Abstract
The tetrasaccharide heparan sulfate (HS) mimetic PG545, a clinical anti-cancer candidate, is an inhibitor of the HS-degrading enzyme heparanase. The kinetics of heparanase inhibition by PG545 and three structural analogues were investigated to understand their modes of inhibition. The cholestanol aglycon of PG545 significantly increased affinity for heparanase and also modified the inhibition mode. For the tetrasaccharides, competitive inhibition was modified to parabolic competition by the addition of the cholestanol aglycon. For the trisaccharides, partial competitive inhibition was modified to parabolic competition. A schematic model to explain these findings is presented.
Keywords: Heparanase, PG545, Inhibitor, Kinetics, Cancer
Abbreviations: ECM, extracellular matrix; HS, heparan sulfate; HSPG, heparan sulfate proteoglycan; SE, standard error; SS, sum of squares; WST-1, 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate
Graphical Abstract
Highlights
-
•
Clinical candidate PG545 inhibits heparanase with parabolic competitive kinetics.
-
•
Analogues of this compound show different modes of heparanase inhibition.
-
•
The cholestanol aglycon of PG545 increases affinity for heparanase.
-
•
Tetrasaccharides have higher affinity for heparanase than trisaccharides.
-
•
A schematic model of inhibition by PG545 and analogues is proposed.
1. Introduction
Heparanase (HPSE) is an endo β-d-glucuronidase that cleaves heparan sulfate (HS) polymers in the extracellular matrix (ECM) and contributes to tissue remodelling. HS is an important structural and chemical component of the ECM where they are attached to core proteins to form HS-proteoglycans (HSPG) such as perlecan, syndecan and glypican. During tissue remodelling, the ECM is degraded by proteases, including matrix metalloproteinases, and heparanase. This breaks down the structure of the ECM and, additionally, the degradation of HS polymers also leads to the release of a variety of HS-binding cytokines including VEGF and FGF-2 which lead to altered physiological responses [1]. Heparanase is the only enzyme known to cleave HS in the ECM, and for this reason it is at the nexus of these structural and physiological transformations.
Malignant cancers co-opt the tissue remodelling functions of heparanase during two processes that are important to tumour growth and spread: angiogenesis and metastasis. Heparanase is important in the tissue remodelling and signalling involved in the generation of the new blood vessels required for the growth of tumours [2]. The ECM-degrading properties of this enzyme allow cancer cells to migrate through the ECM, a crucial step in the metastatic spread of cancer cells around the body to form new tumours [2]. The metastatic spread of tumours to parts of the body remote from the original tumour is a major cause of mortality from this disease [3].
Heparanase cleaves HS chains at specific glucuronic acid residues located within highly sulfated regions along the polymers. Interactions between the sulfate and carboxylate groups in these regions and patches of basic amino acid residues on the surface of heparanase coordinate the HS polymer and allow two glutamate residues in the catalytic site to hydrolyse the glycosidic bond. The carboxylate of the glucuronic acid at the point of cleavage and 2-N- and 6-O-sulfate groups on the glucosamine residues flanking it constitute a trisaccharide sequence proposed to be necessary for heparanase binding and catalysis [4,5]. Although no three dimensional structure of heparanase has been published, homology modelling using similar proteins has suggested that the binding region could interact with a larger sequence of four or five saccharides within the HS chain [6]. Recent experimental evidence also supports the notion that saccharide residues flanking the trisaccharide sequence may be involved in the binding of HS to heparanase [7].
Due to its importance in promoting tumour growth and metastatic spread, heparanase has been the target of numerous therapeutic development programs [8]. Two broad approaches characterise the design of heparanase inhibiting compounds: small molecules binding specifically to the catalytic site and HS mimetics that bind in a similar manner as substrate thus preventing access to the catalytic site. The identification of small molecule heparanase inhibitors has produced some interesting compounds [9–11] though none have advanced to clinical trials and further progress has been hampered by the lack of a structure for heparanase. In contrast, a number of HS mimetics have shown promise and four with heparanase inhibition activity have entered clinical trials: PI-88 [12,13], M402 [14], SST0001 [15] and PG545 [16–18].
