Abstract
Lactococcus lactis can undergo respiration when hemin is added to an aerobic culture. The most distinctive feature of lactococcal respiration is that lactate could be consumed in the stationary phase concomitantly with the rapid accumulation of diacetyl and acetoin. However, the enzyme responsible for lactate utilization in this process has not yet been identified. As genes for fermentative NAD-dependent l-lactate dehydrogenase (l-nLDH) and potential electron transport chain (ETC)-related NAD-independent l-LDH (l-iLDH) exist in L. lactis, the activities of these enzymes were measured in this study using crude cell extracts prepared from respiratory and fermentation cultures. Further studies were conducted with purified preparations of recombinant LDH homologous proteins. The results showed that l-iLDH activity was hardly detected in both crude cell extracts and purified l-iLDH homologous protein while l-nLDH activity was very significant. This suggested that l-iLDHs were inactive in lactate utilization. The results of kinetic analyses and the effects of activator, inhibitor, substrate and product concentrations on the reaction equilibrium showed that l-nLDH was much more prone to catalyze the pyruvate reduction reaction but could reverse its role provided that the concentrations of NADH and pyruvate were extremely low while NAD and lactate were abundant. Metabolite analysis in respiratory culture revealed that the cellular status in the stationary phase was beneficial for l-nLDH to catalyze lactate oxidation. The factors accounting for the respiration- and stationary phase-dependent lactate utilization in L. lactis are discussed here.
Keywords: Lactococcus lactis, Lactate oxidation, Lactate dehydrogenase, Type II IPP isomerase, Proton motive force
Abbreviations: ETC, electron transport chain; LDH, lactate dehydrogenase; iLDH, NAD-independent lactate dehydrogenase; nLDH, NAD-dependent lactate dehydrogenase; FBP, fructose 1,6-bisphosphate; PMF, proton motive force; DCPIP, 2,6-dichlorophenolindophenol; IPP, isopentenyl diphosphate
Highlights
-
•
LutABC proteins do not participate in lactate oxidation in Lactococcus lactis
-
•
Lactococcus lactis has very low NAD-independent lactate dehydrogenase activity
-
•
Fructose-1,6-bisphosphate-dependent lactate dehydrogenase can work in reverse in vivo
-
•
Metabolite concentrations in the stationary phase are favorable for lactate oxidation
-
•
Respiratory metabolism is the basis for continual lactate oxidation in Lactococcus
1. Introduction
As an important industrial microorganism and model lactic acid bacteria, Lactococcus lactis has long been studied with respect to the control of lactate production. Under fermentation conditions, L. lactis produces ATP by substrate-level phosphorylation with l-lactate as the main final product. However, research has shown that L. lactis could form a functional ETC and perform respiration when hemin is added to the aerobic culture [1–3]. Respiratory metabolism yields a number of benefits, such as increased biomass, reduced acid and oxidative stress, and improved long-term survival [1,4,5]. A unique phenomenon frequently observed in L. lactis respiration is that the accumulated lactate can be consumed when glucose becomes limited [1,6,7] concomitantly with the accumulation of the alternative products of pyruvate metabolism (acetate, diacetyl, and acetoin) and an increase in pH. This suggests that lactate is oxidized to pyruvate by an enzyme during such a process.
Based on coenzyme dependence, l-lactate dehydrogenases (l-LDHs) have traditionally been classified as either NAD-dependent l-LDHs (l-nLDHs) or NAD-independent l-LDHs (l-iLDHs) [8,9]. l-nLDHs play an important role in fermentation by producing lactate from pyruvate while regenerating NAD. l-nLDHs can be further divided into fructose 1,6-bisphosphate (FBP)-activated l-nLDHs and FBP-independent l-nLDHs [8]. On the other hand, l-iLDHs include two members of FMN-dependent α-hydroxyacid dehydrogenase [i.e. lactate oxidase (EC1.1.3.15) and flavocytochrome b2 (EC1.1.2.3)], a non-flavin iron–sulfur enzyme complex (encoded by lutABC operon), and a flavin and iron–sulfur cluster containing oxidoreductase [10–12]. Generally, l-lactate is oxidized by l-iLDHs whose activity can be measured using artificial electron acceptors, such as 2,6-dichlorophenolindophenol (DCPIP). In some cases, more than one l-iLDH can be found in l-lactate-utilizing organisms, although some may be inactive in lactate utilization [10–12]. Occasionally, l-lactate is found to be oxidized to pyruvate by FBP-independent l-nLDH [9]; however, FBP-activated l-nLDHs have long been regarded as irreversible in vivo [8,13,14].
Two l-iLDH homologous genes exist in the L. lactis MG1363 genome. In addition to the potential lutABC cluster (expected values <2e−40), the fni gene is the other candidate. FNI, the product of fni gene, is a type II isopentenyl diphosphate (IPP) isomerase and shares a weak but significant homology with lactate oxidase and flavocytochrome b2 (expected values of 1e−8 and 7e−5, respectively). In NCBI, the function of FNI is noted as “l-lactate dehydrogenase (FMN-dependent) and related alpha-hydroxy acid dehydrogenases”. IPP isomerase is essential in organisms that synthesize isoprenoid exclusively via the mevalonate pathway, such as Lactobacillales [15]. The fni gene is constitutively expressed in L. lactis, as evidenced by the fact that menaquinones are produced throughout the growth phase [16].
Four l-nLDH genes exist in the genome of L. lactis MG1363 [17], and l-nLDH activity is FBP dependent [18,19]. Reverse transcription PCR has revealed that besides ldh, ldhB and ldhX are transcribed to some extent in L. lactis MG1363 [19]. However, research has also shown that the product of the ldhX gene has little nLDH activity as well as that ldhB exhibits only leaky transcription and plays a minor role in lactate yield. LDHA encoded by ldh has been found to perform major l-nLDH activity in L. lactis MG1363, with the contribution of other alternative l-nLDHs being small. Whole-genome microarrays and proteome analysis have revealed that l-nLDH genes are not included among the genes differentially expressed in respiratory conditions [20,21], indicating that it is LDHA that plays the major role in respiration.
