Abstract
The human LINE-1/L1 ORF2 protein is a multifunctional enzyme which plays a vital role in the life cycle of the human L1 retrotransposon. The protein consists of an endonuclease domain, followed by a central reverse transcriptase domain and a carboxy-terminal C-domain with unknown function. Here, we explore the nucleic acid binding properties of the 180-amino acid carboxy-terminal segment (CTS) of the human L1 ORF2p in vitro. In a series of experiments involving gel shift assay, we demonstrate that the CTS of L1 ORF2p binds RNA in non-sequence-specific manner. Finally, we report that mutations destroying the putative Zn-knuckle structure of the protein do not significantly affect the level of RNA binding and discuss the possible functional role of the CTS in L1 retrotransposition.
Highlights
-
•
The 180-aa C-terminal segment (CTS) of human L1 ORF2p was expressed and purified from bacteria.
-
•
The nucleic acid binding properties of the CTS of L1ORF2p were examined in vitro.
-
•
The CTS of L1 ORF2p is an RNA binding domain.
-
•
The CTS of L1 ORF2p binds RNA in non-sequence-specific manner in the low nanomolar range.
-
•
Disruption of the putative Zn-knuckle structure does not significantly affect RNA binding.
1. Introduction
Long Interspersed Nuclear Element-1 (LINE-1) or L1 elements are active members of an autonomous family of non-LTR retrotransposons and comprise up to 17% of the human genome. The majority are inactive due to mutations, truncations and recombination, but about one hundred copies are retrotransposition-competent [1].
Full-length human L1 elements are about 6 kb long and contain two non-overlapping open reading frames (ORFs) encoding essential proteins for its re-integration into the genome. The ORF1 encodes a 40 kDa RNA-binding protein, which associates with L1 RNA [2] and functions as a chaperone [3,4]. The product of ORF2 is a 149 kDa multifunctional polymerase with endonuclease [5,6], RNA- and DNA-dependent DNA polymerase activities [7,8]. L1 ORF2p reverse transcriptase (RT) is a highly processive polymerase unlike retroviral RTs [9]. It seems that both proteins interact with their own L1 RNA in a cis preference manner [10] forming ribonucleic acid particles (RNP) that are probable intermediates of retrotransposition [11–13]. It has been proposed that the L1 ORF2 protein uses nicked DNA as a primer to initiate cDNA synthesis on the RNA template in a target-primed reverse transcription (TPRT) reaction [14] originally demonstrated for the reverse transcriptase encoded by the non-LTR retrotransposon R2 element from Bombyx mori [15].
The L1 ORF2 polypeptide forms an AP-like endonuclease (EN) domain at the N-terminus, then an RT domain followed by the cysteine-rich carboxy-terminal part, the C-domain [16], which contains a putative CCHC zinc knuckle structure. The EN, RT and C-domain are essential for successful L1 retrotransposition [17]. Thus, mutation of the C-domain zinc finger severely affects the retrotransposition frequency of L1 elements in the cell culture-based retrotransposition assay [17–19], reduction in the content of ORF2p in RNP and dramatic decrease in RT activity in L1 RNP as well as disperse nuclear localization of L1 RNP [13]. However the function of the C-domain, particularly the putative CCHC zinc knuckle structure remains unclear. Unlike retroviral RTs, where one third of their polypeptide chain at the C-terminus is reserved with an RNaseH domain [20], the long C-terminal portion of human L1 ORF2p bears neither sequence similarities [21,22] nor activities [9] corresponding to RNase H. Thus, an elucidation of C-domain function in L1 retrotransposition is of great interest.
We have previously demonstrated that human L1 RT (L1 ORF2p) is a highly processive polymerase, able to incorporate hundreds of nucleotides per template binding event [9]. At that time, we hypothesized that L1 ORF2p C-domain, enriched with basic amino acid (aa) residues, was able to provide a more stable interaction of the L1 RT with L1 mRNA during DNA synthesis, and contributed to high processivity of L1 RT in vitro.
In the research presented here, we have examined the nucleic acid binding activity of a distal 180 aa C-terminal portion of L1 ORF2 protein, namely the carboxy-terminal segment (CTS). L1 ORF2p CTS was expressed individually in bacteria followed by its purification and examination of nucleic acid binding properties in a series of experiments involving gel shift assay with wild type CTS and its mutant form carrying aa substitutions disrupting CCHC zinc knuckle structure. We have demonstrated that L1 ORF2p CTS has a high non-specific affinity to RNA in the nanomolar range.
