Abstract
The cellulosome is a large extracellular multi-enzyme complex that facilitates the efficient hydrolysis and degradation of crystalline cellulosic substrates. During the course of our studies on the cellulosome of the rumen bacterium Ruminococcus flavefaciens, we focused on the critical ScaA dockerin (ScaADoc), the unique dockerin that incorporates the primary enzyme-integrating ScaA scaffoldin into the cohesin-bearing ScaB adaptor scaffoldin. In the absence of a high-resolution structure of the ScaADoc module, we generated a computational model, and, upon its analysis, we were surprised to discover a putative stacking interaction between an N-terminal Trp and a C-terminal Pro, which we termed intramolecular clasp. In order to verify the existence of such an interaction, these residues were mutated to alanine. Circular dichroism spectroscopy, intrinsic tryptophan and ANS fluorescence, and NMR spectroscopy indicated that mutation of these residues has a destabilizing effect on the functional integrity of the Ca2+-bound form of ScaADoc. Analysis of recently determined dockerin structures from other species revealed the presence of other well-defined intramolecular clasps, which consist of different types of interactions between selected residues at the dockerin termini. We propose that this thematic interaction may represent a major distinctive structural feature of the dockerin module.
Keywords: Stacking interaction, Protein stability, Scaffoldin, Cohesin
Abbreviations: ANS, 8-anilino-1-naphthalenesulfonate; CBM, carbohydrate-binding module family 3a from C. thermocellum; Cc, Clostridium cellulolyticum; Coh, cohesin; Ct, Clostridium thermocellum; Doc, dockerin; cELISA, competitive enzyme-linked interaction assay; HBS, hepes-buffered saline; IPTG, isopropyl-1-thio-β-d-galactoside; TMB, 3,3′,5,5′-tetramethylbenzidine; Xyn, xylanase T6 from Geobacillus stearothemophilus
Graphical abstract
Highlights
-
•
A structural model for the Ruminococcus flavefaciens ScaA dockerin is proposed.
-
•
A stacking interaction between N- and C-terminal residues was derived from the model.
-
•
Mutations of putative interacting residues resulted in reduced stability and binding.
-
•
Similar intramodular “clasp” interactions were observed in other dockerin structures.
1. Introduction
Herbivores and other mammals, including humans, benefit from symbiosis with cellulolytic anaerobic bacteria that populate their gastrointestinal tract [1]. In fact, in herbivores these bacteria are responsible for the degradation plant-cell wall polysaccharides, which cannot be achieved alone by the host enzymes. The multi-enzyme molecular machine used by anaerobic bacteria to efficiently degrade cellulose and other plant cell-wall polysaccharides is termed the cellulosome [2]. Since the discovery of cellulosome, dockerin and cohesin modules have been shown to form calcium (Ca2+)-dependent heterocomplexes that incorporate the various cellulolytic enzymes onto a non-catalytic subunit, coined scaffoldin [3]. The scaffoldin is attached to the bacterial cell via anchoring proteins and, in most cases, to its substrate via a carbohydrate-binding module (CBM).
The cellulosome of cow rumen bacteria Ruminococcus flavefaciens is one of the most elaborate described thus far [4,5]. In particular, the cellulosome is composed of three major cohesin-containing non-catalytic scaffoldin subunits (ScaA, ScaB and ScaC), which are encoded by linked genes within the sca gene cluster [6,7]. The ScaA dockerin module (ScaADoc) serves an important role in integrating the cellulases and other carbohydrate-active enzymes onto the central ScaB scaffoldin via binding to ScaB cohesins. This adaptor scaffoldin subunit is attached to the cell surface through its C-terminal dockerin module binding to the single cohesin module of ScaE, which is covalently attached to the peptidoglycan of the bacterial cell surface [8]. As opposed to other cellulosomes that have been characterized, the R. flavefaciens cellulosome appears to rely on the CBMs of a separate cell-associated protein, termed CttA, for binding to cellulosic substrates, rather than a defined, centralized scaffoldin-borne CBM.
Due to its importance in the assembly of the cellulosome in R. flavefaciens and as a follow-up to our previous studies describing its Ca2+-binding properties [9] we focused on ScaADoc, the critically important and unique dockerin of this species (the single member of group 5 according to a newly defined dockerin classification system [10]). In the absence of a three-dimensional structure, a structural model of ScaADoc was created using the I-TASSER server [11,12] based on existing crystal structures, from which we identified a putative intramolecular stacking interaction between a tryptophan and a proline residue located at the N- and C-termini, respectively. To examine the significance of this putative interaction, we proceeded to investigate the impact of alanine mutation of these residues on the Ca2+-binding properties, structural stability, and ScaB cohesin recognition. The results indicated the importance of the intramolecular clasp on all three properties. The formation of intramolecular clasps is further supported by their detection in known cellulosomal and non-cellulosomal dockerin crystal structures, derived from other bacteria. The importance of this interaction for dockerin structure and stability is discussed along with its implications on cellulosome assembly and function.
2. Materials and methods
2.1. I-TASSER modeling
The ScaA dcokerin structural model (Fig. 1A) was produced using the I-TASSER online server http://zhanglab.ccmb.med.umich.edu/I-TASSER [12,13]. The following dockerin module structures were used to build the model: 2CCL:B, 2VN6:B, 2B59:B, 2CCL:A. 1DAV and 1DAQ templates were precluded from threading by a restraint file, since the latter structures were solved by NMR and may not properly represent the dockerin structure, based on other available dockerin structures from the complexes.