In this study, l-iLDH and l-nLDH activities were determined using crude cell extracts from L. lactis MG1363 aerobic fermentation and respiratory cultures. fni and ldh genes were separately cloned from the L. lactis MG1363 genome and expressed in Escherichia coli as His-tagged proteins to evaluate their lactate oxidation capacity and the relationship between lactate oxidation activity and their native activity. Enzyme activities, kinetics, and the effects of various factors on equilibrium were closely studied with purified recombinant proteins. Finally, targeted cellular metabolites were determined to test if they were beneficial for the oxidation of lactate.
2. Materials and methods
2.1. Strains, growth conditions, and crude enzyme preparation
L. lactis MG1363 fermentation and respiratory cultures were performed as described by Duwat [1] but with the modifications that, where indicated, hemin was added to a final concentration of 2.5 mg/L and the shaking speed was kept at 220 rpm.
E. coli M15 was grown at 37 °C in M9 minimal medium supplied with 0.5% glucose or 0.5% l-sodium lactate under aerobic conditions.
Cells from 50 ml cultures were collected by centrifugation and washed twice with 0.85% NaCl to prepare crude cell extracts. The pellets were resuspended in a 5 ml buffer containing 20 mM potassium phosphate (pH 7.0) and 10% glycerol, and the cells were disrupted by sonication on ice (12 cycles of 10 s sonication with intervals of 10 s). Intact cells were removed by centrifugation.
2.2. Preparation of recombinant proteins
Based on the entire nucleotide sequence of fni from L. lactis MG1363 (GenBank Accession No. AM406671.1), two oligonucleotides, 5′-GGATCCATGATG-AAAAGTGAAAAAG-3′ and 5′-CTGCAGTTATTTTTTTCTTTGTTGG-3′, as primers were synthesized, and L. lactis MG1363 chromosomal DNA was used as a template to amplify fni. The PCR product was purified using an AxyPrep PCR Clean-up Kit (Axygen), cloned into the pMD19-T vector and sequenced.
Plasmid DNA with the correct fni gene sequence was digested with BamHI and PstI and ligated into the N-terminal His tag expression vector pQE30Xa (Qiagen), which had been treated with the same enzymes. The resulting plasmid pQEFNI5 was transformed into E. coli strain M15 (pREP4).
The recombinant E. coli M15 (pREP4, pQEFNI5) was cultured at 37 °C in 2× YT broth containing 25 μg/ml kanamycin (Amresco) and 100 μg/ml ampicillin (Sigma) with shaking. When the optical density at 600 nm reached 0.5–0.7, IPTG was added to a final concentration of 0.5 mM, and the culture was incubated at 30 °C for an additional 4 h. Cells were harvested by centrifugation and stored at −20 °C.
After thawing, the frozen cells were resuspended with buffer A composed of 300 mM NaCl, 10% glycerol, and 50 mM NaH2PO4 (pH 7.0), lysed by incubation with lysozyme (1.5 mg/ml for 20 min at room temperature), and then subjected to sonication. After centrifugation, the supernatant was applied on a Ni–nitrilotriacetic acid agarose resin (Qiagen, China) previously equilibrated with 20 mM imidazole in buffer A. The resin was washed with 20 mM imidazole in buffer A, after which the protein was eluted with 1 M imidazole in the same buffer.
The oligonucleotides 5′-ATGGCTGATAAACAACGTAAGAAAG-3′ and 5′-GTTTTTAACTGCAGAAGCAAATTCT-3′ were used as the forward and reverse primers, respectively, to amplify ldh gene. The PCR product was inserted into the C-terminal His tag expression vector pEASY-E2 (Transgen, Beijing, China) and transformed into E. coli JM109. Colony PCR was used to screen the insert with the correct orientation using the vector-specific T7 promoter primer (5′-TAATACGACTCACTATA-3′) and ldh-specific reverse primer. The recombinant plasmid pEASYLDH4 with the correct orientation was verified by sequencing and then transformed into E. coli Transetta (DE3) (Transgen).
The recombinant E. coli Transetta (DE3) (pRARE, pEASYLDH4) was cultured in 2× YT broth containing 34 μg/ml chloramphenicol (Amresco) and 100 μg/ml ampicillin with shaking. Other procedures for the expression and purification of recombinant LDHA were done in the same way as those of FNI.
2.3. FNI product analysis by 1H NMR
The FNI protein used for type II IPP isomerase assay was previously desalted by gel filtration through a PD-10 column (Amersham Pharmacia) previously equilibrated with 25 mM K2HPO4 (pH 7.0).
A 500 μl reaction mixture containing 0.1 M K2HPO4 (pH 7.0), 5 mM MgCl2, 2 mM DTT, 10 μM FMN, 5 mM NADH, 6.73 mM IPP (Isoprenoids, LC), and 0.2 mg of FNI protein was incubated at 30 °C for 15 h. After the addition of D2O to a final concentration of 10% (v/v), the samples were analyzed by 1H NMR spectroscopy (Bruker) at 500 MHz. A reaction mixture without the FNI protein was analyzed as a control.
2.4. Enzyme analysis
All enzymes were assayed spectrophotometrically by measuring the decrease (or increase) in the absorbance of NADH at 340 or 380 nm [18] or the reduction of DCPIP at 600 nm at 30 °C in 100 mM Tris–maleate (pH 7.0). One unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol product per minute or the consumption of 1 μmol substrate per minute. Approximately 40–200 ng/ml of protein was used to assay LDHA, and 66 μg/ml of protein was used to assay FNI.
NADH dehydrogenase activity was assayed in a volume of 1.2 ml containing 0.15 mM NADH.
iLDH activity was assayed using a slightly modified form of the procedure described by Hao [22]. The 1.2 ml reaction mixture contained 40 μM DCPIP, 30 μM phenazine methosulfate, and 10 mM l- or dl-sodium lactate.
nLDH activities were assayed by measuring pyruvate reduction and lactate oxidation reactions. Unless indicated in the text, pyruvate reduction activity was assayed by the method described by Koebmann [23]. FBP was omitted from the reaction mixture used for FNI. The lactate oxidation activity of LDHA was assayed in a 1.2 ml reaction mixture containing 5 mM NAD, 1 mM FBP, and 200 mM l-sodium lactate. The lactate oxidation activity of FNI was assayed under similar conditions but with 3.2 mM NAD and 10 mM l-sodium lactate.
Km and Vm values were estimated using Origin (Microcal Software).