This data allowed us to suggest that the carboxy-terminal segment provides high processivity for L1 ORF2p reverse transcriptase in comparison with that of retroviral RTs. Furthermore, non-specific RNA-binding activity of L1 ORF2p CTS may contribute to both cis-preference of L1 ORF2 protein to its own RNA, and a move of non autonomous mobile elements.
2. Materials and methods
pSM42, containing the ORF2 of the active human L1 element LRE1, was a gift from Prof. H. Kazazian. The numbering of the L1.2 sequence is that of Genbank: M80343.
A polypeptide sequence of the ORF2 protein from 1096 to 1275 aa was cloned as a fusion with thioredoxin (TRX). For this purpose, a 0.54 kb fragment of ORF2 corresponding to the 180 aa sequence was amplified with PCR using primers listed in Supplementary data SD1; and ligated into pET32b (Novagen) within EcoRI and XhoI sites. The resulting plasmid pET-TRX-CTS coded for a fusion of TRX and a C-terminal 21.3 kDa polypeptide of L1 ORF2p, referred as rCTS.
Additional materials and methods including protein expression and purification, protein analysis, RNA and DNA template synthesis, electrophoretic mobility shift assay and statistics are detailed in SD1.
3. Results
3.1. Purification of recombinant carboxy-terminal segment of L1 ORF2p
Human L1 ORF2p is 149 kDa multifunctional enzyme comprising of distinguished domains: EN and RT, both of which may contribute to nucleic acid binding activity (Fig. 1A). Therefore, we decided that individual domain characterization would be the preferred option. In this manner EN has been successfully expressed, purified and characterized [5]. The exact boundaries of the RT domain and C-domain are not yet defined, but the localization of the putative Zinc knuckle structure is predicted. Therefore, the cystein-rich carboxy-terminal polypeptide sequence of L1 ORF2 was analyzed for prediction of RNA binding residues/motifs using the packages BindN [23] and RNABindR [24]. Results of this prediction demonstrated a cluster distribution of RNA binding residues in the C-domain of the protein (Fig. 1B). Taking into consideration both the aa distribution and the C-domain size, one would suggest a subdomain organization of the C-domain.
Fig. 1.
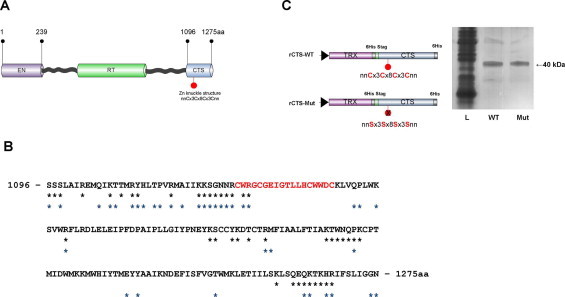
(A) Schematic representation of the full length polypeptide encoded by human L1 ORF2 (aa numbering according to GenBank sequence M80343). The endonuclease domain, (EN), reverse transcriptase, (RT), and the 180 aa carboxy-terminal segment (CTS) are shown. The putative Zn-knuckle structure is marked by a red pin within the CTS. (B)The results of the BindN [23] and RNABindR [24] prediction software for the 180 aa CTS polypeptide. The black and blue asterisks indicate predicted RNA-binding amino acid residues identified with the BindN and RNABindR, respectively. The putative Zn-knuckle sequence is in red. (C) Schematic diagrams of expression plasmids (left). SDS–PAGE of the purified recombinant fusion proteins containing CTS (rCTS, 40 kDa) visualized by silver staining (right). The Zn-knuckle sequence and aa substitutions are presented. The position of 6His tags is indicated. Theoredoxin, TRX, lysate, L, wild type of rCTS, WT, Zn finger mutant form of rCTS, Mut.
The distal C-terminal polypeptide sequence of the ORF2 protein (1096–1275 aa) containing both the putative Zn-knuckle structure and multiple predicted RNA binding residues was selected and referred to as CTS (carboxy-terminal segment) (Fig. 1C). L1 ORF2p CTS was inserted into pET32b plasmid DNA resulting in a fusion protein of ≈40 kDa. This recombinant protein, referred to as rCTS, is fusion protein of CTS with TRX and flanked with 6 His tag.
rCTS was expressed in soluble form and purified using metal affinity chromatography to near homogeneity (Fig. 1C) as detailed in Supplementary material (SD1). The integrity and the concentration of all recombinant proteins were verified by Silver staining gel and Pierce BSA protein assay, respectively.