Despite the interest in developing heparanase inhibitors, few have been subjected to a thorough analysis of their inhibition mode and, as a result, linear competitive inhibition of heparanase by many HS mimetics is assumed and not verified. Until recently this was partly due to the lack of a heparanase assay suitable to conduct such analyses, but with the publication of a simple colorimetric assay based on the substrate fondaparinux, such an assay is now available [19]. Of the HS mimetics in clinical use, PG545 is the most appropriate for kinetic examination because it is a single molecular entity. The study presented here has characterised the inhibition mode of PG545 and compared it with three structural analogues to improve our understanding of how these molecules bind to heparanase (Fig. 1). Such data will also lead to a better understanding of the active site of this important enzyme.
Fig. 1.
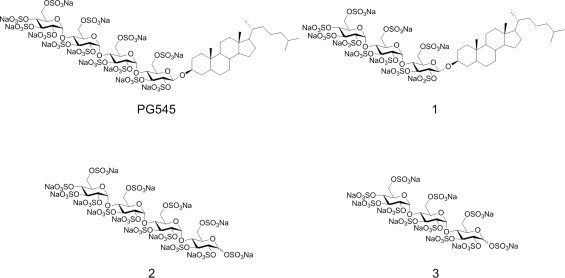
Structures of PG545 and the analogues examined here. Compound 1 is the trisaccharide analogue of PG545, while compounds 2 and 3 are the tetra- and trisaccharides sulfated at the site occupied by the cholestanol aglycon.
2. Materials and methods
PG545 and compound 1 were synthesised as previously reported [20,21]. Compound 2 and compound 3 were synthesised from maltotetraose and maltotriose respectively, which were purchased from Hayashibara Biochemical Laboratories, and sulfated according to the general procedure reported previously [21].
Recombinant human heparanase was expressed in insect cells [22] and then purified from the cell media into which it had been secreted according to a previously described method [19]. The tetrazolium salt WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) was obtained from Dojindo. Unless otherwise stated, all other reagents were obtained from Sigma–Aldrich.
2.1. Fondaparinux heparanase assays
Fondaparinux heparanase assays were conducted as described previously [19]. Assays were carried out in 96 well microplates (Costar 9018 EIA/RIA, Corning) pre-treated with a solution of 4% BSA in phosphate-buffered saline containing 0.05% Tween 20 (PBST) for 2 h at 37 °C. The plates were then washed three times with PBST and shaken dry. Pre-treated plates were stored at 4 °C for up to 2 weeks before use. Assays (100 μL) typically contained 40 mM sodium acetate buffer (pH 5.0), 0.25 nM heparanase and varying concentrations of fondaparinux and study compounds. The assays were prepared minus fondaparinux and allowed to equilibrate for 10 min before adding the substrate to start catalysis. Incubation at 37 °C for 18 h was followed by development with 100 μL of 1.69 mM WST-1 solution in 0.1 M NaOH at 60 °C for 1 h after which the absorbance at 584 nm was measured (Fluostar Optima platereader, BMG Labtech). For IC50 analysis, data were converted into % inhibition values by comparing to control assays containing no study compound.
2.2. Ultrafiltration heparanase assays
Heparanase activity was also measured using an ultrafiltration assay based on a previously described method [23]. Reactions were set up in 1.5 mL microtubes in a volume of 200 μL containing 40 mM sodium acetate buffer (pH 5.0), 0.005% Tween 20 (w/v), 4 nM heparanase, 5 μM [3H]-HS and various concentrations of study compounds. Initially, all components except the [3H]-HS substrate were allowed to equilibrate for 10 min. The assays were then started by adding [3H]-HS and immediately 40 μL was taken, quenched with 160 μL of 10 mM phosphate (pH 7.0) and the 200 μL transferred to a Microcon YM-10 ultrafiltration device (Millipore) which was centrifuged at 14,000g for 5 min. The solution that passed through the membrane (filtrate) was retained. This sample was considered the t0 sample. The assays were aliquoted into 3 tubes of 40 μL each and these were allowed to react at 37 °C for 18 h. The filtration step was then repeated after quenching with 160 μL of 10 mM phosphate (pH 7.0). The t0 filtrate and the three filtrate samples taken after 18 h (t18h) were counted for 3H using Optiphase HiSafe 2 scintillant (Perkin Elmer) in a Microbeta 1450 counter (Wallac, Perkin Elmer). The difference between the t0 and the averaged t18h samples gave the amount of heparanase activity. Relative inhibition assays were run with a heparanase standard assay which was identical in composition except no inhibitor was present, and the amount of heparanase inhibition in the other assays was determined by comparison with this standard.
