Abstract
The primary cilium, an organelle that transduces extracellular signals important for development and tissue homeostasis, is typically assembled upon cell cycle exit and disassembled upon cell cycle re-entry. Cilium assembly is thought to be suppressed in cycling cells, however the extent of suppression is not clear. For example, primary cilia are present in certain proliferating cells during development, and a period of reciliation has been reported to occur in late G1 in murine 3T3 cells released from serum starvation-induced quiescence. Human retinal pigmented epithelial (hTERT-RPE1; herein, RPE1) cells are commonly used to investigate pathways regulating cilium disassembly, however the ciliary disassembly profile of these cells remains uncertain. A period of reciliation has not been observed. Here, we analyse the ciliary disassembly profile of RPE1 cells by immunofluorescence microscopy. The results suggest a profile similar to 3T3 cells, including a period of reciliation in late G1 and a second wave of deciliation in S phase. We present evidence that arresting cells in early S phase with hydroxyurea or excess thymidine prevents the second wave of deciliation, and that deciliation is initiated shortly after release from a thymidine block, consistent with coupling to DNA replication. These findings support the often overlooked notion that cilium formation can occur in late G1, and suggest that RPE1 cells could serve as a model system for studying the molecular pathways that direct this process, in addition to those that stimulate cilium disassembly. We also present immunofluorescence data indicating that cyclin B1 localises to primary cilia.
Keywords: Primary cilia, Cyclin B1, CDK1, Aurora A, DNA replication, Cilium disassembly
Abbreviations: AurA, Aurora kinase A; BrdU, bromodeoxyuridine; CDK, cyclin-dependent kinase; DAPI, 4’,6-diamidino-2-phenylindole; FBS, fetal bovine serum; HU, hydroxyurea; Mim, mimosine; siRNA, short interfering RNA; SS, serum-starved; Thy, thymidine
Highlights
-
•
Ciliary disassembly profile of human RPE1 cells was analysed by immunofluorescence.
-
•
After release from serum-starvation, RPE1 cells re-assemble cilia in late G1 phase.
-
•
Cilium disassembly is not essential for S phase entry, but is coupled to DNA replication.
-
•
Cyclin B1 localises to primary cilia.
1. Introduction
The primary cilium is a membrane-bound microtubule-based organelle that projects from the cell surface and has key roles in transducing signals from the extracellular environment. Disruption of primary cilium structure or function is thought to underlie many of the clinical features of numerous human genetic disorders, reflecting both the widespread tissue distribution of this organelle and its involvement in signalling pathways required for normal development and tissue homeostasis [1]. Cilium assembly and disassembly are coordinated with the cell cycle by mechanisms that remain incompletely understood. These mechanisms are of considerable interest because of their importance for the cell's responsiveness to external signals, and because their deregulation could also disrupt mitosis [2].
Primary cilia are typically formed upon cell cycle exit and disassembled upon cell cycle re-entry [2]. Pathways that suppress ciliogenesis in cycling cells appear to exist [3–5]. However, the presence or formation of primary cilia is not restricted to quiescent or terminally differentiated cells [6]. For example, early studies using murine 3T3 fibroblasts revealed that, following release from serum starvation-induced quiescence and the accompanying rapid disassembly of cilia, a wave of reciliation occurred in late G1, followed by deciliation coupled to DNA synthesis [7,8]. In hamster BHK 21/C13 fibroblasts, primary cilia are lost some time between the end of S phase and early prophase, and can begin to reappear soon after the completion of mitosis [9], while PtK1 (rat kangaroo kidney epithelial) cells appear to remain ciliated until early mitosis [10]. Primary cilia are also present in highly proliferative cells during vertebrate development, for example in the neural tube [11] and limb bud [12], consistent with involvement in sensing and processing morphogenetic signals.
Primary cilia are not normally present during mitosis, possibly because the mature (mother) centriole of the centrosome, which nucleates cilium formation and in the process attaches to the cell membrane, needs to be liberated to fulfil its role in spindle formation. Activation of the mitotic kinase AurA (Aurora kinase A) at the base of cilia has been implicated in triggering cilium disassembly at the G2/M transition [13]. Surprisingly, AurA was also shown to be transiently activated during the G0/G1 transition, and to be required for cilium disassembly at this time [13]. Several other factors have been implicated in stimulating cilium disassembly at the G0/G1 transition, including Plk1 (polo-like kinase 1), Dvl2 (Dishevelled 2), TcTex-1 and Pifo (Pitchfork) [14–17]. In comparison, the possibility that a period of cilium assembly occurs in late G1 [7], has received little attention.
It has recently been reported that depletion of factors that promote cilium disassembly, including AurA, impairs cell cycle re-entry and possibly S phase entry, specifically in cells able to form cilia [14,18]. These reports suggest that cilium disassembly must be initiated to allow cell cycle re-entry and the G1/S transition. Notably, in addition to promoting cilium disassembly, AurA has been implicated in suppressing ciliogenesis in cycling cells, in conjunction with trichoplein [3]. Depletion of either protein led to cilium formation and G0/G1 arrest, consistent with the idea that cilia act as a brake to prevent cell cycle progression. However, depletion of two other suppressors of ciliogenesis, Cep97 and CP110, has been reported to induce cilium formation without G0/G1 arrest [4]. Moreover, another study found that cell cycle re-entry and progression through interphase occurred normally in AurA-deficient cells, despite a block to cilium disassembly [13]. Thus, the cilium's ability to act as a brake on the cell cycle remains uncertain.
hTERT-RPE1 (human telomerase-immortalised retinal pigmented epithelial; herein RPE1) cells are commonly used to study molecular pathways regulating cilium disassembly, yet the ciliary disassembly profile of these cells remains uncertain. Two waves of cilium disassembly have been reported to occur, the first associated with cell cycle re-entry and the second with mitotic entry [13]. However, other studies have implied that cilium disassembly is completed before S phase entry in this cell line [18], or proceeds gradually over a 24 h period post-serum (in lentivirus-infected hTERT-RPE cells) [15]. Reciliation in G1 has not been observed, and is therefore generally assumed not to occur.