Fig. 1.
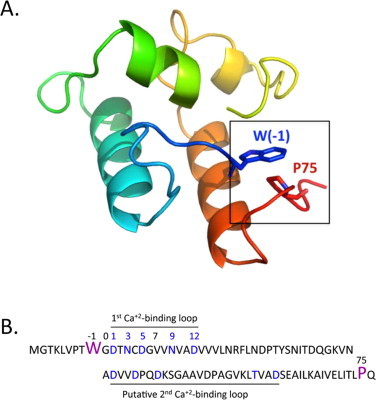
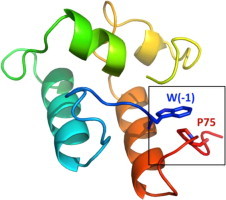
Intramolecular clasp between the N- and C-termini of R. flavefaciens ScaADoc. (A) Model of ScaADoc produced by the I-TASSER server, represented in Pymol (version 1.0r 1, Delano Scientific, LLC). (B) Sequence of the recombinant ScaADoc protein used in this experiment. Residues are numbered relative to the highly conserved glycine (designated 0), which is positioned adjacent to the initial calcium-binding aspartate (D1) of the F-hand motif. W(−1) thus interacts with P75, forming a putative intramolecular clasp in the ScaADoc module.
2.2. Cloning and mutagenesis
A hexahistidine-ScaADoc fusion construct was encoded by the ScaADoc gene inserted between NcoI and XhoI restriction sites of the pET28a plasmid [9]. All the mutants were prepared according to QuickChange site-direct mutagenesis protocol (Stratagen, La Jolla, CA). For the purposes of the present work, we define position 0 (zero) in ScaADoc as the highly conserved glycine that precedes the first coordinating aspartate residue of the initial 12-residue Ca2+-binding motif (Fig. 1B), which is highly conserved among dockerins from various species [14–18] and represents the traditional start point of the dockerin sequence [19]. Consequently, residues (not strictly conserved), positioned before the conserved glycine, will receive negative values. In order to prepare the mutant clones, two complementary primers containing the desired mutation in the middle were used: P75A; W(−1)A and their reverse complements. The double mutant was produced in a sequential manner, in which one mutant served as a template for the subsequent one. To exclude the possibility that more mutations were generated outside of the sequenced region, the mutated module was extracted and then recloned into the original cassette.
2.3. Dockerin expression and purification
All dockerin constructs were expressed in Escherichia coli BL21 cells and purified via a nickel-affinity chromatography similar to that previously described [9]. Proteins were quantified using either absorption spectra or amino acid analysis and stored either in 50% glycerol at −20 °C or at −80 °C after freezing in liquid nitrogen. For ScaADoc mutants lacking a Trp (i.e., W(−1)A and W(−1)A/P75A) the concentration was measured using the Quant-iT™ Protein Assay Kit (Invitrogen, Eugene, OR), which were comparable with that of the wild-type ScaADoc construct. ScaB cohesin 7 was prepared as described previously [9].
2.4. Circular dichroism
CD measurements were performed and analyzed as described previously [9]. Briefly, far-UV spectra (260 to 190 nm) were collected on an Applied Photophysics Chirascan Spectropolarimeter using a 0.1-cm path-length quartz cuvette. All proteins were measured in the absence and presence of 3 mM CaCl2 at 25 °C.
2.5. NMR spectroscopy
Uniformly-labeled 15N-W(−1)A/P75A-ScaADoc was expressed and purified as previously described for the wild-type ScaADoc construct [9] and reconstituted as a 0.1 mM protein solution in 20 mM Tris–HCl, pH 7.5. 2D 1H–15N HSQC experiments were used to assess the response of the W(−1)A mutation to addition of 500 μM Ca2+. NMR spectra were processed using NMRPipe [20] and analyzed in Sparky [21].
2.6. Thermal unfolding monitored by ANS fluorescence
Thermal denaturation experiments using ANS as a fluorescent probe were performed as previously described [22] with the following modifications. An excitation wavelength of 350 nm was used and emission spectra were recorded over a range from 400 to 550 nm on a JHY Spectrofluorometer (Horiba, Japan). Solutions (20 μM) of the wild-type, W(−1)A, P75A, and W(−1)A/P75A ScaADoc proteins containing 20 mM Tris–HCl, pH 7.5, 50 mM NaCl, 2 mM CaCl2 and 300 μM ANS were subjected to incremental changes in the temperature by 5 °C, starting from 25 °C, up to a maximum of 75 °C. Protein solutions were equilibrated for 3 min prior to data collection. The relative intensities at 550 nm and 400 nm were calculated and plotted as a function of the temperature, and were fitted to a one-site model using Origin 7.0 [23].
2.7. Tryptophan fluorescence
Intrinsic tryptophan fluorescence was used to follow the changes in the environment of tryptophan (in ScaADoc Trp9 is located close to the beginning of the first Ca2+-binding loop). Spectral scans were measured on a Cary Eclipse fluorescence spectrometer (Varian, Victoria, Australia) with 10 μM protein in HBS, pH 7.0, at room temperature in a 3-ml 10-mm cuvette. Each sample was initially supplemented with 1 mM EDTA. For measuring samples in presence of Ca2+, the solution was brought to 10 mM Ca2+. Excitation was at 295 nm, and the emission spectrum was scanned from 300 to 450 nm with slit widths of 5 nm.