3.2. The CTS of L1 ORF2p is an RNA binding domain
Wild type rCTS was used for studying CTS–nucleic acid (NA) interactions. This C-terminal region of L1 ORF2p has a potential Zn-knuckle structure as well as being rich in basic aa residues; about 14% of the total aa are either lysines or arginines, and the overall pI value of CTS is 9.1.
To examine the hypothesis that CTS is a nucleic acid binding domain within L1 ORF2p, we performed an electrophoretic mobility shift assay (EMSA) using various templates, dsDNA and ssRNA. The complex rCTS:NA was observed only in the presence of RNA as template in physiological concentrations of monovalent cation and pH (Fig. 2A) demonstrating that CTS has RNA binding activity.
Fig. 2.
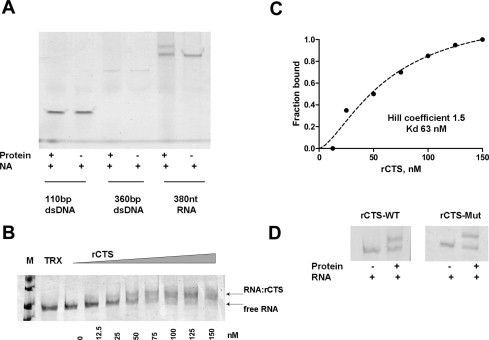
(A) Determination of rCTS binding specificity in physiological conditions using EMSA with various templates, dsDNA and ssRNA. The complex(es) rCTS:NA was observed only in the presence of RNA as template. (The molar ratio of protein:NA in reaction mix was similar.) (B) Determination of the binding affinity of rCTS to ssRNA. The concentration of rCTS was in the range 0–150 nM, whilst the amount of 380 nt RNA was constant at 30nM. The complexes RNA:protein and free probes are marked. An excess of purified TRX was used in a control EMSA reaction to eliminate any contribution of TRX into NA binding. pET32b (Novagen) was used to express TRX which consists of TRX-6Histag-Stag-6Histag. RNA markers, M. (C) Examination of the RNA binding by rCTS using the Hill model. The data are plotted as the fraction of bound RNA versus molar concentration of rCTS. The Hill coefficient and Kd were calculated by fitting a non-linear least squares regression model to the experimental data points using the GraphPad Prism 4 software. The Hill coefficient was 1.5, the apparent Kd was 63 nM. (D) Evaluation of contribution of Zn-knuckle structure on the RNA binding properties of CTS. EMSA was performed with wt and mutant form of rCTS (∼70 nM) in the presence of the same amount of 380 nt RNA.
3.3. The CTS of L1 ORF2p has a high non-sequence specific affinity to RNA
The binding affinity of CTS to ssRNA was analyzed by EMSA. In this experiment, a known amount of rCTS was titrated into a constant amount of tested RNA. To exclude a contribution of TRX or 6 Histag into RNA binding, we performed a control EMSA reaction in the presence of the purified TRX-6Histag-Stag-6Histag at the same conditions (Fig. 2B). In order to assess the percentage of RNA binding, band intensities were measured by densitometric scanning of unbound and bound RNA. The Kd value for binding of rCTS to RNA is apparently in the nanomolar range as could be visualized from the binding pattern on Fig. 2B. Based on the amount of free RNA in each lane, the dissociation constant value for rCTS bound to RNA appears in the range of 50–100 nM. We also examined RNA binding by rCTS applying the Hill binding model. The Hill coefficient was calculated by fitting a non-linear least squares regression model to the experimental data points using the GraphPad Prism 4 software (Fig. 2C). The best fit value for the Hill coefficient was 1.5, suggesting that more than one rCTS molecule bound to RNA under the experimental conditions. The apparent dissociation constant was calculated to be Kd = 63 nM. A discrete oligomerization of rCTS was observed when amounts of RNA and rCTS were increased in the reaction (SD1). The size of RNA tested, the presence of more than one binding site on RNA molecule and protein–protein interactions could contribute to multiply non-specific binding.
To study the RNA-binding specificity of rCTS, we replaced the 3′UTR L1 RNA in the binding assay with different RNA fragments in the size range of 100–500 nt, which were derived from the different locations within the L1 3′UTR and the pET32b plasmid DNA. We found that each of these RNA fragments had a CTS binding profile similar to that of the full length 3′UTR of L1 RNA (data not shown). These results demonstrated that CTS has a high affinity to RNA and possesses a broad RNA-binding specificity. The minimal size of RNA bound to rCTS was 100 nt under given experimental conditions.