Here, we sought to gain a better picture of the ciliary disassembly profile of RPE1 cells in late G1 and during S phase entry, using immunofluorescence microscopy. The results provide evidence for a period of reciliation in late G1, and suggest that deciliation is not required for S phase entry but is coupled to DNA replication.
2. Results
2.1. Time course analysis of serum-induced cilium disassembly in RPE1 cells
We were interested in the ciliary disassembly profile of RPE1 cells around the time of S phase entry. To first determine the timing of S phase entry following release from serum starvation-induced quiescence, we fixed cells at various times post-serum and stained them with an antibody to cyclin B1. We found that detectable levels of cyclin B1, indicative of progression into S phase [19], began to appear between 14 and 16 h post-serum, and that cells began to enter mitosis between 20 and 24 h post-serum, based on DAPI staining of nuclear DNA and cyclin B1 immunostaining (Fig. 1A; data not shown). A similar timing of S phase entry was obtained using BrdU incorporation to identify cells undergoing DNA replication (Fig. 2A).
Fig. 1.
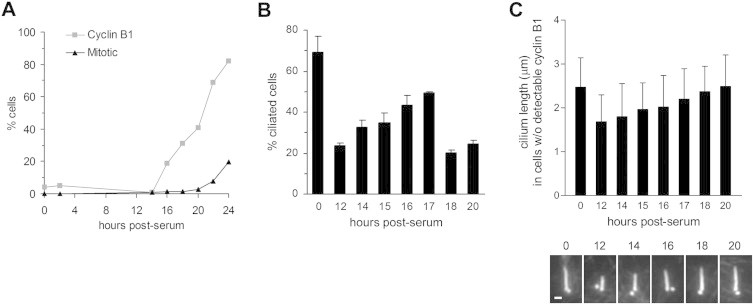
Time course analysis of serum-induced cilium disassembly in RPE1 cells. (A) Quantification of cyclin B1-positive and mitotic cells at various time points after release from serum starvation, based on immunofluorescence microscopy analysis (n = 100 cells). (B) Percentage of cells with cilia at 0 and 12–20 h post-serum. Data represent the mean + s.e.m. from two independent experiments (n > 200 cells). (C) Cilium length in cyclin B1-negative cells at 0–20 h post-serum (mean + s.d.; 0–18 h, n = 50; 20 h, n = 24). Examples of primary cilia in cells fixed at the indicated times post-serum are shown. Cells were stained with an antibody to acetylated α-tubulin. Bar, 1 μm.
Fig. 2.
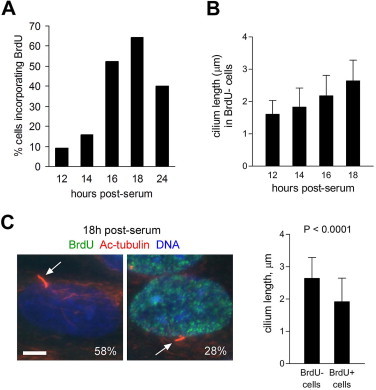
Cilium length increases before, and decreases during, DNA replication. (A) Percentage of cells positive for BrdU immunostaining at different times after release from serum starvation. BrdU was added 30 min before fixation (n > 200 cells). (B) Cilium length in BrdU-negative (BrdU−) cells at different times after release from serum starvation (mean + s.d.; n = 50). (C) Immunofluorescence analysis of ciliation and cilium length with respect to BrdU incorporation status at 18 h post-serum. Examples of BrdU− and BrdU+ cells are shown. An antibody to acetylated α-tubulin was used to mark cilia (arrows); the percentage of ciliated cells in each class is indicated (BrdU−, n = 73; BrdU+, n = 82). Quantification of cilium length is shown (mean + s.d.; n = 50; P value, unpaired two-tailed t-test). Bar, 5 μm.
Next, we released cells from serum starvation for 12–20 h and co-stained them with antibodies to acetylated α-tubulin (to mark cilia and centrioles) and either cyclin B1 or AurA phosphorylated at position T288 (phospho-AurA-T288), a marker of AurA activation [20]. In separate experiments, we confirmed that significant deciliation occurred within 2 h of serum addition (data not shown), as reported previously [13]. As shown in Fig. 1B, we found that ciliation increased between 12 and 17 h post-serum, and decreased between 17 and 18 h post-serum. A wave of reciliation has been reported to occur in late G1 in 3T3 cells [7]. Thus, our data raise the possibility that a similar event occurs in RPE1 cells. In support of this, we found that cilium length increased steadily between 12 and 20 h post-serum in cells that lacked detectable cyclin B1 immunostaining (Fig. 1C). This correlation was not apparent when total cell populations were analysed, probably due to wider variations in cell cycle stage (data not shown). Cyclin B1-negative cells were likely in G1 of the first cell cycle post-serum, because few would have passed through mitosis (and thus lack cyclin B1 due to its destruction at the metaphase–anaphase transition) within 20 h (Fig. 1A). We obtained similar results using BrdU incorporation as a marker of S phase entry (Fig. 2B). It seems reasonable to assume that most cells negative for BrdU at 12–18 h post-serum were still in G1 or early S phase, prior to the onset of detectable DNA replication, since the duration of S phase in human cultured cells is typically at least 5 h [21,22], and the number of cells incorporating BrdU first began to rise significantly between 14 and 16 h post-serum (Fig. 2A).
The time course data suggest that the second wave of deciliation coincides broadly with the appearance of cyclin B1 expression and BrdU incorporation (Fig. 1A, B; Fig. 2A), and therefore that it begins as RPE1 cells enter S phase, rather than mitosis. In support of this conclusion, analysis of cells fixed at 18 h post-serum, when BrdU incorporation peaked, showed that BrdU-positive cells were less likely to be ciliated, and typically possessed shorter cilia, than BrdU-negative (presumptive G1) cells (Fig. 2C). Collectively, these results suggest that the ciliary disassembly profile of RPE1 cells closely resembles that of 3T3 cells, in which a period of reciliation occurs in late G1, followed by deciliation coupled to DNA synthesis [7]. Importantly, this profile differs from previous analyses of RPE1 cells [13,15].