2.3. Data analysis
To determine IC50 values, % inhibition data were fitted to the four parameter logistic equation. Kinetic constants, apparent Km and apparent Vmax, were determined by fitting v versus [S] data to the Michaelis–Menten equation. Curve fitting was performed by nonlinear regression using Prism 6.02 software (GraphPad Software) and R2 and sum of squares (SS) analysis used to compare goodness of fit of different models. Estimated kinetic parameters are quoted with standard errors (SE).
3. Results
The interaction of PG545 with the active site of heparanase was investigated by measuring the inhibition of this enzyme by PG545 and three structural analogues using two heparanase assays that have different substrates and measurement methods. PG545 and its three analogues all inhibit heparanase activity in a concentration-dependant manner when measured using the fondaparinux assay (Fig. 2A) or the ultrafiltration assay (Fig. 2B).
Fig. 2.
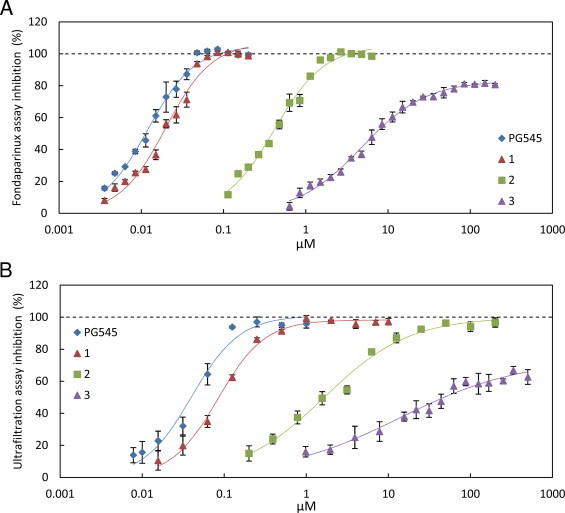
Inhibition of heparanase by PG545 and analogues measured using the fondaparinux assay (panel A) and the ultrafiltration assay (panel B). Fondaparinux assays contained 40 mM sodium acetate buffer (pH 5.0), 0.25 nM heparanase, 100 μM fondaparinux and the indicated concentrations of inhibitors. Ultrafiltration assay conditions as per Materials and methods. Data are means of 4 measurements (panel A) or 3 measurements (panel B) and error bars are standard errors.
In terms of affinity for heparanase, PG545 was the tightest binding followed by 1, 2 and 3 which had the lowest affinity (IC50 data in Table 1). This affinity ranking indicates that tetrasaccharide based compounds (PG545 and 2) have higher affinity than their corresponding trisaccharide compounds. Also, the cholestanol-functionalised compounds (PG545 and 1) have higher affinity than their corresponding tetra- and trisaccharides. The increased affinity conferred by the cholestanol group suggests that this hydrophobic modification allows PG545 and 1 bind to a hydrophobic patch or pocket near or within the heparanase active site in addition to binding to the basic amino acid residues involved in substrate binding. The IC50 values for compound 2 show reasonable agreement with a previously published IC50 value of 10 μg/mL (4700 nM) given the different assays and substrates [24].
Table 1.
Heparanase IC50 values for PG545 and analogues measured using fondaparinux and ultrafiltration assays. Errors are SE.
| Compound | Fondaparinux assay | Ultrafiltration assay |
|---|---|---|
| PG545 | 12.0 ± 0.7 nM | 40.1 ± 6.0 nM |
| 1 | 19.8 ± 1.7 nM | 84.0 ± 4.5 nM |
| 2 | 416 ± 25 nM | 1650 ± 220 nM |
| 3 | 4820 ± 230 nM | 13,500 ± 4100 nM |
All inhibitors except compound 3 showed complete inhibition of heparanase at saturating concentrations. In contrast, 3 inhibited approximately 80% of heparanase activity at concentrations approximately 40 times the measured IC50 values in both assays. Competitive inhibition of heparanase by HS mimetics is typically assumed, thus incomplete inhibition, as observed with 3, is unexpected.
Given the significant differences in affinity and the incomplete inhibition by compound 3, the inhibition modes of PG545 and its analogues were investigated further. The fondaparinux assay was used for this analysis because it utilises an homogenous substrate which has only one heparanase cleavage site per molecule. While this assay does not measure initial reaction velocities, an assumption inherent in the models used to characterise the data, it does conform to Michaelis–Menten kinetics [19]. The Km of 3.6 ± 0.5 μM was measured for the interaction of fondaparinux with heparanase and used in subsequent analyses. The value is lower than previously reported which reflects the improved handling of this assay since its development and initial publication [19].