2.8. Stability in urea
Urea concentrations ranging between 0 and 7 M were prepared (in steps of 0.25 M). Each well contained 40 μM of the respective ScaADoc construct in either 10 mM EDTA or 10 mM CaCl2. Proteins constructs were incubated for at least half an hour with Ca2+ and then dissolved in equal volumes of urea solutions. The intrinsic fluorescence of the protein was measured on Tecan infinite® 200 (Tecan GmbH, Austria), a multifunctional microplate reader, with 295 nm excitation. Fluorescence was measured at an emission wavelength of 325 nm. The data were normalized to the initial absorbance of each sample (at 0 M urea). Three independent experimental replicates were averaged for the final result.
2.9. Competitive ELISA (cELISA)
Maxisorp 96-well plates were coated with 10 μM CBM-CohB7, quenched with blocking buffer containing BSA, and exposed to different concentrations of either the wild-type or mutant ScaADoc constructs, all in competition with equimolar amounts (100 pM) of wild-type ScaADoc fused to xylanase T6. The bound Xyn-ScaADoc was determined through the interaction with primary (rabbit anti-xylanase) and then secondary (HRP-goat anti-rabbit) antibodies. HRP was assayed with TMB, and the reaction was stopped with 1 N H2SO4. Optical density was measured at 450 nm using a microplate reader. The result was obtained by averaging five independent experiment repeats. The data were fitted into non-linear regression competitive one-site binding curve using in GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA).
3. Results
3.1. Structural model of ScaADoc
The accumulated wealth of structural information on dockerin modules was leveraged to generate a structural model of Ca2+-bound ScaADoc using the I-TASSER server [24,25] (Fig. 1A), which was evaluated by the following parameters: C-score (a confidence score, based on the quality of the threading alignments and the convergence of the I-TASSER's structural assembly refinement simulations) and TM-score (template modeling score, which assesses the topological similarity of protein structure pairs) [12]. For our best model, the C-score and TM-score of −0.85 and 0.6 ± 0.1, respectively, were both above their empirical thresholds that were determined to be −1.5 and 0.5, respectively.
Inspection of the ScaADoc model indicated that W(−1) and P75 (numbered relative to the conserved glycine as position 0, as explained in Section 2), at the respective N- and C-termini of ScaADoc, form a stacking interaction or intramolecular clasp (Fig. 1B). W(−1)A and P75A mutations in ScaADoc are peripheral to the first Ca2+-binding loop so they were not expected to interfere with the Ca2+-induced conformational changes in the ScaADoc structure. As such, these clasp mutants provided an opportunity to assess their potential in contributing to the structural stability of the Ca2+-bound form of the ScaADoc module.
3.2. The effect of clasp mutants on Ca2+-binding
To initially assess the solution behavior of the wild-type and mutant ScaADoc constructs, size exclusion chromatographic elution profiles in the absence of Ca2+ were obtained (not shown) and confirmed that the mutations themselves did not affect the elution volume of ScaADoc.
Previously, ScaADoc was shown to undergo a Ca2+-dependent conformational change that accompanied the tertiary folding of the molecule [9]. In order to determine whether the mutations of putative clasp residues affected the Ca2+-binding properties of ScaADoc, CD spectra were collected to measure the secondary structural properties of ScaADoc in the absence and presence of Ca2+. Relative to the wild-type ScaADoc [9], the extent of Ca2+-induced structural changes expected for the double mutant was not observed (Fig. 2A) and is consistent with the NMR data presented (Fig. 2B).
Fig. 2.
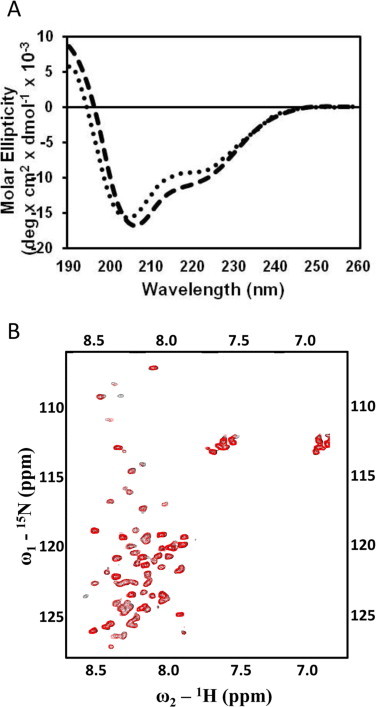
The W(−1)A/P75A double mutant does not undergo complete Ca2+-induced conformational change. (A) The CD spectra of the wild-type ScaADoc (dashed line) and W(−1)A/P75A (dotted line) proteins showcase how complete abrogation of the clasp interaction interferes with the folding of the molecule in the presence of Ca2+. (B) Overlay of the 2D 1H–15N HSQC spectra of the Ca2+-bound (red) and apo-form (black) of the double mutant, W(−1)A/P75A-ScaADoc, wherein the divalent metal cation has little effect on its tertiary structure. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To determine whether there existed a correlation between the clasp formation and the structural stability of ScaADoc we measured the melting temperature (Tm) by ANS fluorescence for wild-type ScaADoc and the set of clasp mutants (Table 1). The P75A mutation decreased thermal stability significantly (55 °C versus 61 °C for the wild-type ScaADoc), while the W(−1)A mutation had a more deleterious effect on thermal stability with a measured Tm value of 41 °C, the same as the double mutant. These data are consistent with the tryptophan fluorescence data, wherein the previously anticipated blue shift [9] for the W(−1) in the P75A ScaADoc mutant did not occur (Fig. 3A). In monitoring the effect of exposing the P75A ScaADoc mutant to increasing urea concentration (Fig. 3B), 1.8 M urea was sufficient to achieve a 50% unfolding of the mutant ScaADoc as opposed to 2.5 M for wild-type ScaADoc.