2.5. Metabolite determinations
In total, 1.5 ml cultures were taken at indicated time points and cells were removed by centrifugation at 12,000g for 10 min to measure the end products of L. lactis MG1363 cultures. The supernatant was frozen at −20 °C until further analysis. Metabolites were separated using a 300 mm Aminex HPX-87H column (Bio-Rad) equipped with a refractive index detector at 65 °C with 5 mM H2SO4 as the mobile phase at the flow rate of 0.5 ml/min.
The L. lactis MG1363 respiratory cultures were harvested at the mid-log (6.5 h after cultivation) and stationary (10 h after cultivation) phases, quenched in liquid nitrogen, and thawed on ice to measure the intracellular metabolites. Cells from 9 ml culture were collected by centrifugation and washed once with 20 mM HEPES (pH 7.5) containing 10% glycerol. Intracellular metabolites were extracted with 5 or 7 ml of boiling buffered ethanol solution according to the protocol described by Villas-Bôas [24]. After centrifugation, the ethanol extracts were evaporated to dryness under vacuum at 45 °C, resuspended in 1 ml of double-distilled water, and stored at −80 °C until analysis. A C-18 reversed-phase column was used to analyze NADH, NAD, and ADP with 0.1 M KH2PO4 (pH 6.5), 5 mM tetrabutylammonium hydroxide, and 10% methanol as the mobile phase at the flow rate of 0.7 ml/min. Cellular pyruvate and lactate were analyzed using an Aminex HPX-87H column equipped with a UV detector at 40 °C. Cellular volume was estimated according to Poolman [25].
Supernatants from thawed cultures and the wash solutions were also collected and analyzed for leaked metabolites; however, these materials were completely obscured by the complex medium components. Although leakage was likely to happen, it was non-specific and the trend of metabolite concentrations was not severely affected.
3. Results
3.1. Metabolite profiling and LDH activities in respiratory and fermentation cultures
The levels of glucose and its metabolites in respiratory and fermentation cultures were determined by HPLC (Fig. 1). Lactate was similarly accumulated in both cultures when glucose was available. However, significant metabolic alterations could be observed when glucose became limited. Lactate production in respiratory cultures was frequently found to decrease at the onset of glucose exhaustion, whereas it increased slightly in fermentation cultures. Concomitant with the reduction in lactate accumulation, the alternative products of pyruvate metabolism (acetate, diacetyl, and acetoin) greatly accumulated in the respiratory culture. Changes were not observed in the levels of the above-described products in the fermentation culture.
Fig. 1.
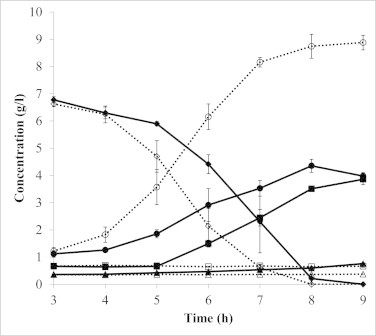
Time course of glucose consumption and end-product accumulation under respiratory and anaerobic fermentation conditions. Solid lines, respiration; dotted lines, anaerobic fermentation; diamond, glucose; circle, lactate; square, acetoin and diacetyl; triangle, acetate. Error bars represent SD of two independent experiments.
Crude enzyme extracts were prepared from L. lactis MG1363 respiratory and fermentation cultures at the mid-log and stationary phases, and their l-nLDH and l-iLDH activities were measured (Table 1). 2 mM sodium azide was used in the l-iLDH reaction mixture to partly inhibit cytochrome oxidase activity. The l-iLDH activity was always hardly detected, regardless of the growth phase and culture conditions. As a positive control, the l-iLDH activity of E. coli M15 (a derivative of E. coli K12) was much higher. When grown on l-lactate, the l-iLDH activity of E. coli M15 was hundreds-fold higher than that of L. lactis MG1363. The faint l-iLDH activity suggested that the potential l-iLDHs may not be involved in the lactate utilization in L. lactis.
Table 1.
l-LDH activities of L. lactis MG1363 and E. coli M15. The l-iLDH and l-nLDH activities of L. lactis MG1363 were determined using the cell extracts from respiratory and fermentation cultures at the logarithmic and stationary phases. The l-iLDH activity of E. coli M15 was used as a positive control. Data are the mean ± SD of three independent measurements. ND indicates not detected.
| Strain and growth conditions | Phase | l-iLDH (nmol min−1 mg−1) | l-nLDH (μmol min−1 mg−1) |
|---|---|---|---|
| L. lactis MG1363 | |||
| Fermentation | Mid log | ND | 12.2 ± 1.2 |
| Stationary | 0.2 ± 0.1 | 11.7 ± 0.6 | |
| Respiration | Mid log | ND | 10.0 ± 1.0 |
| Stationary | 0.4 ± 0.0 | 9.3 ± 0.3 | |
| E. coli M15 | |||
| In M9-glucose medium | Stationary | 5.5 ± 1.1 | |
| In M9-l-lactate medium | Stationary | 45.1 ± 1.8 | |
By contrast, FBP-activated l-nLDH activity was constantly very significant throughout the growth period, even when lactate was consumed in the stationary phase of respiration. Apparently, during the lactate utilization period, l-nLDH activity was regulated by some factors and the pyruvate reduction activity was completely inhibited or masked.
3.2. LDH activities of type II IPP isomerase
3.2.1. Type II IPP isomerase activity of recombinant FNI validation by 1H NMR
The fni gene of L. lactis MG1363 was cloned into the N-terminal His-tagged expression vector and overexpressed in E. coli. The purified recombinant protein was golden yellow and showed an absorption peak at approximately 450 nm, which indicated that it was a flavoprotein. SDS–PAGE analysis showed that its purity was higher than 95% and that it had a subunit molecular mass of 41 kDa, which was consistent with the estimated value. The native type II IPP isomerase activity of FNI was analyzed by 1H NMR. After overnight incubation, conversion of IPP to dimethylallyl diphosphate was observed as evidenced by the appearance of signals for dimethylallyl diphosphate at δ 1.53 (C-4 methyl protons), 1.57 (C-5 methyl protons), 4.27 (C-1 methylene protons) and 5.25 (C-2 methine proton) ppm (Fig. 2). These findings confirm that the purified recombinant FNI was biologically active.
Fig. 2.