Phospho-AurA-T288 has been detected at the base of shortened cilia at 2 h post-serum in RPE1 cells, in line with its involvement in stimulating ciliary resorption at this time [13]. We were unable to detect phospho-AurA-T288 at the base of shortened cilia at 18 h post-serum (Fig. 3). However, we also failed to detect it at 2 h post-serum, and therefore cannot rule out the possibility that very low levels were present at the later time point (Fig. 3). We note that, under the same fixation conditions, relatively low levels of phospho-AurA-T288 were detectable at centrioles in cells that appeared to be in late G2 (based on the presence of well-developed procentrioles, prior to centrosome separation) (Fig. 3).
Fig. 3.
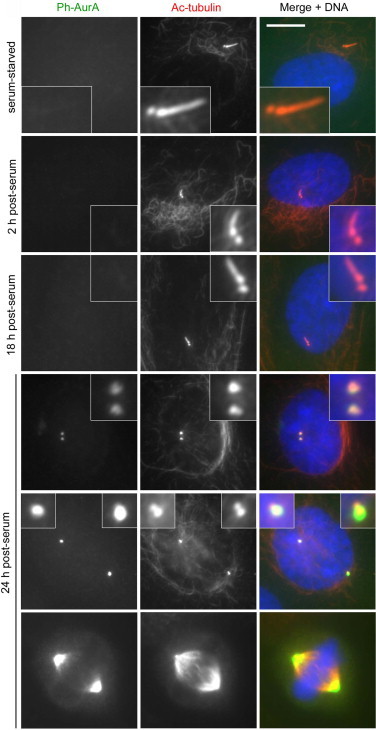
Activated AurA is undetectable at the base of shortened cilia during S phase. RPE1 cells were released from serum starvation for the indicated times and stained with antibodies to AurA phosphorylated at position T288 (Ph-AurA) and acetylated α-tubulin. DNA was stained with DAPI. Ph-AurA was not detectable at the base of shortened cilia during S phase (18 h post-serum), or during the initial wave of deciliation (2 h post-serum), but became detectable in late G2 phase (24 h post-serum), as indicated by the presence of at least one clearly visible, well-developed procentriole (less intense dot of acetylated α-tubulin adjacent to the more brightly stained centriole; both procentrioles were not always clearly visible, most likely due to differing orientations of each centriole–procentriole pair). Examples of cells in prophase and metaphase are included to show that the conditions used were suitable for detection of Ph-AurA. Bar, 10 μm.
2.2. Cells arrested in late G1 or early S phase show high levels of ciliation
To further examine the timing of the second wave of deciliation, and the preceding period of reciliation, with respect to S phase entry in RPE1 cells, we arrested cells in either late G1 (using mimosine) or early S phase (using hydroxyurea or excess thymidine) [23–26]. Cells were released from serum starvation in the presence or absence of each chemical for 4 or 24 h, then fixed and stained with antibodies to acetylated α-tubulin and cyclin B1. At 24 h post-serum, cyclin B1 immunostaining was extremely weak or undetectable in mimosine-treated cells, consistent with G1 arrest, and present at low levels in HU- and thymidine-treated cells, consistent with early S phase arrest (Fig. 4A). It was more intense in most cells treated with serum alone, or serum plus vehicle, consistent with progression into G2 and M phase in the absence of cell cycle inhibitors (Fig. 4A and data not shown). Immunoblot analysis of total cellular levels of cyclin B1 and cyclin A (a second marker of S phase entry [27]), at 24 h post-serum corroborated the immunofluorescence data (Fig. 4B).
Fig. 4.
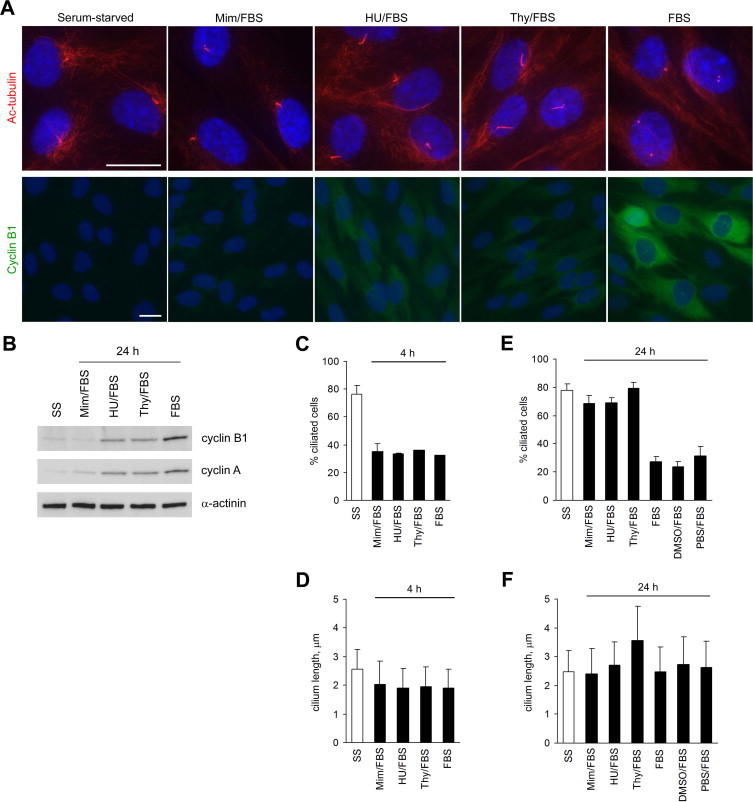
Effect of cell cycle inhibitors on serum-induced cilium disassembly. (A) RPE1 cells were serum-starved for 24 h to induce cilium formation, then either fixed or incubated in medium containing 10% FBS and mimosine (Mim), hydroxyurea (HU) or thymidine (Thy) for 24 h, before fixation. Cells were stained with an antibody to acetylated tubulin (red), to mark cilia, and an antibody to cyclin B1 (green), to confirm cell cycle stage. DNA was stained with DAPI (blue). Bars, 20 μm. (B) Immunoblot analysis of cyclin B1 and cyclin A expression in cells treated as in (A), with α-actinin as a loading control. (C) Percentage of cells with cilia following serum starvation and subsequent release for 4 h with the indicated treatments. SS, serum starved. Data represent the mean + s.e.m. from two independent experiments (n > 200 cells). (D) Cilium length in cells treated as in (C) (mean + s.d.; n = 50). (E) Percentage of cells with cilia following serum starvation and subsequent release for 24 h with the indicated treatments. Data represent the mean + s.e.m. from three independent experiments (n > 100 cells). (F) Cilium length in cells treated as in (E) (mean + s.d.; n = 40). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The first wave of cilium disassembly occurred normally in the presence of each chemical, as judged by analysis of ciliation and cilium length at 0 and 4 h post-serum (Fig. 4C and D). At 24 h post-serum, in the absence of inhibitors, the level of ciliation was low, as expected (Fig. 4E). In contrast, at 24 h post-serum in the presence of mimosine, the level of ciliation was similar to that before serum stimulation (Fig. 4E). Cilium length had also increased compared to the 4 h time point, and was similar to that before serum stimulation (Fig. 4F). Since mimosine arrests cells in late G1, this result is consistent with a period of reciliation occurring in late G1, as suggested by our initial time course data (Fig. 1).