Table 1.
Stability measured as Tm in ANS fluorescence experiments in presence of Ca2+.
| ScaADoc or mutant | Tm, °C |
|---|---|
| Wild-type | 61 |
| W(−1)A | 41 |
| P75A | 55 |
| W(−1)A/P75A | 41 |
Fig. 3.
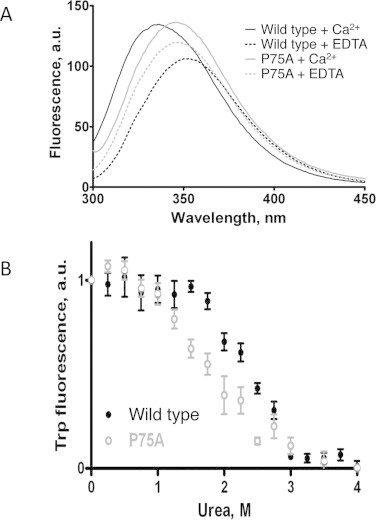
Elimination of proline from the C-terminus of ScaADoc (P75A mutant) effects folding and stability of the protein determined by intrinsic tryptophan fluorescence. (A) Structural changes upon Ca2+-binding to R. flavefaciens ScaADoc in tryptophan fluorescence wavelength scan. In the wild-type protein a blue fluorescence shift is observed as a result of Ca2+-induced folding. In the P75A mutant no change in hydrophobicity of the tryptophan environment occurred upon folding. Peaks of tryptophan fluorescence: 351 nm, wild type + EDTA (- - -); 335 nm, wild type + Ca2+ (—); 347 nm, P75A + EDTA ( ); 346 nm, P75A + Ca2+ (
); 346 nm, P75A + Ca2+ ( ). (B) Decreased stability of the P75A mutant R. flavefaciens ScaADoc. Assay of stability under conditions of increasing urea concentrations. Urea at a concentration of 1.8 M causes unfolding of half of the mutant ScaADoc constructs (○) versus 2.5 M for the wild-type ScaADoc (•).
). (B) Decreased stability of the P75A mutant R. flavefaciens ScaADoc. Assay of stability under conditions of increasing urea concentrations. Urea at a concentration of 1.8 M causes unfolding of half of the mutant ScaADoc constructs (○) versus 2.5 M for the wild-type ScaADoc (•).
3.3. Effect of the W(−1)A and P75A mutants on the recognition of ScaB cohesin
We have previously developed a correspondence between the effects of the mutations on the Ca2+-bound form of ScaADoc with its ability to recognize its cognate binding partner, ScaB cohesin [9]. Using competitive ELISA-based experiments, we determined that the interaction was impaired in a similar manner for all of the mutants by approximately one order of magnitude (Fig. 4). These observations are consistent with the differential thermal stabilities obtained by ANS fluorescence, such that the formation of the ScaB cohesin-binding site in ScaADoc is highly dependent on the clasp interaction.
Fig. 4.
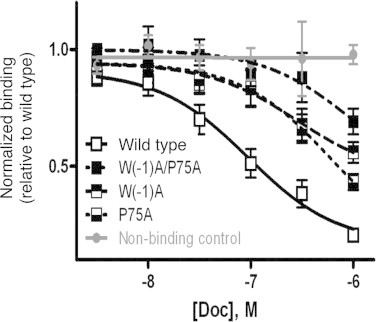
Interaction of the R. flavefaciens ScaADoc mutants with a ScaB cohesin measured by competitive ELISA. Binding was impaired in all of the indicated mutants as compared to wild-type ScaADoc. Non-binding control: substitution of dockerin from unrelated bacterial species (i.e., C. thermocellum).
3.4. Structural evidence for an intramolecular clasp in other dockerins
The putative presence of an intramolecular clasp in ScaADoc was based on a predicted structural model. In order to verify whether this phenomenon represents a general feature of dockerin modules, we evaluated whether similar interactions between residues at the termini exist in other dockerins with known crystal structures. To date, most of the known dockerin structures were achieved as part of a complex with their respective cohesin partner, and their evaluation indicated that all of the intact dockerin modules indeed possessed an intramolecular clasp.
In the first reported crystal structure of the Clostridium thermocellum type-I dockerin in complex with its cohesin partner [14], the dockerin construct commenced at the conserved glycine (i.e., the zero position, G0, as defined in Fig. 1B). The three residues at positions (−3) to (−1) that could potentially interact with appropriate C-terminal residues were, however, lacking in this construct, which would preclude clasp formation in this structure. In a subsequent structure of a mutated form of the same dockerin [15], the three residues prior to the conserved glycine were present, but the C terminus was unfortunately shortened to exclude the putative clasp-forming residues (e.g. FPV sequence). Similarly, the structure of a Clostridium cellulolyticum dockerin [26] retained the three residues at the (−3) to (−1) positions on its N terminus, but also had a truncated C terminus that exempted clasp formation.