Thus, the C-terminal sequence (1096–1275 aa) of L1 ORF2p binds to RNA with high affinity but without apparent sequence specificity in vitro.
3.4. The Zn-knuckle structure does not affect the RNA binding affinity level of the CTS of L1 ORF2p
To evaluate the contribution of the Zn-knuckle structure to the RNA binding affinity of CTS, four Cys in its aa sequence were replaced by Ser (Fig. 1C) using the GeneTailor mutagenesis system. This mutant form of CTS was expressed and purified as described above (Fig. 1C, SD1). We hypothesized that if Zn-knuckle structure affects RNA binding, then no binding by rCTS-mut will be detected at the observed Kd for rCTS. The RNA binding of rCTS-mut was tested in EMSA under the same conditions as wild type rCTS and both purified rCTS-mut and rCTS at ∼70 nM were incubated with same amount of 380 nt RNA (Fig. 2D). No significant loss in RNA binding activity was observed. These results demonstrated that the Zn-knuckle structure does not affect the RNA binding affinity level by CTS of L1 ORF2p. Unfortunately, we were not able to register any specific interactions of L1 ORF2p with L1 RNA, probably due to low sensitivity of the technique.
4. Discussion
In the present work, we have further elucidated the properties of the carboxy-terminal segment of the L1 ORF2 protein. In a series of experiments involving gel shift assay with a recombinant CTS and different nucleic acid templates, we demonstrated that the C-terminal 180 residues of L1 ORF2p are sufficient for a strong interaction with RNA in vitro, in an apparently non-specific manner. This RNA binding was cooperative and saturable, with an apparent dissociation constant of the low nanomolar range (Fig. 2B and C). Several factors could contribute to this observation: the size of the RNA molecule, a non-specific RNA binding activity of rCTS and/or protein–protein interactions. Importantly, comparable high affinity has been observed for mouse L1 ORF1 protein, which binds RNA with nanomolar affinity and little regard for nucleotide sequence in physiological concentrations of monovalent cation [4].
The high affinity of L1 ORF2p CTS to RNA suggests that it may play an important role in cDNA synthesis, nucleic acid interactions and in the formation of L1 RNP. We have previously demonstrated that L1 ORF2p reverse transcriptase alone is able to polymerize hundreds of nucleotides per template binding event and hypothesized that the unique positively charged C-domain of L1 ORF2 protein may be responsible for this phenomenon [9]. Such an intrinsic domain can provide a tight, but flexible binding with template, promoting long chain DNA synthesis without accessory proteins [25]. RNA-binding properties of CTS make it an attractive candidate for the role of an intramolecular processivity factor of L1 RT. Moreover, the RNA binding activity of L1 ORF2p CTS may also protect L1 RNA, stabilize L1 RNA-target primer complex and facilitate the first steps in TPRT during integration of L1 element into host genome.
At the same time our data suggest that the RNA-binding C-terminus of L1 ORF2p is liable for the cis-preference of the protein to its own RNA [10,26], as well as formation of an L1 ribonucleoprotein particle [11,13]. Apparently, the newly translated L1 ORF2 protein immediately binds to a native template with its high affinity C-terminal tail. One could not exclude that, not only non-specific binding occurs but also specific interactions of L1 ORF2p with cis-sites of L1 template. However, in our experiments with the L1 3′UTR RNA fragment, such binding either masked by non-specific interaction or cis-sites are not presented. It is also possible that CTS within the full length ORF2p can bind L1 RNA in a sequence-specific manner.
Finally, non-specific RNA-binding activity of L1 ORF2p CTS is most likely involved in a move of non-autonomous mobile elements, such as Alu and SVAs. These elements lack RT-encoded sequences and utilize RT activities provided by autonomous retrotransposons for their transposition. It is believed that Alu and SVA elements are mobilized by the L1 retrotransposition machinery via the TPRT mechanism [27,28]. The RNA-binding CTS of L1 ORF2p may directly participate in this process, capturing distant transcripts of non-autonomous elements for subsequent reverse transcription and insertion in the host genome.
We found no evidence for contribution of the Zn-knuckle structure in RNA recognition and binding. This is possible explained by the non-specific nature of electrostatic interactions between the basic residues of CTS involved and the RNA phosphate groups. Therefore, mutations in the CTS destroying the Zn-knuckle structure, did not significantly affect the affinity level to RNA because non-specific interactions occurred even in the absence of the Zn-knuckle structure.