Somewhat surprisingly, given recent data suggesting that primary cilia inhibit S phase entry [14,18], HU- and thymidine-treated cells also displayed high levels of ciliation at 24 h post-serum (Fig. 4E). The cilia of HU-treated cells were similar in length to before serum stimulation, while those of thymidine-treated cells were significantly longer (Fig. 4F). The persistence of cilia in HU- and thymidine-treated cells suggests that the second wave of cilium disassembly is initiated downstream of S phase entry, since both chemicals cause cell cycle arrest by inhibiting DNA synthesis, rather than S phase entry.
Next, to further investigate if deciliation occurs during S phase progression, we analysed ciliation and BrdU incorporation in cells fixed at 0, 3 and 6 h after release from a thymidine block. We found that ciliation and cilium length both decreased within 3 h of thymidine washout (Fig. 5A). Ciliary loss/shortening was associated with DNA synthesis, based on analysis of individual cells at 3 h post-washout (Fig. 5B), and therefore was not simply due to removal of excess thymidine from the growth medium. It seems reasonable to assume that most cells negative for BrdU at 3 h post-washout were still in early S phase, prior to the onset of detectable DNA replication, since, as noted above, the duration of S phase in human cells is typically at least 5 h, and ∼50% of cells were still incorporating BrdU at 6 h post-washout (Fig. 5A). In summary, these data indicate that deciliation coincides with DNA replication following release from a thymidine block, in line with the data for cells released from serum starvation in the absence of cell cycle inhibitors (Fig. 2C).
Fig. 5.
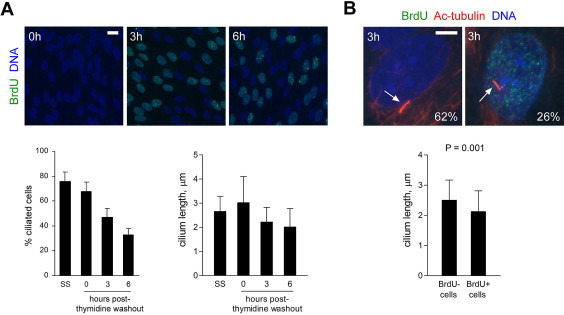
Cilium disassembly coincides with DNA synthesis following release from a thymidine block. (A) Immunofluorescent detection of BrdU incorporation and primary cilia following thymidine washout. Cells were initially released from serum-starvation in the presence of excess thymidine for 24 h, then incubated in thymidine-free medium (with serum) for 3 or 6 h prior to fixation. In parallel, cells were fixed immediately after serum-starvation (SS) or thymidine treatment (0 h post-washout). BrdU was added 30 min prior to fixation. Cells were co-stained with antibodies to acetylated α-tubulin (not depicted) and BrdU; DNA was stained with DAPI. Charts show the percentage of cells with cilia (mean + s.e.m. from three independent experiments; n = 150–300 cells) and cilium length (mean + s.d.; n = 50–75). (B) Ciliation status of individual cells, with respect to the presence or absence of detectable BrdU immunostaining, 3 h after thymidine-washout. Examples of ciliated BrdU-negative (BrdU−) and BrdU-positive (BrdU+) cells are shown. Arrows indicate primary cilia; the percentage of cells with cilia is indicated (BrdU− cells, n = 50; BrdU+ cells, n = 70). The chart shows cilium length for cells in each class (mean + s.d.; BrdU− cells, n = 64; BrdU+ cells, n = 79; P value, unpaired two-tailed t-test). Bars, 20 μm (A) and 5 μm (B).
2.3. Ciliary localisation of cyclin B1
In agreement with our previous findings [28], we noted that a minority (8.4%, n = 131) of late G2 cells were ciliated (late G2 was defined here as the presence of intense cyclin B1 immunostaining at unseparated, duplicated centrosomes [29]), 20–24 h after release from serum starvation in the absence of cell cycle inhibitors (Fig. 6A). Also in agreement with the previous study, following release from serum starvation, cilia were typically absent in cells undergoing centrosome separation (duplicated centrosomes separated by >2 μm, prior to prometaphase; 0.6% cells ciliated, n = 155; Fig. 6A), and in cells beginning to undergo chromatin condensation in prophase, as indicated by weak phospho-histone H3 immunostaining throughout the nucleus (1.7% cells ciliated, n = 60; data not shown). These observations suggest that primary cilia which remain in late G2 are typically resorbed shortly before, or coincident with, mitotic entry in RPE1 cells.
Fig. 6.