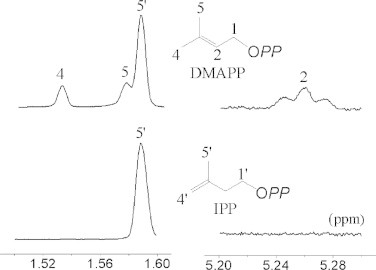
1H NMR assay of the type II IPP isomerase activity of FNI. The upper panel shows part of the 1H NMR spectrum of the reaction mixture, whereas the lower panel shows the control without enzyme addition.
3.2.2. LDH activities of FNI
The nLDH and iLDH activities of FNI are listed in Table 2. The oxidoreductase activities of FNI were extremely low, which was in good agreement with the faint l-iLDH activity of L. lactis MG1363 cell extracts. Divalent metal ions (Mg2+, Mn2+, Ca2+, and Zn2+), which are essential for type II IPP isomerase activity [26], were separately added to the reaction system, but they did not increase activity. In fact, l-iLDH activity was further reduced such that it could no longer be detected. dl-Lactate sodium was found to be a better substrate because its dl-iLDH activity was approximately seven-fold higher than the l-iLDH activity. The nLDH activities of FNI were validated in this study, but they were determined to be extremely low, only approximately 2–7 nmol min−1 (mg protein)−1, similar to reported results [15].
Table 2.
LDH activities of FNI. The iLDH and nLDH activities were determined using the recombinant FNI. M2+, divalent metal ions (Ca2+, Mg2+, Mn2+, or Zn2+) were added to the l-iLDH reaction mixture to a final concentration of 2 mM; nLDH (pyruvate reduction) activity was measured as pyruvate-dependent NADH oxidation activity, with its intrinsic NADH dehydrogenase activity eliminated; ND, not detected. Data are the mean ± SD of five independent measurements.
| Activity (nmol min−1 mg−1) | |
|---|---|
| l-iLDH | 2.9 ± 0.1 |
| l-iLDH (M2+) | ND |
| dl-iLDH | 22.1 ± 0.9 |
| nLDH (pyruvate reduction) | 7.0 ± 1.2 |
| l-nLDH (l-lactate oxidation) | 2.2 ± 0.5 |
3.3. Analysis of fermentative nLDH activities and its regulatory mechanism
3.3.1. Purification and biological activity of recombinant LDHA
Recombinant LDHA was expressed as a His-tagged protein and purified as a soluble protein under native conditions. SDS–PAGE showed a single band with an apparent molecular weight of 37 kDa, which was in good agreement with the calculated value. The NAD(H)-dependent pyruvate reduction (forward reaction) and lactate oxidation (reverse reaction) activities of LDHA were measured in the absence of FBP under the same conditions as those for FNI. The forward reaction activity was only 0.14± 0.01 μmol min−1 (mg protein)−1, whereas the reverse reaction activity was not detected. However, when FBP was added to a final concentration of 1 mM, the forward reaction activity sharply increased to 1008± 25.2 μmol min−1 (mg protein)−1 and the reverse reaction rate increased to 15.5± 0.5 μmol min−1 (mg protein)−1. Both values were tens of thousands higher than those of FNI. These results validated that recombinant LDHA was successfully expressed and that it catalyzed reactions in a manner strongly dependent on FBP.
3.3.2. Catalytic capacity of LDHA for the forward and reverse reactions
Km (Michaelis constant) values for different substrates and Vm (maximum enzymatic activity) values for the forward and reverse reactions were determined to objectively evaluate the catalytic capacity of LDHA for the forward and reverse reactions.
As shown in Table 3, the Vm value for the reverse reaction was nearly 20% of that for the forward reaction, and the apparent Km values for the reverse substrates were much higher than those for the forward substrates. In particular, the apparent Km value for lactate was nearly eighty-fold higher than that for pyruvate. Obviously, LDHA is prone to catalyzing pyruvate reduction. Its lactate oxidation activity will not be observed unless the forward reaction activity is severely decreased.
Table 3.
Kinetic constants for pyruvate reduction and lactate oxidation reactions of LDHA. Kinetic constants were determined in 0.1 M Tris–maleate (pH 7.0) with 1 mM FBP at 30 °C.
| Substrate | Km value (mM) | Vm value (μmol min−1 mg−1) |
|---|---|---|
| NADH | 0.066 ± 0.008 | 1534 ± 45 |
| Pyruvate | 1.95 ± 0.12 | |
| NAD | 0.38 ± 0.01 | 313 ± 3 |
| l-Lactate | 158 ± 4 |
3.3.3. Influence of effectors on LDHA
FBP and inorganic phosphate are known as the activator and inhibitor of LDHA, respectively [18,27]. Their effects on both reaction rates were investigated in this study (Table 4).
Table 4.
Effects of FBP and phosphate on the rates of reactions catalyzed by LDHA. Values are presented as percentage rate relative to assays carried out under standard conditions described in Section 2. The standard pyruvate reduction and lactate oxidation rates were 1086.1 ± 53.2 and 174.4 ± 6.2 μmol min−1 (mg protein)−1, respectively. The values are expressed as the mean ± SD of at least three independent measurements.
| Concentration (mM) | Pyruvate reduction (%) | Lactate oxidation (%) |
|---|---|---|
| FBP | ||
| 1 | 100 ± 4.1 | 100 ± 1.9 |
| 0.5 | 84 ± 0.6 | 89 ± 2.7 |
| 0.2 | 66 ± 4.7 | 62 ± 4.5 |
| 0.01 | 1.7 ± 0.2 | 1.9 ± 0.2 |
| Phosphate | ||
| 10 | 96 ± 0.5 | 97 ± 0.1 |
| 20 | 94 ± 0.7 | 96 ± 0.5 |
| 30 | 78 ± 0.8 | 77 ± 1.0 |
FBP and inorganic phosphate were observed to have modulating effects on LDHA activities, although the changes observed differ from previously reported results [18], as their effects were dependent on the buffer species [13]. More importantly, the extent of activation or inhibition on both reaction directions were nearly equal such that they would not affect the reaction equilibrium. This can be attributed to the fact that FBP and phosphate are allosteric effectors of LDHA and affect the activity by acting upon the regulatory site but not the substrate binding site of LDHA.