Double reciprocal analysis of a matrix of fondaparinux heparanase assays at a range of substrate and PG545 concentrations indicates that this compound is a competitive inhibitor of heparanase (Fig. 3A). Interestingly, when the slopes of the double reciprocal plot are replotted against inhibitor concentration, the resulting curved response indicates that PG545 is a parabolic competitive inhibitor (Fig. 3B). These data were compared to the parabolic competitive inhibition model [25].
| (1) |
Fig. 3.
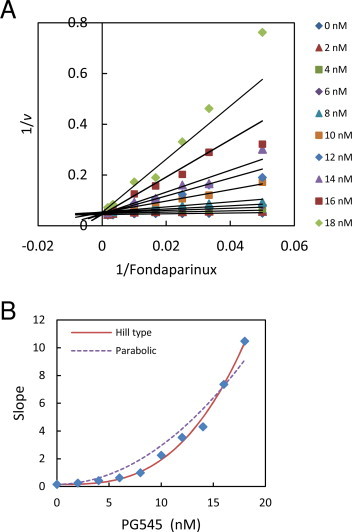
Double reciprocal analysis of PG545 inhibition of heparanase (panel A). Fondaparinux assays conducted as per Section 2. Data are means of 2 measurements. Lines were generated from the apparent Km and Vmax values. The slopes from panel A were replotted as a function of PG545 concentration in panel B. The solid line in panel B is the fit of Eq. (4) to the data (R2 = 0.993, SS = 0.735) and the dotted line is the fit of Eq. (2) (R2 = 0.950, SS = 5.450) for comparison.
Eq. (1) was converted into the following form for analysis of the slope data (Fig. 3B, dotted line, R2 = 0.950, SS = 5.450).
| (2) |
A better fit to the data, however, was achieved using the Hill-type model of Cao et al. [26].
| (3) |
This equation was also rearranged for analysis of the slope data (Fig. 3B, solid line, R2 = 0.993, SS = 0.735).
| (4) |
The Ki and Hill coefficient (n) determined from the curve fit were 4.44 ± 0.30 nM and 2.98 ± 0.16 respectively.
The kinetics of the three PG545 analogues were analysed in a similar manner. Compound 1, which shares the cholestanol functional group with PG545, shows similar parabolic competitive inhibition (Fig. 4A and B). Once again, the Hill-type inhibition model (R2 = 0.964, SS = 0.972) showed a better fit to the data than the conventional parabolic competitive model (R2 = 0.880, SS = 3.281). The resulting Ki and n determined from the curve fit were 9.82 ± 1.12 nM and 3.62 ± 0.44 respectively.
Fig. 4.
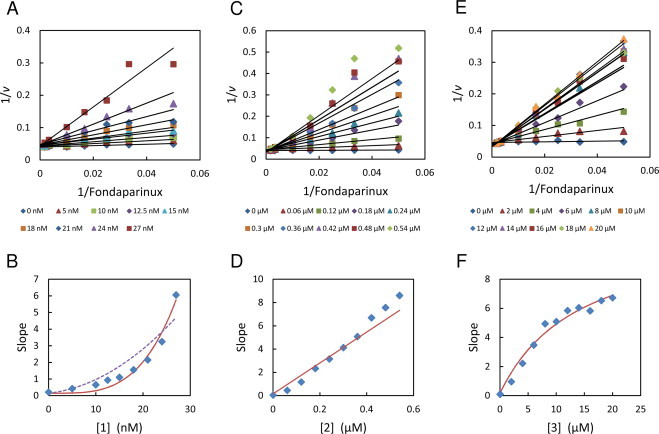
Double reciprocal analysis of heparanase inhibition by compounds 1, 2 and 3 (panels A, C and E). Fondaparinux assays conducted as per Section 2. Data are means of 2 measurements. Lines were generated from the apparent Km and Vmax values. The slopes from panels A, C and E were replotted as a function of inhibitor concentration in panels B, D and F respectively. The solid line in panel B is the fit of Eq. (4) to the slopes data (R2 = 0.964, SS = 0.972) and the dotted line is the fit of Eq. (2) (R2 = 0.880, SS = 3.281) for comparison. The solid line in panel D represents the global fit of the competitive inhibitor rate Eq. (5) to the velocity data set. The solid line in panel F is the fit of Eq. (6) to the slopes data.