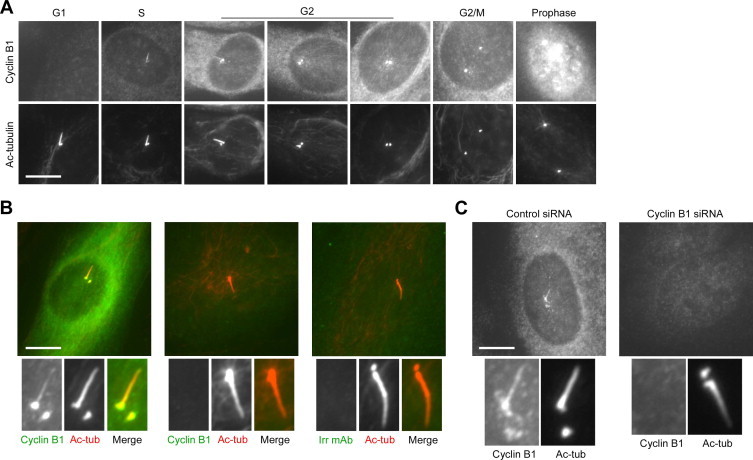
Cyclin B1 localises to the primary cilium. (A) RPE1 cells were released from serum starvation for up to 24 h, then fixed and stained with antibodies to cyclin B1 (GNS-1) and acetylated α-tubulin. Cell cycle stage, based on cyclin B1 immunostaining, is indicated. (B) 142BR human fibroblasts were stained with antibodies to acetylated α-tubulin and cyclin B1 (GNS-1) or an irrelevant antibody, as indicated. Main images are merges. (C) RPE1 cells were transfected with a negative control siRNA or cyclin B1-directed siRNA for 24 h, then fixed and stained with antibodies to cyclin B1 (GNS-1) and acetylated α-tubulin. Main images show GNS-1 staining. The presence of procentrioles (dots of acetylated α-tubulin immunostaining, adjacent to the more intensely stained centrioles) was used as a marker of entry to S/G2 phase. Bars, 10 μm.
Surprisingly, in the course of this work we noticed that the cyclin B1 antibody (termed GNS-1) stained primary cilia, in addition to the expected staining of centrosomes [30] (Fig. 6A). In support of antibody specificity, we did not observe ciliary GNS-1 immunostaining in cells in which cytoplasmic GNS-1 staining was undetectable (presumed to be in G0 or G1 phase), nor did a negative control antibody detectably stain primary cilia (Fig. 6A and B). Ciliary immunostaining was apparent using an alternative cyclin B1 antibody, methanol or PFA fixation, and other cell lines (Fig. 7A; data not shown; Fig. 6B), and was abolished by siRNA-mediated depletion of cyclin B1 (Fig. 6C). An antibody to cyclin B2, in contrast, did not detectably stain primary cilia (Fig. 7B). These results strongly suggest that cyclin B1 localises to primary cilia. Notably, we observed weak ciliary immunofluorescence with an antibody to CDK1, the cyclin-dependent kinase partner of cyclin B1 (Fig. 7C), suggesting that CDK1 also localises to primary cilia.
Fig. 7.
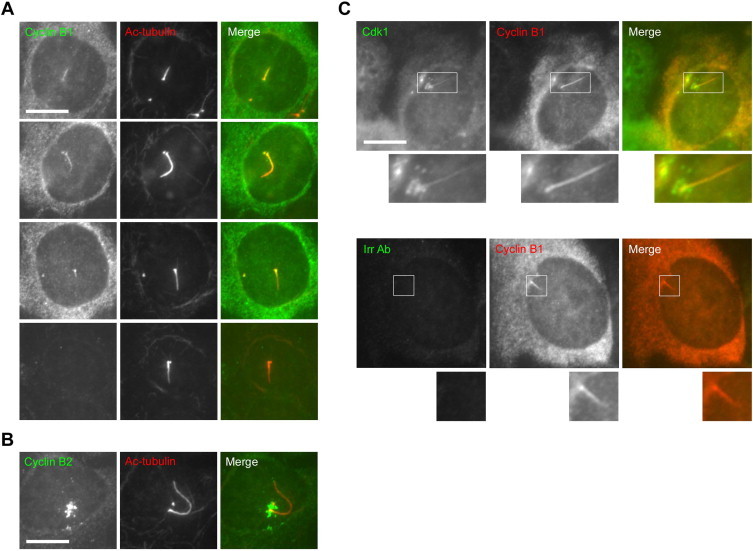
Antibodies to cyclin B1 and CDK1, but not cyclin B2, stain primary cilia in murine IMCD3 cells. (A) Cells were fixed in methanol and stained with a rabbit polyclonal antibody to cyclin B1 and a mouse monoclonal antibody to acetylated α-tubulin. Examples shown include cells in which cyclin B1 was detectable only in the proximal portion of the cilium, and a ciliated cell lacking detectable cyclin B1 expression, demonstrating that the cyclin B1 antibody does not stain cilia non-specifically. (B) Cells were fixed in methanol and stained with a rabbit polyclonal antibody to cyclin B2 and a mouse monoclonal antibody to acetylated α-tubulin. (C) Cells were fixed in methanol and stained with a rabbit polyclonal antibody to cyclin B1, to mark cilia, and either a mouse monoclonal antibody to Cdk1 or an irrelevant mouse antibody, as indicated. Bars, 10 μm.
3. Discussion
Our results suggest that the ciliary disassembly profile of RPE1 cells resembles that of 3T3 cells [7,8], with a wave of deciliation at the G0/G1 transition, a period of reciliation in late G1 and a second wave of deciliation coincident with DNA synthesis. Evidence of reciliation is particularly notable because it is generally assumed not to occur in RPE1 cells. Selection of different time points may explain why it was not seen in previous time course analyses [13,15]. Time course analyses of the type used here are hampered by the semi-synchronous nature of cell cycle re-entry and progression in response to serum stimulation [31]. However, two further observations are consistent with reciliation in late G1: after the initial deciliation, cilium length increased with time in cyclin B1/BrdU-negative cells, and cells arrested in late G1 by mimosine showed high levels of ciliation. A notable difference between 3T3 and RPE1 cells is that in asynchronously growing populations, approximately one third of 3T3 cells have cilia [7], compared to <5% [3] or ∼10% [4] of RPE1 cells. Thus, it appears that cilium assembly in G1 is less common in RPE1 cells. However, our results suggest that a significant proportion of RPE1 cells can form a cilium in G1, at least in the first cell cycle following release from serum starvation.
Evidence for reciliation in G1 can be reconciled with the existence of pathways that suppress ciliogenesis in cycling cells if suppressors such as trichoplein [3] are displaced temporarily from the mother centriole in late G1. There is a precedent for this type of behaviour, in that the key suppressor of ciliogenesis CP110 has been reported to disappear from the mother centriole, but not the adjacent centriole/procentrioles, in ciliated 3T3 cells in G1 and S/G2 phase [4]. Late G1 in particular may be permissive for cilium assembly, because activation of AurA and Plk1 at cell cycle re-entry appears to be transient [13,17], and because expression of CP110 is low in G1 [32].