Our first success in locating a clasp occurred when we reexamined the type-II dockerin structure of C. thermocellum [27], where the clasp comprised a stacking interaction between a tryptophan at an N-terminal position (−2 relative to the G0 position) and a tyrosine (Y60) located at the C terminus (Fig. 5A). More recently, another type-I dockerin from C. thermocellum was solved in a ternary complex, which contained both type-I and type-II cohesin–dockerin interactions [28]. The type-I dockerin possessed a hydrophobic interaction between a methionine in position (−1) and a proline in position 64 (Fig. 5B), and the clasp of the type-II dockerin matched that of the previously described structure [27]. Another dockerin structure of relevance is part of an enzymatic toxin complex produced by the opportunistic pathogen, Clostridium perfringens [29,30], which does not contain a cellulosome, but uses a cohesin–dockerin interaction to associate two complementary enzymes [16]. In this case, however, instead of an aromatic stacking interaction, there is an electrostatic interaction between a glutamic acid in position (−3) and an arginine towards the C-terminus of the dockerin module that together form an alternative type of intramolecular clasp (Fig. 5C). In the recently available cohesin–dockerin complex structure from Bacteroides cellulosolvens (Viegas et al., PDB ID 2Y3N), we identified a new kind of clasp between an aromatic (tyrosine) residue and positively charged lysine in the dockerin module (Fig. 5D) that interacts through the hydrophobic part of its chain. Finally, a new cohesin–dockerin complex structure from R. flavefaciens [31], which is responsible for cellulosome attachment to the bacterial cell, was solved and also exhibits a clasp interaction between a lysine in position (−1) and two glutamic acids on the C-terminus (Fig. 5E). Together, these findings demonstrate the existence of a diverse range of clasp interactions in dockerins from various bacterial sources and support the generality of this phenomenon in dockerin modules.
Fig. 5.
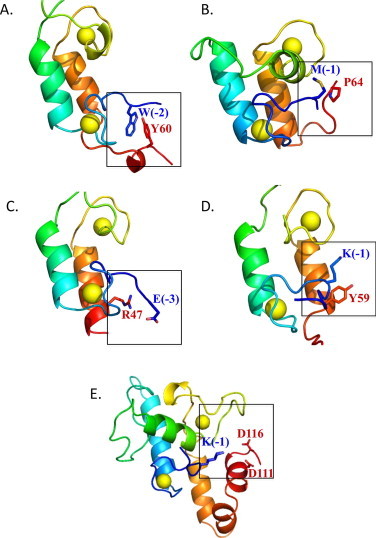
Intramolecular clasps identified in known dockerin structures (represented in Pymol). (A) C. thermocellum type-II dockerin (clasp between tryptophan and tyrosine, PDB: 2B59). (B) C. thermocellum type-I dockerin (clasp between methionine and proline, PDB: 4FL4). (C) C. perfringens dockerin (clasp between arginine and glutamic acid, PDB: 2OZN). (D) B. cellulosolvens dockerin (clasp between lysine and tyrosine, PDB: 2Y3N). (E) R. flavefaciens CttA dockerin (clasp between lysine and two aspartic acids, PDB: 4IU2). Backbones are represented as cartoons, interacting clasp residues as sticks. Calcium ions are shown as yellow spheres. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In the wake of the profusion of new bacterial genome sequences that have recently become available, we have become exposed to an enormous collection of novel dockerin modules from a variety of cellulosomal and non-cellulosomal species from the Bacteria and Archaea [32–38], some of which are distinguished by significant sequence variability. However, structural information regarding the dockerins cannot be easily achieved, especially not in the absence of cohesin, and in most cases we have to rely on the information we can glean from sequence analysis. In the case of R. flavefaciens ScaADoc, for which a structure is currently unavailable, sequence analysis alone is a limited resource, owing to the atypical sequence of this particular dockerin [9]. Therefore, in order to obtain a better understanding of how the unusual sequence would affect its structure, particularly for the region containing the second F-hand Ca2+-binding loop we employed the I-TASSER server [24,25] to provide a structural model for ScaADoc, (Fig. 1A).
Upon careful examination of the putative intramolecular contacts among the ScaADoc residues in the model, we noted a particularly conspicuous stacking interaction between W(−1) at the N-terminus of the ScaADoc module and P75 at its C terminus. Such a prominent non-covalent bond between the two termini could add considerable stability to the Ca2+-bound form of the protein. How would the stability be provided by an intramolecular clasp? Ca2+-induced folding of wild-type ScaADoc would presumably bring positions W(−1) and P75 into close proximity, thereby allowing formation of an intramolecular clasp, which would stabilize the overall dockerin structure and facilitating the formation and stabilization of the cohesin-binding site. Thus, calcium binding is essential for clasp formation and for a proper binding of the dockerin.
In comparing the stability characteristics of the different mutants, one would presumably expect a similar Tm measurement for both the W(−1)A and P75A ScaADoc mutants, since we assume that the stacking interaction would have a role in stability. However, it is clear from the data that mutating W(−1) and P75 impose different effects on ScaADoc. The more pronounced decrease in stability of W(−1)A most likely reflects the loss of additional hydrophobic contacts with surrounding residues. Based on a similar Tm value for the W(−1)A/P75A mutant, the absence of the tryptophan is clearly a more dominant factor in the stability of the protein. Taken together, these results provide the first evidence that the putative stacking interaction between W(−1) and P75 is important to stabilize the Ca2+-bound form of the ScaADoc module. In this spirit, the consequence of mutating one or both W(−1)A and P75A residues interferes also with the recognition of the mutant by the ScaB cohesin, because the structural distortions of ScaADoc molecule would presumably affect the topography of the interface required for complex formation.