The two analogues without the cholestanol group, compounds 2 and 3, showed different kinetics. The tetrasaccharide (2) showed linear competitive inhibition of heparanase (Fig. 4C and D) whereas the trisaccharide (3) showed partial competitive inhibition as indicated by the hyperbolic response of the double reciprocal slopes when plotted against inhibitor concentration (Fig. 4E and F). The competitive inhibition rate Eq. (5) was fitted to velocity data set for compound 2 using global nonlinear regression. From this fit, the Ki of heparanase inhibition by compound 2 was estimated to be 12.4 ± 0.4 nM.
| (5) |
The conclusion that compound 3 is a partial competitive inhibitor of heparanase is supported by the earlier observation that it failed to completely inhibit the enzyme even as inhibitor concentration increased to approximately 40 times the IC50: such behaviour is characteristic of partial competitive inhibitors. Because partial competitive inhibitors can bind to both free enzyme and enzyme-substrate complex, the relative affinities of compound 3 for these two forms of heparanase were estimated by fitting a modified form of the partial competitive inhibitor (Eq. (6)) to the data in Fig. 4F.
| (6) |
From the curve fit the Ki for compound 3 was estimated to be 197 ± 27 nM and the α value 2.8 ± 1.1 indicating that this compound has considerably higher affinity for the unoccupied heparanase active site compared to the substrate bound active site.
4. Discussion
Heparanase is an important protein involved in cancer spread and malignancy that has been the target of drug development programs since its discovery. A variety of HS mimetics have been employed as inhibitors of this enzyme, both experimentally and in clinical trials. PG545 has already entered cancer clinical trials and is expected to re-commence trials in the near future. Understanding the binding modes of HS mimetics to heparanase is, therefore, of considerable importance. The lessons learned from studying the interaction of heparanase with these inhibitors and its substrate may also be applicable to other important enzymes that have polymeric substrates.
PG545 and its three structural analogues possess three distinct modes of heparanase inhibition. This diversity is unusual considering, firstly, their similarity and, secondly, that heparanase is thought to exist as a heterodimer with one active site, thus precluding interaction between active sites. A structure of heparanase has not been published although information about the three-dimensional arrangement of important parts of the protein has been gleaned from comparative modelling of the sequence based upon the structures of related proteins [27,6,28]. The enzymatic domain of heparanase comprises an (α/β)8 TIM-barrel with two catalytic glutamate residues located at the surface, close to the rotational axis of this motif. Basic amino acid residues, which are involved in HS binding, are located in two patches either side of the active site.
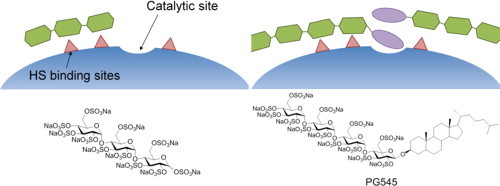
Although the exact positions and distances between these residues is unknown, heparanase interacts with at least three and possibly five saccharide units of substrate HS polymer indicating that the substrate binding region is quite large. Given the three modes of inhibition kinetics observed here and our knowledge of the substrate-binding region, a schematic model of the binding of these ligands to heparanase can be proposed (Fig. 5). Conventional competitive HS mimetic inhibitors, such as compound 2, are able to bridge the two patches of HS binding basic residues and, although they probably do not have any inherent affinity for the catalytic site, they obstruct it preventing substrate from docking and completely inhibit catalysis (Fig. 5A). If the HS mimetic is not sufficiently long enough to bridge the two binding patches, it may bind to only one and, leaving the other binding patch and the catalytic site unobstructed, allow catalysis to proceed even when the population of enzyme active sites is fully occupied by inhibitors (Fig. 5B). It is this behaviour – partial competitive inhibition – that was observed with the shorter of the two-saccharide chain analogues, the trisaccharide 3. Similar behaviour has been seen previously with sulfated di- and trisaccharide inhibitors of heparanase [29]. These latter compounds have a mannose backbone in contrast to the glucose based compounds presented here, indicating that this inhibition behaviour is not specific to the current compounds and it is likely, therefore, that the same behaviour would be observed with other similar sized HS-mimetics.
Fig. 5.
