRNA. After incubation of the cells with [3H]HA for 24 h, HA fragments in the media were analyzed by size exclusion chromatography. Efficiency of the knockdown was evaluated by immunoblotting (insets). Representative data from two siRNAs are shown. (D) Double immunostaining of mKiaa1199 (green) and CHC (red) with anti-KIAA1199 and anti-CHC antibodies in mKiaa1199/HEK293 cells. The inset shows a high-power view of the boxed area. Dotted lines show the plasma membrane. N, nucleus. Scale bar, 5 μm; scale bar for inset, 1 μm. (E) HA localization in mKiaa1199/HEK293 cells after incubation with biotin-labeled HA, followed by reaction with streptavidin conjugated to Alexa-Fluor 488. Arrows, HA-positive structures; N, nucleus. Scale bar, 5 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The current study provides the first evidence that like hKIAA1199, mKiaa1199 has the capability of binding specifically to HA, leading to HA depolymerization. The HA depolymerization by mKiaa1199 was almost identical to that by hKIAA1199, although slight differences were found in the elution profiles and peak sizes of the minimum degradates of HA. Note also that, like the cell lysates of hKIAA1199 [6], the cell lysates of stable transfectants of mKiaa1199 lacked HA depolymerizing activity. hKIAA1199 is reported to be widely expressed in human organs including the brain, skin, lung, testis and ovary, but it is notably absent in the liver [12]. We observed a similar tissue distribution of mRNA expression of mKiaa1199 by real-time PCR using commercially available mouse tissue RNA library (Okada et al., unpublished data). Mouse Hyal1 and Hyal2 are also broadly expressed in various mouse tissues, with peak expression in the liver and negligible expression in the brain [15,16]. The tissue expression patterns of these mouse molecules suggest that the mKiaa1199 and Cd44/Hyals systems may have different or cooperative roles in the physiological HA catabolism in mouse organs. Previous studies showed that mice deficient in the Hyal1 or Hyal2 gene exhibit no significant accumulation of HA locally within tissues [16,17]. These observations tempt us to speculate that mKiaa1199 might degrade HA in a compensatory fashion in these mice. KIAA1199 was originally reported as a candidate gene for the congenital or childhood-onset bilateral nonsyndromic sensorineural hearing loss [13]. In addition, our previous study demonstrated that two mutations at the Arg187 residue (R187C and R187H) out of the four reported mutations result in loss of the HA depolymerization [6]. Since mKiaa1199 is highly expressed in the mouse inner ear [13] and the Arg187 residue in the molecule is conserved in both humans and mice (Supplementary Fig. 1), the mutations at this residue in mKiaa1199 may be related to hearing loss in mice. hKIAA1199 is highly expressed in rheumatoid or osteoarthritic synovium [6], human cancer tissues such as carcinomas of the colorectum and stomach [12,18] and also skin fibroblasts from a patient with Werner syndrome, an adult-onset progeroid disease [12], although these studies on cancer tissues and Werner syndrome provided no data on the involvement in HA catabolism or the biological functions. Since the present study has disclosed that mKiaa1199 shares the functional activity with hKIAA1199, development of knockout and transgenic mice and disease models using the mice would be useful to study roles of this gene in physiological HA catabolism in vivo and investigate detailed pathogenesis of the KIAA1199-linked human diseases such as hearing loss, arthritis and cancers.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2013.08.003.
Appendix. Supplementary materials
Supplementary Fig. 1 Sequence alignment of the mouse and human KIAA1199 proteins. Identical amino acids and conserved Arg187 residue in mKiaa1199 and hKIAA1199 are shaded in yellow and red, respectively. Two GG domains and one G8 domain found in hKIAA1199 are underlined by solid and dashed lines, respectively. Four regions within boxes denote PbH1 domains (shaded in green). The alignment was generated by GENETYX (ver. 9).