Notably, treatment of quiescent 3T3 cells with different components of serum has revealed some of the requirements for deciliation and DNA synthesis. Tucker et al. [8] reported that PDGF (platelet-derived growth factor) induced an initial rapid deciliation, followed by a period of reciliation, but that plasma components were required to efficiently induce the second wave of deciliation and DNA synthesis. Treatment with plasma alone did not induce the initial deciliation or DNA synthesis. It is now known that ciliary localisation of PDGFRα (PDGF receptor α) is important for the cellular response to its ligand PDGF-AA [33]. Moreover, one of the plasma components that induces DNA synthesis, IGF1 (insulin-like growth factor 1) [34], has been shown to act at least partly through the primary cilium during differentiation of 3T3-L1 preadipocytes [35]. These findings raise the possibility that reciliation in G1 could serve to sensitize cells to IGF1 and other plasma components that induce progression into S phase. It is also noteworthy that the mTOR (mammalian target of rapamycin)-dependent, nutrient-sensing ‘cell growth checkpoint’ is situated in late G1 [36]. The mTOR pathway is regulated by cilium-mediated sensation of fluid flow in renal cells, and it is possible that cilia have a role in mTOR signalling in other contexts [37].
A previous study has suggested that, after the wave of cilium disassembly associated with cell cycle re-entry, a second wave of cilium disassembly coincides with mitotic entry in RPE1 cells [13], consistent with stimulatory roles for the mitotic kinases AurA, Plk1 and Nek2 [13,15,17,28,38]. In partial agreement, our results suggest that some cells undergo deciliation at the time of centrosome separation. Overall, however, our results suggest that the second wave of ciliary resorption begins in S phase. Interestingly, we found that cilia persisted in conditions of early S phase arrest. Since the chemicals used to induce this arrest (HU and thymidine) inhibit DNA synthesis, this result extends the notion that deciliation is coupled to DNA synthesis [7], and implies that any experimental condition that inhibits DNA synthesis will delay the second wave of cilium disassembly. It also suggests that cyclin-CDK activities sufficient to trigger S phase entry are insufficient to trigger deciliation. However, it may be argued that replication stress or another effect of HU/thymidine treatment could promote ciliation. For example, hydroxyurea treatment can lead to DNA damage [39], which may in turn inhibit AurA, Plk1 and Nek2 [40–42]. Thus, it remains possible that in unperturbed cells deciliation is controlled by the same activities that promote the G1/S transition. Notably, the presence of full length (or in the case of thymidine-treated cells, unusually long) cilia in S phase-arrested cells appears to conflict with data suggesting that ciliary resorption must be initiated to allow S phase entry [14,18].
In summary, the results presented here support the notion that cilium formation can occur in G1 phase in cycling cells, and suggest that cilium disassembly is not essential for S phase entry, but is coupled to DNA replication. Thus, RPE1 cells could serve as a model system in which to investigate the assembly and function of primary cilia in G1, as well as the pathways that regulate their disassembly.
In the course of this work, we noticed that cyclin B1 antibodies specifically stained primary cilia. An antibody to CDK1 also produced ciliary staining, consistent with a recent proteomic analysis of IMCD3 primary cilia [43]. To our knowledge, ciliary immunolocalisation of mammalian cyclins/CDKs has not been previously reported. It is uncertain if there is a selective barrier for soluble proteins at the base of cilia in mammalian cells, although there is mounting evidence for a mechanism similar to that governing entry to the nucleus [44,45]. Thus, it may be interesting to investigate if importin-β1, which is thought to mediate the relatively slow nuclear import of cyclin B1 in interphase [46], also mediates its localisation to primary cilia. The immunofluorescence data are intriguing, considering the key cell cycle regulatory functions of these proteins, together with data indicating that cyclin B1 targets CDK1 to microtubules, and that CDK1 activity can negatively influence microtubule stability and tubulin polymerisation [47,48]. It is also noteworthy that CDK1 activity has recently been implicated in promoting pericentriolar material 1 (PCM1)-mediated centrosomal recruitment of Plk1, which in turn promotes cilium disassembly before mitotic entry [38]. We have found that treatment of RPE1 cells with a CDK1 inhibitor increases cilium length (unpublished data). However, a similar effect apparent in serum starved quiescent cells suggests that inhibition of another CDK, or a non-specific effect, was at least partly responsible. Further work is required to determine if CDK1 influences cilium length, either via PCM1 or more directly.
4. Materials and methods
4.1. Cell culture and synchronisation
hTERT-RPE1 (LGC Standards, Middlesex, UK) and IMCD3 (ECACC, Porton Down, UK) cells were maintained in DMEM/Ham's F12 supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37 °C and 5% CO2 (reagents from Invitrogen, Paisley, UK). 142BR human skin fibroblasts (ECACC) were maintained in DMEM with the same supplements and conditions. For immunofluorescence microscopy, cells were seeded in Lab-Tek II chamber slides (VWR, Lutterworth, UK), or on cover slips in 12-well plates. For time course analyses, cells were incubated in serum-free medium for 24 h to induce primary cilium formation, then fixed at various time points after addition of 10% FBS. For cell cycle synchronisation, cells were incubated in serum-free medium for 24 or 48 h to induce primary cilium formation, then in medium containing 10% FBS and either 0.5 mM mimosine, 4 mM hydroxyurea or 5 mM thymidine (or vehicle) for a further 24 h. For thymidine-washout, cells were rinsed three times with thymidine-free medium containing 10% FBS and then incubated in this medium for 3 or 6 h before being processed for immunofluorescence. Chemicals were from Sigma–Aldrich (Dorset, UK). Stock solutions of mimosine, hydroxyurea and thymidine were prepared with culture medium, dimethyl sulfoxide (DMSO) and phosphate buffered saline (PBS), respectively.
4.2. Bromodeoxyuridine (BrdU) incorporation assay
Cells were incubated in growth medium containing BrdU (10 μg/ml; Sigma–Aldrich) for the last 30 min prior to methanol fixation (5 min). Fixed cells were rinsed twice with PBS, incubated in 1.5 M HCl for 20 min at room temperature, then rinsed three times with PBS before incubation with primary antibodies for immunofluorescence, as described below.