3.3.4. Effects of adenine nucleotides on LDHA activities
ADP and ATP have been reported as competitive inhibitors of nLDH [27,28], and their effects on both reaction directions were determined (Fig. 3). The inhibitory effects of ATP on both directions were weak and similar as both rates remained above 80% in the presence of 8 mM ATP. ADP was shown to be a more severe inhibitor and to have a more severe inhibitory effect on the reverse reaction. However, its relative inhibition on reverse reactions was weak at low ADP concentrations. On the other hand, the competitive inhibitory effect would be greatly affected by the ratio of NADH (or NAD) to ADP. At a fixed ADP concentration, the increase in NAD or decrease in NADH concentration would also help reduce the relative inhibitory effect on the reverse reaction and may even reverse it.
Fig. 3.
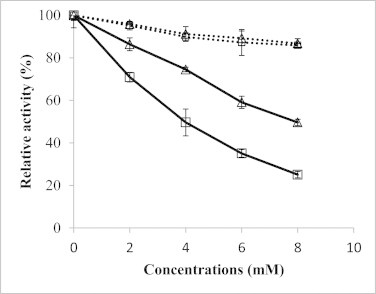
Inhibitory effects of ATP and ADP on LDHA activities. ATP and ADP were separately added to pyruvate reduction and lactate oxidation mixtures. Dotted line, ATP; solid line, ADP; square, forward reaction activity; triangle, reverse reaction activity. Error bars represent SD of at least three independent measurements.
3.3.5. Effects of substrate and product concentrations on LDHA activities
As a primary factor regulating nLDH activity, the effects of NADH/NAD ratios on both reaction rates were measured at a constant substrate concentration and varying product concentrations (Fig. 4).
Fig. 4.
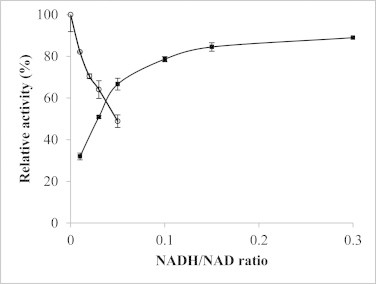
Effects of NADH/NAD ratios on LDHA activities. Solid square, pyruvate reduction activity (NADH at 0.15 mM); circle, lactate oxidation activity (NAD at 5 mM). Error bars represent SD of three independent measurements.
Pyruvate reduction activity was slightly influenced over a wide range of NADH/NAD ratios. It was mildly reduced when the NADH/NAD ratio was above 0.05. When the ratio reached 0.03, it was still approximately 50% of the control. On the other hand, the reverse reaction (i.e. lactate oxidation) was significant only in a narrow range of NADH/NAD ratios. When the ratio reached 0.05, the rate was reduced by nearly 50%.
In addition, the affinity of LDHA for pyruvate was found to be NADH concentration dependent. The apparent Km value for pyruvate was 1.95 ± 0.12 mM at 0.6 mM NADH and increased to 2.50 ± 0.20 mM at 0.1 mM NADH. At 0.05 mM NADH, the Km value reached 2.64 ± 0.19 mM. These observations suggested that the affinity of LDHA for pyruvate decreased with decreasing NADH concentration, which may be related to the ordered bi–bi kinetic mechanism of nLDH with the coenzyme binding first [29].
As lactate can be intracellularly accumulated at high levels [18,30] and its severe inhibitory effect has been reported for LDHB [18], its inhibitory effect on the forward reaction of LDHA was investigated. Its inhibition was weak as the activity was 95 ± 1.6% of the control in the presence of 50 mM lactate. When the lactate concentration increased to 150 and 200 mM, the activity remained at 83 ± 2.2% and 78 ± 2.8%, respectively. A high concentration of lactate thus has a weak inhibitory effect on the forward reaction activity but is meaningful for a significant lactate oxidation rate as the Km value of LDHA for l-lactate is very high.
By contrast, the inhibitory effect of pyruvate on the reverse reaction (lactate oxidation) was much stronger. Even 0.5 mM pyruvate could reduce the reverse reaction activity to 82 ± 4.3% of the control. It was further reduced to 50 ± 2.9% and 25 ± 0.8% of the control when pyruvate was added at 2 and 5 mM, respectively. These observations suggested that lactate oxidation would not be observed unless pyruvate and especially NADH dropped to low levels.
3.4. Intracellular metabolites in respiratory culture
A L. lactis MG1363 respiratory culture was collected at the mid-log and stationary phases and the concentrations of cellular NADH, NAD, ADP, pyruvate and lactate were determined (Table 5). Cellular ADP concentration was always kept at low levels and would not have a significant impact on the reaction equilibrium. When the respiratory culture entered stationary phase, cellular NAD concentration remained almost the same while the NADH content decreased several fold. Thus the NADH/NAD ratio decreased sharply from 0.10 to 0.02, which would be greatly beneficial for lactate oxidation. The concentration of lactate was found to decrease, which could be attributed to the inactivation of glycolysis after glucose exhaustion. However, it was high enough for a significant lactate oxidation rate. Pyruvate concentration decreased significantly when the culture entered stationary phase. In the mid-log phase, the concentration of pyruvate was very significant. However, it decreased to an undetectable level in stationary phase, because an obvious peak for pyruvate could not be detected. Although diacetyl and acetoin rapidly accumulated during this period, the concentration of pyruvate was not as high as expected. This may be attributed to the extremely low available NADH which would reduce the affinity of LDHA for pyruvate and make α-acetolactate synthase for diacetyl and acetoin synthesis become more competitive for pyruvate.
Table 5.
Cellular metabolites in L. lactis MG1363 respiratory culture. The L. lactis MG1363 respiratory culture was collected at the mid-log and stationary phases. After quenching and extraction, targeted metabolites were measured by HPLC. Two extracts were made from the same batch culture. ND, not detected.
| Growth phase | ADP (mM) | NAD (mM) | Lactate (mM) | NADH (μM) | Pyruvate (mM) |
|---|---|---|---|---|---|
| Mid-log | 0.82 ± 0.05 | 3.60 ± 0.12 | 80.62 ± 9.11 | 358 ± 25 | 7.19 ± 1.17 |
| Stationary | 0.74 ± 0.01 | 3.49 ± 0.01 | 62.08 ± 7.56 | 77 ± 3 | ND |
Overall, the cellular status at the stationary phase under respiratory conditions is beneficial for LDHA to catalyze the oxidation of lactate.