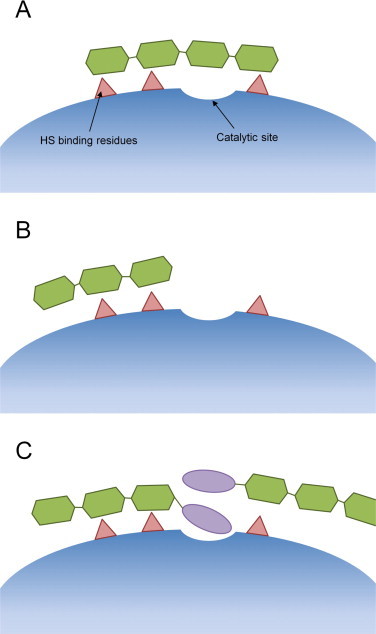
Schematic model of the three modes of heparanase inhibition displayed by PG545 and its three analogues. Competitive inhibitors, such as compound 2, straddle the entire substrate binding region and completely occlude the catalytic site (panel A). Partial competitive inhibitors which are not large enough to bridge the two HS binding patches, such as compound 3, allow substrate access to the catalytic site (panel B). Parabolic competitive inhibitors, such as PG545 and compound 1, bind two molecules, the second molecule binding with higher affinity (panel C).
In contrast to compounds 2 and 3, PG545 and its trisaccharide analogue 1 inhibit heparanase with parabolic competitive kinetics. Several prerequisites must be accommodated when formulating a mechanistic model to describe this behaviour. Firstly, the sulfated trisaccharide is transformed by the addition of a cholestanol group from the partial competitive inhibitor 3 into a higher affinity ligand that completely inhibits heparanase at 100% occupancy. This suggests that the cholestanol group interacts with, or obscures, the catalytic site and that this interaction contributes significantly to the affinity of compound 1. Homology modelling has identified a channel of hydrophobic residues surrounding the catalytic residues forming a possible binding site for the cholestanol group [28].
Secondly, parabolic competitive inhibitors bind to two sites on the enzyme and each interaction is sufficient to completely prevent catalysis. The two patches of HS binding residues flanking the catalytic site of heparanase are likely candidates with molecules of PG545 or 1 binding to these patches in such a way that one or both of the cholestanol groups obscures the catalytic site.
Thirdly, the binding of the first parabolic competitive inhibitor molecule increases the affinity of the subsequent molecule's interaction with the enzyme. This could result from a conformational change in heparanase induced by the binding of the cholestanol group which increases the affinity between the transiently modified second site and another inhibitor molecule. An alternative explanation is that the bound cholestanol group of the first inhibitor molecule serves as a binding site for the same moiety of the second inhibitor and once bound, the first cholestanol group becomes buried and inaccessible to solvent (Fig. 5C). Such an arrangement, given the hydrophobic nature of this functional group, is probably energetically favourable. This latter explanation also provides a mechanism to explain the increased affinity for the second inhibitor's binding and it is, therefore, considered the more likely of the two.
Parabolic competitive and partial competitive are generally quite rare modes of enzyme inhibition. Even rarer and more interesting is the finding that structurally related compounds can show such a range of divergent modes of enzyme inhibition. This discovery highlights the added complexity of an interaction between an enzyme and a polymeric substrate. The absence of a three dimensional heparanase structure, long lamented by heparanase researchers, continues to hamper our understanding of the interactions between heparanase and substrate. This gap in our knowledge presents difficulties to those attempting to target this enzyme using HS mimetics, an approach which has, nevertheless, shown some promise with several of these compounds entering clinical trials for the treatment of cancer. The improved understanding of the interaction of heparanase with HS mimetics gained from the data presented here can only increase our ability to construct more efficient inhibitors of this important enzyme.
Conflict of interest
E.H. and P.H. are current employees of Progen Pharmaceuticals. K.D. was previously employed by Progen Pharmaceuticals and is currently a paid consultant. I.B. was previously employed by Progen Pharmaceuticals.