4.3. RNA interference
RPE1 cells were transfected with siRNA duplexes at a final concentration of 50 nM using HiPerFect reagent (Qiagen, Crawley, UK). Cells were fixed in methanol 24 h later and processed for immunofluorescence microscopy as described below. An siRNA duplex targeting cyclin B1 (siRNA3 in Ref. [49]) and AllStars negative control siRNA were purchased from Qiagen.
4.4. Immunoblotting
Cells synchronised as above were rinsed with PBS and lysed in RIPA buffer (Sigma–Aldrich) supplemented with Roche Complete Mini protease inhibitors. Cell lysates were cleared by centrifugation at 13,000 rpm at 4 °C for 10 min. Proteins were denatured and reduced, separated on NuPAGE 4–12% Bis–Tris gels (Invitrogen) and transferred to Hybond ECL nitrocellulose membrane (VWR). Antibody incubations and ECL detection were done as described [50]. Primary antibodies were: rabbit anti-cyclin B1 (H-433; Insight Biotechnology, Wembley, UK), mouse anti-cyclin A (Sigma–Aldrich) and mouse anti-α-actinin (BM-75.2; Sigma–Aldrich).
4.5. Immunofluorescence microscopy
Cells were rinsed with PBS and fixed in chilled methanol for 5 min or 4% paraformaldehyde (PFA) for 10 min. We included an antigen retrieval step that we have found improves staining of centrosomal proteins in both methanol and paraformaldehyde fixed cells: slides/coverslips were immersed in 10 mM sodium citrate (85–90 °C) and allowed to cool at room temperature for 30 min, then rinsed in PBS for 5 min. Immunofluorescence was otherwise performed as described [50]. Cells were mounted in Vectashield with DAPI (Vector Labs, Peterborough, UK) and images captured using an Axio Observer Z1 microscope equipped with an AxioCam MRm digital camera (Carl Zeiss Ltd., Welwyn Garden City, UK).
4.6. Antibodies
Primary antibodies for indirect immunofluorescence were: mouse anti-acetylated α-tubulin (6-11B-1) and anti-BrdU (Sigma–Aldrich); mouse anti-cyclin B1 (GNS-1) and anti-CDK1 (BD Biosciences, Oxford, UK); rabbit anti-cyclin B1 (H-433) and anti-cyclin B2 (H-105) (Insight Biotechnology); mouse anti-phospho-histone H3 (Ser10) (6G3) and rabbit monoclonal anti-phospho-Aurora A (Thr288) (New England Biolabs, Hitchin, UK). Goat secondary antibodies were: FITC anti-rabbit (Sigma–Aldrich), Alexa Fluor 594 anti-rabbit, Alexa Fluor 488 anti-mouse IgG1 and Alexa Fluor 594 anti-mouse IgG2b (Invitrogen).
Acknowledgements
This work was supported by the University of Southampton. T.H. was also partly supported by a Wellcome Trust Value in People Award.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Oh E.C., Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi T., Dynlacht B.D. Regulating the transition from centriole to basal body. J. Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoko A., Matsuyama M., Goto H., Ohmuro-Matsuyama Y., Hayashi Y., Enomoto M., Ibi M., Urano T., Yonemura S., Kiyono T. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J. Cell Biol. 2012;197:391–405. doi: 10.1083/jcb.201106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spektor A., Tsang W.Y., Khoo D., Dynlacht B.D. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T., Tsang W.Y., Li J., Lane W., Dynlacht B.D. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145:914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 6.Seeley E.S., Nachury M.V. The perennial organelle: assembly and disassembly of the primary cilium. J. Cell Sci. 2010;123:511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker R.W., Pardee A.B., Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- 8.Tucker R.W., Scher C.D., Stiles C.D. Centriole deciliation associated with the early response of 3T3 cells to growth factors but not to SV40. Cell. 1979;18:1065–1072. doi: 10.1016/0092-8674(79)90219-8. [DOI] [PubMed] [Google Scholar]
- 9.Archer F.L., Wheatley D.N. Cilia in cell-cultured fibroblasts. II. Incidence in mitotic and post-mitotic BHK 21/C13 fibroblasts. J. Anat. 1971;109:277–292. [PMC free article] [PubMed] [Google Scholar]
- 10.Rieder C.L., Jensen C.G., Jensen L.C. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J. Ultrastruct. Res. 1979;68:173–185. doi: 10.1016/s0022-5320(79)90152-7. [DOI] [PubMed] [Google Scholar]
- 11.Louvi A., Grove E.A. Cilia in the CNS: the quiet organelle claims center stage. Neuron. 2011;69:1046–1060. doi: 10.1016/j.neuron.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonte V.G., Searls R.L., Hilfer S.R. The relationship of cilia with cell division and differentiation. J. Cell Biol. 1971;49:226–229. doi: 10.1083/jcb.49.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., Golemis E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li A., Saito M., Chuang J.Z., Tseng Y.Y., Dedesma C., Tomizawa K., Kaitsuka T., Sung C.H. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat. Cell Biol. 2011;13:402–411. doi: 10.1038/ncb2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K.H., Johmura Y., Yu L.-R., Park J.-E., Gao Y., Bang J.K., Zhou M., Veenstra T.D., Yeon Kim B., Lee K.S. Identification of a novel Wnt5a-CK1ɛ-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J. 2012;31:3104–3117. doi: 10.1038/emboj.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinzel D., Boldt K., Davis E.E., Burtscher I., Trumbach D., Diplas B., Attie-Bitach T., Wurst W., Katsanis N., Ueffing M. Pitchfork regulates primary cilia disassembly and left–right asymmetry. Dev. Cell. 2010;19:66–77. doi: 10.1016/j.devcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seeger-Nukpezah T., Liebau M.C., Hopker K., Lamkemeyer T., Benzing T., Golemis E.A., Schermer B. The centrosomal kinase Plk1 localizes to the transition zone of primary cilia and induces phosphorylation of nephrocystin-1. PLoS ONE. 2012;7:e38838. doi: 10.1371/journal.pone.0038838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S., Zaghloul N.A., Bubenshchikova E., Oh E.C., Rankin S., Katsanis N., Obara T., Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat. Cell Biol. 2011;13:351–360. doi: 10.1038/ncb2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pines J., Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter A.O., Seghezzi W., Korver W., Sheung J., Lees E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene. 2000;19:4906–4916. doi: 10.1038/sj.onc.1203847. [DOI] [PubMed] [Google Scholar]