Interestingly, the dockerins’ binding partners, the cohesins, were found to be remarkably mechanostable [39], i.e., resistant to mechanical stress such as strong stirring. These authors suggested that the property of mechanostability is essential for scaffoldins to be able to stretch between the cell and the substrate. This hypothesis can now be extended to include the dockerins, notably those that are responsible for connecting different scaffoldins, and among them the ScaA dockerin described here.
The general tendency of the N- and C-termini of proteins to appear in close proximity to one another and the contribution of this phenomenon to stability have been known for decades [40]. In dockerin modules this tendency appears to be further developed to form distinctive interactions between residues at the two termini and appears to represent a defined characteristic of the dockerin module as a self-folding modular component of the parent protein. The nature of these interactions may differ as we observed in the dockerins that originate from different bacterial sources (Fig. 5). Aromatic interactions are known to be very common in the stabilization of protein tertiary structures [41]. In the available dockerin structures clasp interactions involving proline and charged residues also appear to be common. In previous studies, it was shown that the interaction between proline and aromatic residues could be as strong as the interaction between two aromatic residues [42–44]. Computational studies show that the strength is mostly contributed by the interaction with the N heteroatom, but also by the cyclic nature of proline [44].
Observation of intramolecular clasps in the published dockerin structures highlights the singularity of the three amino acids preceding the first Ca2+-binding loop in the sequence, which can both participate directly in formation of the clasp and provide additional intramolecular stabilizing contacts. Tyrosine is very prominent at the (−1) position in the sequences of many dockerins. Moreover, in all three positions preceding Ca2+-binding loop (−1, −2, −3) there is a clear preference either for hydrophobic residues (Leu, Ile, Val) or for lysine. One of these residues typically interacts with one of the residues at the C terminus of the dockerin (commonly 2–8 residues from the end of the module).
The conclusions of this study should be taken into account when producing dockerin-anchored enzymes by synthetic biology [45–47]. For example, in designing fusion protein tags, full-length dockerin sequences should be employed in order to avoid low levels of protein expression, solubility and stability and their consequent effects on cohesin binding. Moreover, this finding raises many interesting questions regarding the relationship between sequence and structure of the dockerins to their function. For example, do different types of clasp (aromatic, hydrophobic or electrostatic) provide different levels of stability and do they correlate in some way with dockerin function? Finally, the exploitation of these properties can potentially to impact the activity and efficiency of designer cellulosomes [45–47]. In any case, it is clear that the intramolecular clasp is key characteristic of the dockerins that affects its performance, along with the Ca2+ binding function and internal dockerin symmetry.
Acknowledgements
This research was supported by the Israel Science Foundation (ISF; Grant No. 1349/13 to E.A.B.) and by the Natural Sciences and Engineering Research Council of Canada (Grant No. RGPIN 398508 to S.P.S.). Additional support was obtained by a grant (No. 24/11) issued to R.L. by The Sidney E. Frank Foundation through the ISF and grants to E.A.B. and R.L. from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel. A grant was awarded to E.A.B. from the F. Warren Hellman Grant for Alternative Energy Research in Israel in support of alternative energy research in Israel administered by the Israel Strategic Alternative Energy Foundation (I-SAEF). This research was also supported by the establishment of an Israeli Center of Research Excellence (I-CORE Center No. 152/11, EAB) managed by the Israel Science Foundation, by the Weizmann Institute of Science Alternative Energy Research Initiative (AERI) and the Helmsley Foundation. The authors acknowledge the technical support from Kim Munro of the Protein Function Discovery Facility. E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Flint H.J., Bayer E.A., Rinco M.T., Lamed R., White B.A. Polisaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008;6:1–11. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 2.Bayer E.A., Chanzy H., Lamed R., Shoham Y. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 3.Bayer E.A., Belaich J.-P., Shoham Y., Lamed R. The cellulosomes: multi-enzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 4.Vodovnik M., Duncan S.H., Reid M.D., Cantlay L., Turner K., Parkhill J., Lamed R., Yeoman C.J., Berg Miller M.E., White B.A., Bayer E.A., Marinsek-Logar R., Flint H.J. Expression of cellulosome components and type IV pili within the extracellular proteome of Ruminococcus flavefaciens 007. PLoS One. 2013;8:e65333. doi: 10.1371/journal.pone.0065333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rincon M.T., Cepeljnik T., Martin J.C., Barak Y., Lamed R., Bayer E.A., Flint H.J. A novel cell surface-anchored cellulose-binding protein encoded by the sca gene cluster of Ruminococcus flavefaciens. J. Bacteriol. 