Supplementary Fig. 2 Time course digestion of HA by stable transfectants with different expression levels of mKiaa1199, and the peak size of HA degradates by mKiaa1199/HEK293-L cells. (A) The expression of mKiaa1199 proteins in mKiaa1199 transfectant was assessed by immunoblotting using anti-KIAA1199 monoclonal antibody. GAPDH, loading control. mKiaa1199 transfectants were cultured in the presence of FA-HA H1 (1760 kDa) for 3, 6, 12, 24, 48 and 72 h. Conditioned media were harvested and FA-HA in the media were fractionated on a Sepharose CL-6B column. Dotted lines indicate FA-HA H1 without incubation. (B and C) HPLC pattern (B) and the peak size (C) of HA fragments from FA-HA H1 depolymerized by mKiaa1199/HEK293-L cells. The cells were incubated with FA-HA H1 for 72 h and the media were applied to a TSK-GEL G5000PW. The solid and dotted lines indicate FA-HA H1 without incubation and HA degradates by mKiaa1199/HEK293-L cells, respectively (B). The molecular weight of the HA degradates by mKiaa1199/HEK293-L cells was determined using the following FA-HA species as a standard: FA-HA H1, M1, L1, S1, T1, U1 and 3K (C).
Supplementary Fig. 3 Expressions of KIAA1199, CHC, and α-adaptin by mKiaa1199/HEK293 and hKIAA1199/HEK293 cells. The levels of KIAA1199, CHC, and α-adaptin protein expression in mKiaa1199/HEK293 and hKIAA1199/HEK293 cells were analyzed by immunoblotting. GAPDH, loading control.
References
- 1.Pandey M.S., Harris E.N., Weigel J.A. The cytoplasmic domain of the hyaluronan receptor for endocytosis (HARE) contains multiple endocytic motifs targeting coated pit-mediated internalization. J. Biol. Chem. 2008;283:21453–21461. doi: 10.1074/jbc.M800886200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh P. The role of hyaluronic acid (hyaluronan) in health and disease: interactions with cells, cartilage and components of synovial fluid. Clin. Exp. Rheumatol. 1994;12:75–82. [PubMed] [Google Scholar]
- 3.Sugahara K.N., Murai T., Nishinakamura H. Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J. Biol. Chem. 2003;278:32259–32265. doi: 10.1074/jbc.M300347200. [DOI] [PubMed] [Google Scholar]
- 4.Harada H., Takahashi M. CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J. Biol. Chem. 2007;282:5597–5607. doi: 10.1074/jbc.M608358200. [DOI] [PubMed] [Google Scholar]
- 5.Csoka A.B., Frost G.I., Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499–508. doi: 10.1016/s0945-053x(01)00172-x. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida H., Nagaoka A., Kusaka-Kikushima A. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc. Natl. Acad. Sci. USA. 2013;110:5612–5617. doi: 10.1073/pnas.1215432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohda D., Morton C.J., Parkar A.A. Solution structure of the link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell. 1996;86:767–775. doi: 10.1016/s0092-8674(00)80151-8. [DOI] [PubMed] [Google Scholar]
- 8.Yang B., Yang B.L., Savani R.C. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J. 1994;13:286–296. doi: 10.1002/j.1460-2075.1994.tb06261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J., Cheng H., Zhao S. GG: a domain involved in phage LTF apparatus and implicated in human MEB and non-syndromic hearing loss diseases. FEBS Lett. 2006;580:581–584. doi: 10.1016/j.febslet.2005.12.076. [DOI] [PubMed] [Google Scholar]
- 10.He Q.Y., Liu X.H., Li Q. G8: a novel domain associated with polycystic kidney disease and non-syndromic hearing loss. Bioinformatics. 2006;22:2189–2191. doi: 10.1093/bioinformatics/btl123. [DOI] [PubMed] [Google Scholar]