- 21.Robinson K., Asawachaicharn N., Galloway D.A., Grandori C. c-Myc accelerates S-phase and requires WRN to avoid replication stress. PLoS ONE. 2009;4:e5951. doi: 10.1371/journal.pone.0005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi T., Garcia-Higuera I., Andreassen P.R., Gregory R.C., Grompe M., D’Andrea A.D. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 23.Bianchi V., Pontis E., Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 1986;261:16037–16042. [PubMed] [Google Scholar]
- 24.Krude T. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 1999;247:148–159. doi: 10.1006/excr.1998.4342. [DOI] [PubMed] [Google Scholar]
- 25.Watson P.A., Hanauske-Abel H.H., Flint A., Lalande M. Mimosine reversibly arrests cell cycle progression at the G1-S phase border. Cytometry. 1991;12:242–246. doi: 10.1002/cyto.990120306. [DOI] [PubMed] [Google Scholar]
- 26.Bjursell G., Reichard P. Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J. Biol. Chem. 1973;248:3904–3909. [PubMed] [Google Scholar]
- 27.Pagano M., Pepperkok R., Verde F., Ansorge W., Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spalluto C., Wilson D.I., Hearn T. Nek2 localises to the distal portion of the mother centriole/basal body and is required for timely cilium disassembly at the G2/M transition. Eur. J. Cell Biol. 2012;91:675–686. doi: 10.1016/j.ejcb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Lindqvist A., van Zon W., Karlsson Rosenthal C., Wolthuis R.M. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5:e123. doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailly E., Pines J., Hunter T., Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome. J. Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- 31.Davis P.K., Ho A., Dowdy S.F. Biological methods for cell-cycle synchronization of mammalian cells. Biotechniques. 2001;30:1322–1331. doi: 10.2144/01306rv01. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z., Indjeian V.B., McManus M., Wang L., Dynlacht B.D. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 33.Schneider L., Clement C.A., Teilmann S.C., Pazour G.J., Hoffmann E.K., Satir P., Christensen S.T. PDGFRαα signaling is regulated through the primary cilium in fibroblasts. Curr. Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Stiles C.D., Capone G.T., Scher C.D., Antoniades H.N., Van Wyk J.J., Pledger W.J. Dual control of cell growth by somatomedins and platelet-derived growth factor. Proc. Natl. Acad. Sci. U.S.A. 1979;76:1279–1283. doi: 10.1073/pnas.76.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu D., Shi S., Wang H., Liao K. Growth arrest induces primary-cilium formation and sensitizes IGF-1-receptor signaling during differentiation induction of 3T3-L1 preadipocytes. J. Cell Sci. 2009;122:2760–2768. doi: 10.1242/jcs.046276. [DOI] [PubMed] [Google Scholar]
- 36.Foster D.A., Yellen P., Xu L., Saqcena M. Regulation of G1 cell cycle progression: distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s) Genes Cancer. 2010;1:1124–1131. doi: 10.1177/1947601910392989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boehlke C., Kotsis F., Patel V., Braeg S., Voelker H., Bredt S., Beyer T., Janusch H., Hamann C., Godel M. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat. Cell Biol. 2010;12:1115–1122. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G., Chen Q., Zhang X., Zhang B., Zhuo X., Liu J., Jiang Q., Zhang C. PCM1 recruits Plk1 to pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J. Cell Sci. 2013;126:1355–1365. doi: 10.1242/jcs.114918. [DOI] [PubMed] [Google Scholar]
- 39.Lundin C., Erixon K., Arnaudeau C., Schultz N., Jenssen D., Meuth M., Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smits V.A., Klompmaker R., Arnaud L., Rijksen G., Nigg E.A., Medema R.H. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- 41.Krystyniak A., Garcia-Echeverria C., Prigent C., Ferrari S. Inhibition of Aurora A in response to DNA damage. Oncogene. 2006;25:338–348. doi: 10.1038/sj.onc.1209056. [DOI] [PubMed] [Google Scholar]
- 42.Fletcher L., Cerniglia G.J., Nigg E.A., Yen T.J., Muschel R.J. Inhibition of centrosome separation after DNA damage: a role for Nek2. Radiat. Res. 2004;162:128–135. doi: 10.1667/rr3211. [DOI] [PubMed] [Google Scholar]
- 43.Ishikawa H., Thompson J., Yates J.R., 3rd, Marshall W.F. Proteomic analysis of mammalian primary cilia. Curr. Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kee H.L., Dishinger J.F., Lynne Blasius T., Liu C.-J., Margolis B., Verhey K.J. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dishinger J.F., Kee H.L., Jenkins P.M., Fan S., Hurd T.W., Hammond J.W., Truong Y.N., Margolis B., Martens J.R., Verhey K.J. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat. Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore J.D., Yang J., Truant R., Kornbluth S. Nuclear import of Cdk/Cyclin complexes: identification of distinct mechanisms for import of Cdk2/Cyclin E and Cdc2/Cyclin B1. J. Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ookata K., Hisanaga S., Bulinski J.C., Murofushi H., Aizawa H., Itoh T.J., Hotani H., Okumura E., Tachibana K., Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J. Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fourest-Lieuvin A., Peris L., Gache V., Garcia-Saez I., Juillan-Binard C.l., Lantez V., Job D. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol. Biol. Cell. 2006;17:1041–1050. doi: 10.1091/mbc.E05-07-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan J., Yan R., Kramer A., Eckerdt F., Roller M., Kaufmann M., Strebhardt K. Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells. Oncogene. 2004;23:5843–5852. doi: 10.1038/sj.onc.1207757. [DOI] [PubMed] [Google Scholar]
- 50.Hearn T., Spalluto C., Phillips V.J., Renforth G.L., Copin N., Hanley N.A., Wilson D.I. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54:1581–1587. doi: 10.2337/diabetes.54.5.1581. [DOI] [PubMed] [Google Scholar]