4. Discussion
Lactate oxidation is usually catalyzed by iLDHs [10,12]. As a paralog of l-iLDH, FNI is likely for several reasons to catalyze lactate oxidation in the stationary phase of respiration. First, it shares significant homology with known l-iLDHs and thus may catalyze the same reaction. Second, as an essential enzyme in L. lactis, it exists throughout the growth period, which is in accordance with the fact that no gene related to lactate metabolism is distinctively expressed under respiratory conditions [20,21]. Third, the mevalonate pathway, to which FNI belongs, has a close relationship with carbohydrate catabolism (Fig. 5). Acetyl-CoA, the first substrate of mevalonate pathway, is derived from the glycolysis product pyruvate [26]. Furthermore, IPP isomerization catalyzed by type II IPP isomerase is strictly dependent on reductive conditions [26,31]. In respiration, NADH can be oxidized via the electron transport chain; in addition, it becomes more unfavorable for FNI to catalyze IPP isomerization when glucose is exhausted and FNI may then be spared to catalyze the oxidation of lactate to pyruvate. However, the extremely low l-iLDH activity of FNI precludes such a possibility. This can be attributed to the fact that the conserved active site residues of the α-hydroxyacid dehydrogenase family are absent in FNI [15].
Fig. 5.
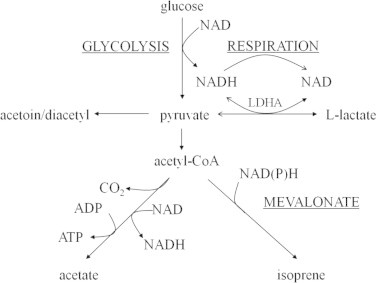
Pyruvate metabolic pathway in Lactococcus lactis under aerobic conditions. Underlined words in uppercase, metabolic pathways. Open arrows, reactions that occur under respiratory conditions.
Similarly, lutABC cluster was proved to be inactive in lactate oxidation, as the cell extracts from L. lactis MG1363 respiratory culture do not exhibit significant l-iLDH activity. In fact, the lutABC operon is widely present in a variety of species, but its physiological role remains unclear [10,12]. In Bacillus subtilis, this operon is required for growth on l-lactate and has an influence on the architectural complexity of biofilm formation on l-lactate [10]. In Neisseria meningitidis, this operon is involved in protecting against oxidative stress [32]. E. coli K12 ΔlldD mutant with lutABC operon cannot grow on l-lactate; however, plasmid-driven expression of the E. coli lutABC operon could successfully restore the ability of Shewanella oneidensis MR-1 Δdld-IIΔlldF double mutant to grow on l-lactate [12]. Further research is needed to determine the physical role of the lutABC operon in L. lactis.
In some cases, fermentative FBP-independent l-nLDHs are involved in lactate utilization [9] but the activity of FBP-dependent l-nLDHs has long been regarded as irreversible under physiological conditions [8,13,14], although FBP-dependent and -independent l-nLDHs share great homology with each other. The thermodynamic equilibrium constant is related to the standard Gibb's free energy of the reaction but independent of enzyme properties per se [33]. Theoretically, FBP-dependent nLDHs can also catalyze the oxidation of lactate.
It was reported that L. lactis IL1403 was one of the few strains in which the benefits of respiration were not significant [34]. However, Arioli et al. [35] have found that the benefits of L. lactis IL1403 respiration will be enhanced if its fermentative nLDH is inhibited, for more NADH and pyruvate fluxes can be redirected to other pathways. It can be seen that fermentative nLDH is a strong competitor for NADH and pyruvate. Their results [35] were in good accordance with our findings. When glucose was sufficient, most pyruvate and NADH were efficiently transformed into lactate and NAD by nLDH (Fig. 1). When glucose was exhausted, the supply of NADH and pyruvate ran short and the pyruvate reduction activity of nLDH was severely restricted. Meanwhile, the respiratory chain continued to actively oxidize NADH to establish the proton motive force (PMF). Consequently, the affinity of nLDH for pyruvate decreased with decreasing NADH concentration, and the α-acetolactate synthase and pyruvate dehydrogenase complex became more competitive for pyruvate (Fig. 5). The continuous removal of NADH and pyruvate revealed the lactate oxidation activity of nLDH, and the significant concentrations of NAD and lactate allowed for it to proceed at a significant rate.
In L. lactis, lactate utilization has only been observed under respiratory conditions [1]. This may be attributed to the lactate transport mechanism. In lactate-utilizing species, the lactate permease gene is always found in the genome and expressed by the induction of lactate [11,12,36]. However, no homologs can be found in Lactococcus. In L. lactis, lactate is transported in symport with one or more protons, and the transport direction is determined by the difference between the inward and outward driving forces [37]. The outward force is provided by the transmembrane lactate gradient, and the PMF exerts an inward force on the protons [38]. As shown in Fig. 6, under fermentation conditions, F1Fo-ATPase is mainly responsible for PMF generation by pumping protons at the expense of ATP [39]. However, under respiratory conditions, a functional respiratory chain is formed and PMF is generated in the process of NADH oxidation via the ETC [6,39,40]. The PMF established by respiration is large enough to promote proton influx through F1Fo-ATPase, and ATP is concomitantly synthesized from ADP. Koebmann [6] confirmed that increased biomass yield in L. lactis respiration can be partly attributed to the benefit of a reverse function of F1Fo-ATPase toward oxidative phosphorylation. Similar to F1Fo-ATPase, lactate translocation is reversible [37]. When the PMF generated by the ETC exceeds the transmembrane lactate gradient, lactate uptake in symport with protons can occur, which enables lactate oxidation to proceed continually. Additional energy may benefit from acetate production and NADH oxidation via the ETC and further growth could be observed [1]. Moreover, lactate oxidation during starvation can further reduce acid stress, which is beneficial for the survival of the cells. Extracellular lactate utilization could not be observed under fermentation conditions, which can be attributed to its inability to drive lactate influx, and then lactate oxidation reaction would not proceed continually. The lactate permease gene is also absent in some respiratory species with the lutABC cluster [10,12]. If they could grow on l-lactate, then lactate may be imported in a similar way as in L. lactis.
Fig. 6.
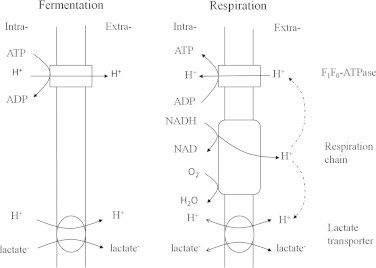
Comparison of proton and lactate transport pathways under fermentation and respiratory conditions.