- 11.Birkenkamp-Demtroder K., Maghnouj A., Mansilla F. Repression of KIAA1199 attenuates Wnt-signalling and decreases the proliferation of colon cancer cells. Br. J. Cancer. 2011;105:552–561. doi: 10.1038/bjc.2011.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michishita E., Garces G., Barrett J.C. Upregulation of the KIAA1199 gene is associated with cellular mortality. Cancer Lett. 2006;239:71–77. doi: 10.1016/j.canlet.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 13.Abe S., Usami S., Nakamura Y. Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters’ cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J. Hum. Genet. 2003;48:564–570. doi: 10.1007/s10038-003-0079-2. [DOI] [PubMed] [Google Scholar]
- 14.Reissig J.L., Storminger J.L., Leloir L.F. A modified colorimetric method for the estimation of N-acetylamino sugars. J. Biol. Chem. 1955;217:959–966. [PubMed] [Google Scholar]
- 15.Csoka A.B., Scherer S.W., Stern R. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics. 1999;60:356–361. doi: 10.1006/geno.1999.5876. [DOI] [PubMed] [Google Scholar]
- 16.Jadin L., Wu X., Ding H. Skeletal and hematological anomalies in HYAL2-deficient mice: a second type of mucopolysaccharidosis IX? FASEB J. 2008;22:4316–4326. doi: 10.1096/fj.08-111997. [DOI] [PubMed] [Google Scholar]
- 17.Martin D.C., Atmuri A., Hemming R.J. A mouse model of human mucopolysaccharidosis IX exhibits osteoarthritis. Hum. Mol. Genet. 2008;17:1904–1915. doi: 10.1093/hmg/ddn088. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzaki S., Tanaka F., Mimori K. Clinicopathologic significance of KIAA1199 overexpression in human gastric cancer. Ann. Surg. Oncol. 2009;16:2042–2051. doi: 10.1245/s10434-009-0469-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Sequence alignment of the mouse and human KIAA1199 proteins. Identical amino acids and conserved Arg187 residue in mKiaa1199 and hKIAA1199 are shaded in yellow and red, respectively. Two GG domains and one G8 domain found in hKIAA1199 are underlined by solid and dashed lines, respectively. Four regions within boxes denote PbH1 domains (shaded in green). The alignment was generated by GENETYX (ver. 9).
Supplementary Fig. 2 Time course digestion of HA by stable transfectants with different expression levels of mKiaa1199, and the peak size of HA degradates by mKiaa1199/HEK293-L cells. (A) The expression of mKiaa1199 proteins in mKiaa1199 transfectant was assessed by immunoblotting using anti-KIAA1199 monoclonal antibody. GAPDH, loading control. mKiaa1199 transfectants were cultured in the presence of FA-HA H1 (1760 kDa) for 3, 6, 12, 24, 48 and 72 h. Conditioned media were harvested and FA-HA in the media were fractionated on a Sepharose CL-6B column. Dotted lines indicate FA-HA H1 without incubation. (B and C) HPLC pattern (B) and the peak size (C) of HA fragments from FA-HA H1 depolymerized by mKiaa1199/HEK293-L cells. The cells were incubated with FA-HA H1 for 72 h and the media were applied to a TSK-GEL G5000PW. The solid and dotted lines indicate FA-HA H1 without incubation and HA degradates by mKiaa1199/HEK293-L cells, respectively (B). The molecular weight of the HA degradates by mKiaa1199/HEK293-L cells was determined using the following FA-HA species as a standard: FA-HA H1, M1, L1, S1, T1, U1 and 3K (C).
Supplementary Fig. 3 Expressions of KIAA1199, CHC, and α-adaptin by mKiaa1199/HEK293 and hKIAA1199/HEK293 cells. The levels of KIAA1199, CHC, and α-adaptin protein expression in mKiaa1199/HEK293 and hKIAA1199/HEK293 cells were analyzed by immunoblotting. GAPDH, loading control.