Taking into consideration our data and the results of the cell culture-based retrotransposition assay where mutation of the zinc knuckle structure resulted in considerably decrease of L1 retrotransposition rates [17–19], diffuse nuclear localization of L1 RNP, reduced content of ORF2p in RNP and consequently a decrease in RT activity in L1 RNP [13], we would speculate that this structure may be primarily responsible for the specific protein–protein and cis-sites interactions as well as L1 RNP formation.
In conclusion, our report demonstrates for the first time RNA binding features of the CTS domain of the human L1 ORF2 protein. We hypothesise that the observed non-specific binding to RNA by the CTS domain is due to electrostatic interactions between the positively charged amino acid residues within this domain and the phosphate groups of the RNA. Without knowing the exact structure of the CTS, it is difficult to explain its interaction with RNA in a non-sequence specific manner and lack of binding with dsDNA. Thus, it would be very interesting to clarify the architecture of CTS, and the nature of its interaction with nucleic acids.
Acknowledgements
We wish to thank Dr. Isabella Bray, Dr. Elaine Kenny and Dr. Suzanne Miller-Delaney for proofreading of the manuscript. The work was supported by Science Foundation Ireland grant to O.P. (08/RFP/BIC1398), DCU Visitor Fellowship to V.S. The funders had no role in study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Olga Piskareva, Email: piskareo@gmail.com.
Vadim Schmatchenko, Email: vadim.schmatchenko@gmail.com.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2013.09.005.
Appendix. Supplementary materials
This document contains a section on 'Materials and methods'.
References
- 1.Goodier J.L., Kazazian H.H., Jr. Retrotransposition revisited: the rehabilitation and restraint of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Hohjoh H., Singer M.F. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin S.L., Bushman F.D. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolosha V.O., Martin S.L. High affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J. Biol. Chem. 2003;278:8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- 5.Feng Q., Moran J.V., Kazazian H.H., Jr., Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 6.Cost G., Boeke J. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 7.Mathias S.L., Scott A.F., Kazazian H.H., Jr., Boeke J.D., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 8.Piskareva O.A., Denmukhametova S.V., Schmatchenko V.V. Functional reverse transcriptase encoded by the human LINE-1 from baculovirus-infected insect cells. Protein Expr. Purif. 2003;28:125–130. doi: 10.1016/s1046-5928(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 9.Piskareva O., Schmatchenko V. DNA polymerization by the reverse transcriptase of the human L1 retrotransposon on its own template in vitro. FEBS Lett. 2006;580:661–668. doi: 10.1016/j.febslet.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 10.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulpa D.A., Moran J.V. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 12.Goodier J.L., Zhang L., Vetter M.R., Kazazian H.H., Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doucet A.J. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6(10) doi: 10.1371/journal.pgen.1001150. pii:e1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 16.Ostertag E.M., Kazazian H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 17.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 18.Lutz S.M., Vincent B.J., Kazazian H.H., Batzer M.A., Moran J.V. Allelic heterogeneity in LINE-1 retrotransposition activity. Am. J. Hum. Genet. 2003;73:1431–1437. doi: 10.1086/379744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farley A.H., Luning Prak E.T., Kazazian H.H., Jr. More active human L1 retrotransposons produce longer insertions. Nucleic Acids Res. 2004;32:502–510. doi: 10.1093/nar/gkh202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Champoux J.J. Roles of ribonuclease H in reverse transcription. In: Skalka A.M., Goff S.P., editors. Reverse Transcriptase. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 103–117. [Google Scholar]
- 21.Xiong Y, Eickbush T.H. Origin and evolution of retroelements based on their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik H.S., Burke W.D., Eickbush T.H. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 23.Wang L., Brown S.J. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terribilini M. RNABindR: a server for analyzing and predicting RNA-binding sites proteins. Nucleic Acids Res. 2007;35:W578–W584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andraos N., Tabor S., Richardson C.C. The highly processive DNA polymerase of bacteriophage T5. Role of the unique N and C termini. J. Biol. Chem. 2004;279:50609–50618. doi: 10.1074/jbc.M408428200. [DOI] [PubMed] [Google Scholar]
- 26.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 27.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 28.Ostertag E.M., Goodier J.L., Zhang Y., Kazazian H.H., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document contains a section on 'Materials and methods'.