Our conclusions would have been strengthened if we had been able to support our conclusions through studying a knockout mutant in the ldh gene. Unfortunately, our attempts using standard procedures using antibiotic insertion methodology have been unsuccessful.
In conclusion, the results presented in this paper show that L-iLDHs (FNI and the products of the lutABC cluster) are not involved in lactate utilization in L. lactis. Fermentative FBP-activated l-nLDH can catalyze lactate oxidation in vivo, provided that NADH and pyruvate are kept at low levels and lactate can be imported into the cell.
Acknowledgements
L. lactis MG1363 was kindly provided by J. Kok (Molecular Genetics University of Groningen, The Netherlands). This study was financially supported by the National 863 grant (No. 2011AA100902) and a grogram for Changjiang Scholars and Innovative Research Team in Northeast Agriculture University (IRT0959).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Duwat P., Sourice S., Cesselin B., Lamberet G., Vido K., Gaudu P., Loir Y.L., Violet F., Loubière P., Gruss A. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 2001;183:4509–4516. doi: 10.1128/JB.183.15.4509-4516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duwat, P., Bravard, A., Sourice, S., and Gruss, A. (1998). Process for preparing starter cultures of lactic acid bacteria. French Patent Application No. FR9809463.
- 3.Sijpesteijn A.K. Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie Leeuwenhoek. 1970;36:335–348. doi: 10.1007/BF02069035. [DOI] [PubMed] [Google Scholar]
- 4.Gaudu P., Vido K., Cesselin B., Kulakauskas S., Tremblay J., Rezaïki L., Lamberet G., Sourice S., Duwat P., Gruss A. Respiration capacity and consequences in Lactococcus lactis. Antonie Leeuwenhoek. 2002;82:263–269. [PubMed] [Google Scholar]
- 5.Rezaïki L., Cesselin B., Yamamoto Y., Vido K., van West E., Gaudu P., Gruss A. Respiration metabolism reduces oxidative and acid stress to improve long-term survival of Lactococcus lactis. Mol. Microbiol. 2004;53:1331–1342. doi: 10.1111/j.1365-2958.2004.04217.x. [DOI] [PubMed] [Google Scholar]
- 6.Koebmann B., Blank L.M., Solem C., Petranovic D., Nielsen L.K., Jensen P.R. Increased biomass yield of Lactococcus lactis during energetically limited growth and respiratory conditions. Biotechnol. Appl. Biochem. 2008;50:25–33. doi: 10.1042/BA20070132. [DOI] [PubMed] [Google Scholar]
- 7.Lan C.Q., Oddone G., Mills D.A., Block D.E. Kinetics of Lactococcus lactis growth and metabolite formation under aerobic and anaerobic conditions in the presence or absence of hemin. Biotechnol. Bioeng. 2006;95:1070–1080. doi: 10.1002/bit.21070. [DOI] [PubMed] [Google Scholar]
- 8.Garvie E.I. Bacterial lactate dehydrogenases. Microbiol. Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffin P., Lorquet F., Kleerebezem M., Hols P. Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase. J. Bacteriol. 2004;186:6661–6666. doi: 10.1128/JB.186.19.6661-6666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai Y.R., Kolter R., Losick R. A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J. Bacteriol. 2009;191:2423–2430. doi: 10.1128/JB.01464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas M.T., Shepherd M., Poole R.K., van Vliet A.H., Kelly D.J., Pearson B.M. Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on l-lactate. Environ. Microbiol. 2011;13:48–61. doi: 10.1111/j.1462-2920.2010.02307.x. [DOI] [PubMed] [Google Scholar]
- 12.Pinchuk G.E., Rodionov D.A., Yang C., Li X.Q., Osterman A.L., Dervyn E., Geydebrekht O.V., Reed S.B., Romine M.F., Collart F.R., Scott J.H., Fredrickson J.K., Beliaev A.S. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc. Natl. Acad. Sci. USA. 2009;106:2874–2879. doi: 10.1073/pnas.0806798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crow V.L., Pritchard G.G. Fructose 1,6-diphosphate-activated l-lactate dehydrogenase from Streptococcus lactis: kinetic properties and factors affecting activation. J. Bacteriol. 1977;131:82–91. doi: 10.1128/jb.131.1.82-91.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Niel E.W.J., Palmfeldt J., Martin R., Paese M., Hägerdal B.H. Reappraisal of the regulation of Lactococcal L-lactate dehydrogenase. Appl. Environ. Microbiol. 2004;70:1843–1846. doi: 10.1128/AEM.70.3.1843-1846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laupitz R., Hecht S., Amslinger S., Zepeck F., Kaiser J., Richater G., Schramek N., Steinbacher S., Huber R., Arigoni D., Bacher A., Eisenreich W., Rohdich F. Biochemical characterization of Bacillus subtilis type II isopentenyl diphosphate isomerase, and phylogenetic distribution of isoprenoid biosynthetic pathways. Eur. J. Biochem. 2004;271:2658–2669. doi: 10.1111/j.1432-1033.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 16.Rezaïki L., Lamberet G., Derré A., Gruss A., Gaudu P. Lactococcus lactis produces short-chain quinones that cross-feed Group B Streptococcus to activate respiration growth. Mol. Microbiol. 2008;67:947–957. doi: 10.1111/j.1365-2958.2007.06083.x. [DOI] [PubMed] [Google Scholar]
- 17.Wegmann U., O’Connell-Motherway M., Zomer A., Buist G., Shearman C., Canchaya C., Ventura M., Goesmann A., Gasson M.J., Kuipers O.P., van Sinderen D., Kok J. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 2007;189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaspar P., Neves A.R., Shearman C.A., Gasson M.J., Baptista A.M., Turner D.L., Soares C.M., Santos H. The lactate dehydrogenases encoded by the ldh and ldhB genes in Lactococcus lactis exhibit distinct regulation and catalytic properties-comparative modeling to probe the molecular basis. FEBS J. 2007;274:5924–5936. doi: 10.1111/j.1742-4658.2007.06115.x. [DOI] [PubMed] [Google Scholar]