2007;189:4774–7283. doi: 10.1128/JB.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rincon M.T., Ding S.-Y., McCrae S.I., Martin J.C., Aurilia V., Lamed R., Shoham Y., Bayer E.A., Flint H.J. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 2003;185:703–713. doi: 10.1128/JB.185.3.703-713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding S.-Y., Rincon M.T., Lamed R., Martin J.C., McCrae S.I., Aurilia V., Shoham Y., Bayer E.A., Flint H.J. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 2001;183:1945–1953. doi: 10.1128/JB.183.6.1945-1953.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rincon M.T., Cepeljnik T., Martin J.C., Lamed R., Barak Y., Bayer E.A., Flint H.J. Unconventional mode of attachment of the Ruminococcus flavefaciens cellulosome to the cell surface. J. Bacteriol. 2005;187:7569–7578. doi: 10.1128/JB.187.22.7569-7578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpol A., Jobby M.K., Slutzki M., Noach I., Chitayat S., Smith S.P., Bayer E.A. Structural and functional characterization of a novel type-III dockerin from Ruminococcus flavefaciens. FEBS Lett. 2013;587:30–36. doi: 10.1016/j.febslet.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Rincon M.T., Dassa B., Flint H.J., Travis A.J., Jindou S., Borovok I., Lamed R., Bayer E.A., Henrissat B., Coutinho P.M., Antonopoulos D.A., Berg Miller M.E., White B.A. Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium, Ruminococcus flavefaciens FD-1. PLoS One. 2010;5:e12476. doi: 10.1371/journal.pone.0012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazzoli A., Tettamanzi A.G., Zhang Y. Computational protein design and large-scale assessment by I-TASSER structure assembly simulations. J. Mol. Biol. 2011;407:764–776. doi: 10.1016/j.jmb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl. 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho A.L., Dias F.M., Prates J.A., Nagy T., Gilbert H.J., Davies G.J., Ferreira L.M., Romão M.J., Fontes C.M. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc. Natl. Acad. Sci. USA. 2003;100:13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho A.L., Dias F.M.V., Nagy T., Prates J.A.M., Proctor M.R., Smith N., Bayer E.A., Davies G.J., Ferreira L.M.A., Romão M.J., Fontes C.M.G.A., Gilbert H.J. Evidence for a dual binding mode of dockerin modules to cohesins. Proc. Natl. Acad. Sci. USA. 2007;104:3089–3094. doi: 10.1073/pnas.0611173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chitayat S., Adams J.J., Furness H.S., Bayer E.A., Smith S.P. The solution structure of the C-terminal modular pair from Clostridium perfringens mu-toxin reveals a noncellulosomal dockerin module. J. Mol. Biol. 2008;381:1202–1212. doi: 10.1016/j.jmb.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 17.Mechaly A., Yaron S., Lamed R., Fierobe H.-P., Belaich A., Belaich J.-P., Shoham Y., Bayer E.A. Cohesin-dockerin recognition in cellulosome assembly: experiment versus hypothesis. Proteins. 2000;39:170–177. doi: 10.1002/(sici)1097-0134(20000501)39:2<170::aid-prot7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 18.Sakka K., Sugihara Y., Jindou S., Sakka M., Inagaki M., Sakka K., Kimura T. Analysis of cohesin–dockerin interactions using mutant dockerin proteins. FEMS Microbiol. Lett. 2011;314:75–80. doi: 10.1111/j.1574-6968.2010.02146.x. [DOI] [PubMed] [Google Scholar]
- 19.Pages S., Belaich A., Belaich J.-P., Morag E., Lamed R., Shoham Y., Bayer E.A. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins. 1997;29:517–527. [PubMed] [Google Scholar]
- 20.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Goddard T.D., Kneller D.G. University of California; San Francisco: 2008. SPARKY 3. [Google Scholar]
- 22.Maier E.M., Gersting S.W., Kemter K.F., Jank J.M., Reindl M., Messing D.D., Truger M.S., Sommerhoff C.P., Muntau A.C. Protein misfolding is the molecular mechanism underlying MCADD identified in newborn screening. Hum. Mol. Genet. 2009;18:1612–1623. doi: 10.1093/hmg/ddp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tandon S., Horowitz P.M. The detection of kinetic intermediate(s) during refolding of rhodanese. J. Biol. Chem. 1990;265:5967–5970. [PubMed] [Google Scholar]
- 24.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl. 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinheiro B.A., Protctor M.R., Martinez-Fleites C., Prates J.A., Money V.A., Davies G.J., Bayer E.A., Fontesm C.M., Fierobe H.P., Gilbert H.J. The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner. J. Biol. Chem. 2008;283:18422–18430. doi: 10.1074/jbc.M801533200. [DOI] [PubMed] [Google Scholar]