Funding source
This study was funded by Progen Pharmaceuticals. There were no external funding sources.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Vlodavsky I, Miao HQ, Medalion B, Danagher P, Ron D. Involvement of heparan sulfate and related molecules in sequestration and growth promoting activity of fibroblast growth factor. Cancer Metastasis Rev. 1996;15:177–186. doi: 10.1007/BF00437470. [DOI] [PubMed] [Google Scholar]
- 2.Vlodavsky I, Elkin M, Abboud-Jarrous G, Levi-Adam F, Fuks L, Shafat I, Ilan N. Heparanase: one molecule with multiple functions in cancer progression. Connect Tissue Res. 2008;49:207–210. doi: 10.1080/03008200802143281. [DOI] [PubMed] [Google Scholar]
- 3.Mina LA, Sledge GW. Rethinking the metastatic cascade as a therapeutic target. Nat Rev Clin Oncol. 2011;8:325–332. doi: 10.1038/nrclinonc.2011.59. [DOI] [PubMed] [Google Scholar]
- 4.Pikas DS, Li JP, Vlodavsky I, Lindahl U. Substrate specificity of heparanases from human hepatoma and platelets. J Biol Chem. 1998;273:18770–18777. doi: 10.1074/jbc.273.30.18770. [DOI] [PubMed] [Google Scholar]
- 5.Okada Y, Yamada S, Toyoshima M, Dong J, Nakajima M, Sugahara K. Structural recognition by recombinant human heparanase that plays critical roles in tumor metastasis. Hierarchical sulfate groups with different effects and the essential target disulfated trisaccharide sequence. J Biol Chem. 2002;277:42488–42495. doi: 10.1074/jbc.M206510200. [DOI] [PubMed] [Google Scholar]
- 6.Gandhi NS, Freeman C, Parish CR, Mancera RL. Computational analyses of the catalytic and heparin-binding sites and their interactions with glycosaminoglycans in glycoside hydrolase family 79 endo-β-d-glucuronidase (heparanase) Glycobiology. 2012;22:35–55. doi: 10.1093/glycob/cwr095. [DOI] [PubMed] [Google Scholar]
- 7.Peterson SB, Liu J. Unraveling the specificity of heparanase utilizing synthetic substrates. J Biol Chem. 2010;285:14504–14513. doi: 10.1074/jbc.M110.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br J Pharmacol. 2007;151:1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courtney SM, Hay PA, Buck RT, Colville CS, Porter DW, Scopes DI, Pollard FC, Page MJ, Bennett JM, Hircock ML, McKenzie EA, Stubberfield CR, Turner PR. 2,3-Dihydro-1,3-dioxo-1H-isoindole-5-carboxylic acid derivatives: a novel class of small molecule heparanase inhibitors. Bioorg Med Chem Lett. 2004;14:3269–3273. doi: 10.1016/j.bmcl.2004.03.086. [DOI] [PubMed] [Google Scholar]
- 10.Ishida K, Hirai G, Murakami K, Teruya T, Simizu S, Sodeoka M, Osada H. Structure-based design of a selective heparanase inhibitor as an antimetastatic agent. Mol Cancer Ther. 2004;3:1069–1077. [PubMed] [Google Scholar]
- 11.Xu YJ, Miao HQ, Pan W, Navarro EC, Tonra JR, Mitelman S, Camara MM, Deevi DS, Kiselyov AS, Kussie P, Wong WC, Liu H. N-(4-{[4-(1H-Benzoimidazol-2-yl)-arylamino]-methyl}-phenyl)-benzamide derivatives as small molecule heparanase inhibitors. Bioorg Med Chem Lett. 2006;16:404–408. doi: 10.1016/j.bmcl.2005.09.070. [DOI] [PubMed] [Google Scholar]
- 12.Basche M, Gustafson DL, Holden SN, O’Bryant CL, Gore L, Witta S, Schultz MK, Morrow M, Levin A, Creese BR, Kangas M, Roberts K, Nguyen T, Davis K, Addison RS, Moore JC, Eckhardt SG. A phase I biological and pharmacologic study of the heparanase inhibitor PI-88 in patients with advanced solid tumors. Clin Cancer Res. 2006;12:5471–5480. doi: 10.1158/1078-0432.CCR-05-2423. [DOI] [PubMed] [Google Scholar]
- 13.Liu CJ, Lee PH, Lin DY, Wu CC, Jeng LB, Lin PW, Mok KT, Lee WC, Yeh HZ, Ho MC, Yang SS, Lee CC, Yu MC, Hu RH, Peng CY, Lai KL, Chang SS, Chen PJ. Heparanase inhibitor PI-88 as adjuvant therapy for hepatocellular carcinoma after curative resection: a randomized phase II trial for safety and optimal dosage. J Hepatol. 2009;50:958–968. doi: 10.1016/j.jhep.