- 19.Gaspar P., Neves A.R., Gasson M.J., Shearman C.A., Santos H. High yields of 2,3-butanediol and mannitol in Lactococcus lactis through engineering of NAD+ cofactor recycling. Appl. Environ. Microbiol. 2011;77:6826–6835. doi: 10.1128/AEM.05544-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedersen M.B., Garrigues C., Tuphile K., Brun C., Vido K., Bennedsen M., Møllgaard H., Gaudu P., Gruss A. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J. Bacteriol. 2008;190:4903–4911. doi: 10.1128/JB.00447-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vido K., Le Bars D., Mistou M.Y., Anglade P., Gruss A., Gaudu P. Proteome analyses of heme-dependent respiration in Lactococcus lactis: involvement of the proteolytic system. J. Bacteriol. 2004;186:1648–1657. doi: 10.1128/JB.186.6.1648-1657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao J., Ma C., Gao C., Qiu J., Wang M., Zhang Y., Cui X., Xu P. Pseudomonas stutzeri as a novel biocatalyst for pyruvate production from dl-lactate. Biotechnol. Lett. 2007;29:105–110. doi: 10.1007/s10529-006-9204-6. [DOI] [PubMed] [Google Scholar]
- 23.Koebmann B., Solem C., Jensen P.R. Control analysis as a tool to understand the formation of the las operon in Lactococcus lactis. FEBS J. 2005;272:2292–2303. doi: 10.1111/j.1742-4658.2005.04656.x. [DOI] [PubMed] [Google Scholar]
- 24.Villas-Bôas S.G., Højer-Pedersen J., Akesson M., Smedsgaard J., Nielsen J. Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast. 2005;22:1155–1169. doi: 10.1002/yea.1308. [DOI] [PubMed] [Google Scholar]
- 25.Poolman B., Smid E.J., Veldkamp H., Konings W.N. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J. Bacteriol. 1987;169:1460–1468. doi: 10.1128/jb.169.4.1460-1468.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneda K., Kuzuyama T., Takagi M., Hayakawa Y., Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. USA. 2000;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonas H.A., Anders R.F., Jago G.R. Factors affecting the activity of the lactate dehydrogenase of Streptococcus cremoris. J. Bacteriol. 1972;111:397–403. doi: 10.1128/jb.111.2.397-403.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao R., Zeidan A.A., Rådström P., van Niel E.W. Inhibition kinetics of catabolic dehydrogenases by elevated moieties of ATP and ADP-implication for a new regulation mechanism in Lactococcus lactis. FEBS J. 2010;277:1843–1852. doi: 10.1111/j.1742-4658.2010.07601.x. [DOI] [PubMed] [Google Scholar]
- 29.Shoemark D.K., Cliff M.J., Sessions R.B., Clarke A.R. Enzymatic properties of the lactate dehydrogenase enzyme from Plasmodium falciparum. FEBS J. 2007;274:2738–2748. doi: 10.1111/j.1742-4658.2007.05808.x. [DOI] [PubMed] [Google Scholar]
- 30.Neves A.R., Ventura R., Mansour N., Shearman C., Gasson M.J., Maycock C., Ramos A., Santos H. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? J. Biol. Chem. 2002;277:28088–28098. doi: 10.1074/jbc.M202573200. [DOI] [PubMed] [Google Scholar]
- 31.Hemmi H., Ikeda Y., Yamashita S., Nakayama T., Nishino T. Catalytic mechanism of type 2 isopentenyl diphosphate: dimethylallyl diphosphate isomerase: verification of a redox role of the flavin cofactor in a reaction with no redox change. Biochem. Biophys. Res. Commun. 2004;322:905–910. doi: 10.1016/j.bbrc.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Grifantini R., Frigimelica E., Delany I., Bartolini E., Giovinazzi S., Balloni S., Agarwal S., Galli G., Genco C., Grandi G. Characterization of a novel Neisseria meningitidis Fur and iron-regulated operon required for protection from oxidative stress: utility of DNA microarray in the assignment of the biological role of hypothetical genes. Mol. Microbiol. 2004;54:962–979. doi: 10.1111/j.1365-2958.2004.04315.x. [DOI] [PubMed] [Google Scholar]
- 33.Bulik S., Grimbs S., Huthmacher C., Selbig J., Holzhütter H.G. Kinetic hybrid models composed of mechanistic and simplified enzymatic rate laws – a promising method for speeding up the kinetic modelling of complex metabolic networks. FEBS J. 2009;276:410–424. doi: 10.1111/j.1742-4658.2008.06784.x. [DOI] [PubMed] [Google Scholar]
- 34.Brooijmans R., Smit B., Santos F., van Riel J., de Vos W.M., Hugenholtz J. Heme and menaquinone induced electron transport in lactic acid bacteria. Microb. Cell Fact. 2009;8:28. doi: 10.1186/1475-2859-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arioli S., Zambelli D., Guglielmetti S., De Noni I., Pedersen M.B., Dedenroth P.D., Pedersen, Dal Bello F., Mora D. Increasing the heme-dependent respiratory efficiency of Lactococcus lactis by inhibition of lactate dehydrogenase. Appl. Environ. Microbiol. 2013;79:376–380. doi: 10.1128/AEM.02734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao C., Hu C., Zheng Z., Ma C., Jiang T., Dou P., Zhang W., Che B., Wang Y., Lv M., Xu P. Lactate utilization is regulated by the FadR-type regulator LldR in Pseudomonas aeruginosa. J. Bacteriol. 2012;194:2687–2692. doi: 10.1128/JB.06579-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto R., Sonnenberg A.S.M., Veldkamp H., Konings W.N. Generation of an electrochemical proton gradient in Streptococcus cremoris by lactate efflux. Proc. Natl. Acad. Sci. USA. 1980;77:5502–5506. doi: 10.1073/pnas.77.9.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konings W.N. The cell membrane and the struggle for life of lactic acid bacteria. Antonie Leeuwenhoek. 2002;82:3–27. [PubMed] [Google Scholar]
- 39.Brooijmans R.J.W., Poolman B., Schuurman-Wolters G.K., de Vos W.M., Hugenholtz J. Generation of a membrane potential by Lactococcus lactis through aerobic electron transport. J. Bacteriol. 2007;189:5203–5209. doi: 10.1128/JB.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blank L.M., Koebmann B.J., Michelsen O., Nielsen L.K., Jensen P.R. Hemin reconstitutes proton extrusion in an H+-ATPase-negative mutant of Lactococcus lactis. J. Bacteriol. 2001;183:6707–6709. doi: 10.1128/JB.183.22.6707-6709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