- 27.Adams J.J., Pal G., Jia Z., Smith S.P. Mechanism of bacterial cell-surface attachment revealed by the structure of cellulosomal type II cohesin-dockerin complex. Proc. Natl. Acad. Sci. USA. 2006;103:305–310. doi: 10.1073/pnas.0507109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Currie M.A., Adams J.J., Faucher F., Bayer E.A., Jia Z., Smith S.P. Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome. J. Biol. Chem. 2012;287:26953–26961. doi: 10.1074/jbc.M112.343897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chitayat S., Greg K., Adams J.J., Ficko-Blean E., Bayer E.A., Boraston A.B., Smith S.P. Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase. J. Mol. Biol. 2008;375:20–28. doi: 10.1016/j.jmb.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Adams J.J., Gregg K., Bayer E.A., Boraston A.B., Smith S.P. Structural bases of Clostridium perfringens toxin complex formation. Proc. Natl. Acad. Sci. USA. 2008;105:12194–12199. doi: 10.1073/pnas.0803154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salama-Alber O., Jobby M.K., Chitayat S., Smith S.P., White B.A., Shimon L.J., Lamed R., Frolow F., Bayer E.A. Atypical cohesin-dockerin complex responsible for cell-surface attachment of cellulosomal components: binding fidelity, promiscuity, and structural buttresses. J. Biol. Chem. 2013;288:16827–16838. doi: 10.1074/jbc.M113.466672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dassa B., Borovok I., Lamed R., Henrissat B., Coutinho P., Hemme C.L., Huang Y., Zhou J., Bayer E.A. Genome-wide analysis of Acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system. BMC Genomics. 2012;13:210. doi: 10.1186/1471-2164-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suen G., Stevenson D.M., Bruce D.C., Chertkov O., Copeland A., Cheng J.F., Detter C., Detter J.C., Goodwin L.A., Han C.S., Hauser L.J., Ivanova N.N., Kyrpides N.C., Land M.L., Lapidus A., Lucas S., Ovchinnikova G., Pitluck S., Tapia R., Woyke T., Boyum J., Mead D., Weimer P.J. Complete genome of the cellulolytic ruminal bacterium Ruminococcus albus 7. J. Bacteriol. 2011;193:5574–5575. doi: 10.1128/JB.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg Miller M.E., Antonopoulos D.A., Rincon M.T., Band M., Bari A., Akraiko T., Hernandez A., Thimmapuram J., Henrissat B., Coutinho P.M., Borovok I., Jindou S., Lamed R., Flint H.J., Bayer E.A., White B.A. Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS One. 2009;4:e6650. doi: 10.1371/journal.pone.0006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feinberg L., Foden J., Barrett T., Davenport K.W., Bruce D., Detter C., Tapia R., Han C., Lapidus A., Lucas S., Cheng J.F., Pitluck S., Woyke T., Ivanova N., Mikhailova N., Land M., Hauser L., Argyros D.A., Goodwin L., Hogsett D., Caiazza N. Complete genome sequence of the cellulolytic thermophile Clostridium thermocellum DSM1313. J. Bacteriol. 2011;193:2906–2907. doi: 10.1128/JB.00322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamaru Y., Miyake H., Kuroda K., Nakanishi A., Kawade Y., Yamamoto K., Uemura M., Fujita Y., Doi R.H., Ueda M. Genome sequence of the cellulosome-producing mesophilic organism Clostridium cellulovorans 743B. J. Bacteriol. 2010;192:901–902. doi: 10.1128/JB.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao G., Wang R., Zhu Y., Dong H., Mao S., Zhang Y., Chen Z., Li Y., Ma Y. Complete genome sequence of Clostridium acetobutylicum DSM 1731, a solvent-producing strain with multireplicon genome architecture. J. Bacteriol. 2011;193:5007–5008. doi: 10.1128/JB.05596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peer A., Smith S.P., Bayer E.A., Lamed R., Borovok I. Noncellulosomal cohesin- and dockerin-like modules in the three domains of life. FEMS Microbiol. Lett. 2009;291:1–16. doi: 10.1111/j.1574-6968.2008.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valbuena A., Oroz J., Hervas R., Vera A.M., Rodriguez D., Menendez M., Sulkowska J.I., Cieplak M., Carrion-Vazquez M. On the remarkable mechanostability of scaffoldins and the mechanical clamp motif. Proc. Natl. Acad. Sci. USA. 2009;106:13791–137916. doi: 10.1073/pnas.0813093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thornton J.M., Sibanda B.L. Amino and carboxy-terminal regions in global proteins. J. Mol. Biol. 1983;167:443–460. doi: 10.1016/s0022-2836(83)80344-1. [DOI] [PubMed] [Google Scholar]
- 41.Waters M.L. Aromatic interactions in model systems. Curr. Opin. Chem. Biol. 2002;6:736–741. doi: 10.1016/s1367-5931(02)00359-9. [DOI] [PubMed] [Google Scholar]
- 42.Riley K.E., Cui G., Merz K.M., Jr. An ab initio investigation of the interactions involving the aromatic group of the set of fluorinated N-(4-sulfamylbenzoyl)benzylamine inhibitors and human carbonic anhydrase II. J. Phys. Chem. B. 2007;111:5700–5707. doi: 10.1021/jp067313m. [DOI] [PubMed] [Google Scholar]
- 43.Morozov A.V., Misura K.M.S., Tsemekhman K., Baker D. Comparison of quantum mechanics and molecular mechanics dimerization energy landscapes for pairs of ring-containing amino acids in proteins. J. Phys. Chem. B. 2004;108:8489–8496. [Google Scholar]
- 44.Biedermannova L., Rilley K.E., Berka K., Hobza P., Vondrasek J. Another role of proline: stabilization interactions in proteins and protein complexes concerning proline and tryptophane. Phys. Chem. Chem. Phys. 2008;10:6350–6359. doi: 10.1039/b805087b. [DOI] [PubMed] [Google Scholar]
- 45.Vazana Y., Moraïs S., Barak Y., Lamed R., Bayer E.A. Designer cellulosomes for enhanced hydrolysis of cellulosic substrates. Methods Enzymol. 2012;510:429–452. doi: 10.1016/B978-0-12-415931-0.00023-9. [DOI] [PubMed] [Google Scholar]
- 46.Bayer E.A., Morag E., Lamed R. The cellulosome – a treasure-trove for biotechnology. Trends Biotechnol. 1994;12:378–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 47.Ohmiya K., Sakka K., Kimura T., Morimoto K. Application of microbial genes to recalcitrant biomass utilization and environmental conservation. J. Biosci. Bioeng. 2003;95:549–561. [PubMed] [Google Scholar]