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Roy S, Cochran E, Zouaoui R, Chu CL, Duffner J, Zhao G, Smith S, Galcheva-Gargova Z, Karlgren J, Dussault N, Kwan RY, Moy E, Barnes M, Long A, Honan C, Qi YW, Shriver Z, Ganguly T, Schultes B, Venkataraman G, Kishimoto TK. M402, a novel heparan sulfate mimetic, targets multiple pathways implicated in tumor progression and metastasis. PLoS One. 2011;6:e21106. doi: 10.1371/journal.pone.0021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie JP, Ramani VC, Ren Y, Naggi A, Torri G, Casu B, Penco S, Pisano C, Carminati P, Tortoreto M, Zunino F, Vlodavsky I, Sanderson RD, Yang Y. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin Cancer Res. 2011;17:1382–1393. doi: 10.1158/1078-0432.CCR-10-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dredge K, Hammond E, Davis K, Li CP, Liu L, Johnstone K, Handley P, Wimmer N, Gonda TJ, Gautam A, Ferro V, Bytheway I. The PG500 series: novel heparan sulfate mimetics as potent angiogenesis and heparanase inhibitors for cancer therapy. Invest New Drugs. 2010;28:276–283. doi: 10.1007/s10637-009-9245-5. [DOI] [PubMed] [Google Scholar]
- 17.Dredge K, Hammond E, Handley P, Gonda TJ, Smith MT, Vincent C, Brandt R, Ferro V, Bytheway I. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br J Cancer. 2011;104:635–642. doi: 10.1038/bjc.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond E, Brandt R, Dredge K. PG545, a heparan sulfate mimetic, reduces heparanase expression in vivo, blocks spontaneous metastases and enhances overall survival in the 4T1 breast carcinoma model. PLoS One. 2012;7:e52175. doi: 10.1371/journal.pone.0052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond E, Li CP, Ferro V. Development of a colorimetric assay for heparanase activity suitable for kinetic analysis and inhibitor screening. Anal Biochem. 2010;396:112–116. doi: 10.1016/j.ab.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Ferro V, Liu L, Johnstone KD, Wimmer N, Karoli T, Handley P, Rowley J, Dredge K, Li CP, Hammond E, Davis K, Sarimaa L, Harenberg J, Bytheway I. Discovery of PG545: a highly potent and simultaneous inhibitor of angiogenesis, tumor growth, and metastasis. J Med Chem. 2012;55:3804–3813. doi: 10.1021/jm201708h. [DOI] [PubMed] [Google Scholar]
- 21.Ferro V, Karoli T, Liu L, Handley PN, Johnstone KD, Wimmer N and Hammond ET (2009) Novel sulfated oligosaccharide derivatives. WO/2009/049370.
- 22.McKenzie E, Young K, Hircock M, Bennett J, Bhaman M, Felix R, Turner P, Stamps A, McMillan D, Saville G, Ng S, Mason S, Snell D, Schofield D, Gong H, Townsend R, Gallagher J, Page M, Parekh R, Stubberfield C. Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. Biochem J. 2003;373:423–435. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karoli T, Liu L, Fairweather JK, Hammond E, Li CP, Cochran S, Bergefall K, Trybala E, Addison RS, Ferro V. Synthesis, biological activity, and preliminary pharmacokinetic evaluation of analogues of a phosphosulfomannan angiogenesis inhibitor (PI-88) J Med Chem. 2005;48:8229–8236. doi: 10.1021/jm050618p. [DOI] [PubMed] [Google Scholar]
- 24.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- 25.Leskovac V. Kluwer; New York: 2003. Comprehensive Enzyme Kinetics. [Google Scholar]
- 26.Cao R, Zeidan AA, Rådström P, van Niel EW. Inhibition kinetics of catabolic dehydrogenases by elevated moieties of ATP and ADP – implication for a new regulation mechanism in Lactococcus lactis. FEBS J. 2010;277:1843–1852. doi: 10.1111/j.1742-4658.2010.07601.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z, Bates M, Madura JD. Structure modeling, ligand binding, and binding affinity calculation (LR-MM-PBSA) of human heparanase for inhibition and drug design. Proteins. 2006;65:580–592. doi: 10.1002/prot.21065. [DOI] [PubMed] [Google Scholar]
- 28.Sapay N, Cabannes E, Petitou M, Imberty A. Molecular model of human heparanase with proposed binding mode of a heparan sulfate oligosaccharide and catalytic amino acids. Biopolymers. 2012;97:21–34. doi: 10.1002/bip.21696. [DOI] [PubMed] [Google Scholar]
- 29.Fairweather JK, Hammond E, Johnstone KD, Ferro V. Synthesis and heparanase inhibitory activity of sulfated mannooligosaccharides related to the antiangiogenic agent PI-88. Bioorg Med Chem. 2008;16:699–709. doi: 10.1016/j.bmc.2007.10.044. [DOI] [PubMed] [Google Scholar]


