Abstract
Overexpression of the ATP-binding cassette (ABC) drug efflux proteins P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) on malignant cells is associated with inferior chemotherapy outcomes. Both, ABCB1 and ABCG2, are substrates of the serine/threonine kinase Pim-1; Pim-1 knockdown decreases their cell surface expression, but SGI-1776, the first clinically tested Pim inhibitor, was shown to reverse drug resistance by directly inhibiting ABCB1-mediated transport. We sought to characterize Pim-1-dependent and -independent effects of SGI-1776 on drug resistance. SGI-1776 at the Pim-1-inhibitory and non-cytotoxic concentration of 1 μM decreased the IC50s of the ABCG2 and ABCB1 substrate drugs in cytotoxicity assays in resistant cells, with no effect on the IC50 of non-substrate drug, nor in parental cells. SGI-1776 also increased apoptosis of cells overexpressing ABCG2 or ABCB1 exposed to substrate chemotherapy drugs and decreased their colony formation in the presence of substrate, but not non-substrate, drugs, with no effect on parental cells. SGI-1776 decreased ABCB1 and ABCG2 surface expression on K562/ABCB1 and K562/ABCG2 cells, respectively, with Pim-1 overexpression, but not HL60/VCR and 8226/MR20 cells, with lower-level Pim-1 expression. Finally, SGI-1776 inhibited uptake of ABCG2 and ABCB1 substrates in a concentration-dependent manner irrespective of Pim-1 expression, inhibited ABCB1 and ABCG2 photoaffinity labeling with the transport substrate [125I]iodoarylazidoprazosin ([125I]IAAP) and stimulated ABCB1 and ABCG2 ATPase activity. Thus SGI-1776 decreases cell surface expression of ABCB1 and ABCG2 and inhibits drug transport by Pim-1-dependent and -independent mechanisms, respectively. Decrease in ABCB1 and ABCG2 cell surface expression mediated by Pim-1 inhibition represents a novel mechanism of chemosensitization.
Keywords: SGI-1776, Pim-1 kinase, ABCG2, ABCB1, multidrug resistance, leukemia
1. Introduction
Intrinsic and acquired resistance to chemotherapy drugs is a formidable barrier to the success of cancer chemotherapy, and ATP-binding cassette (ABC) proteins that function as drug efflux pumps are strongly implicated as an important mechanism of chemotherapy resistance [1]. Overexpression of ABCB1 [P-glycoprotein (Pgp); MDR1] and ABCG2 [breast cancer resistance protein (BCRP); mitoxantrone resistance protein (MXR)] is associated with inferior chemotherapy treatment outcomes in acute myeloid leukemia (AML) [2, 3] and in other malignancies. While inhibition of ABCB1-mediated drug transport is highly effective in sensitizing ABCB1-overexpressing cells to ABCB1 substrate chemotherapy drugs in vitro, administration of clinically applicable inhibitors of ABCB1-mediated drug transport, including PSC-833 and zosuquidar, in conjunction with chemotherapy in clinical trials in AML unfortunately did not improve treatment outcomes [4-8]. One possible reason is lack of inhibition of ABCG2 [9, 10]. Cyclosporine A, which inhibits both ABCB1 and ABCG2, as well as other drug resistance proteins [11], improved treatment outcomes in some [12-14], but not all [15], clinical trials. Given the strong association of ABCB1 and ABCG2 overexpression with treatment failure in AML, exploration of inhibitors of both proteins and of novel approaches to overcoming drug resistance mediated by both proteins is warranted.
The serine/threonine kinase Pim-1, encoded by a proto-oncogene originally identified as the proviral integration site in Moloney murine leukemia virus lymphomagenesis and a member of the Pim kinase family, which also includes Pim-2 and Pim-3 [16], has been implicated in drug resistance mediated by ABCB1 and ABCG2. Our group and our collaborators found that Pim-1 phosphorylates cytoplasmic, non-glycosylated 150 kda ABCB1 on serine 683 [17] and ABCG2 on threonine 362 [18] and that phosphorylation by Pim-1 promotes ABCG2 multimerization as well as translocation of both ABCB1 and ABCG2 to the cell surface, where they mediate drug efflux. Specifically, ABCB1 Pim-1 phosphorylation consensus sequence, QDRKLS, located at serine 683, between nucleotide-binding domain 1 and transmembrane domain 1, and Pim-1 phosphorylates non-glycosylated 150 kDa ABCB1 and thereby protects it from proteolytic and proteasomal degradation and enables its glycosylation to 170 kDa ABCB1, which translocates to the cell surface and mediates drug efflux [17]. Additionally, Pim-1 phosphorylates ABCG2 at threonine 362, knocking down Pim-1 abolishes ABCG2 multimer formation and ABCG2 T362A mutation decreases ABCG2 expression on the cell surface [18]. Pim-1 is expressed in AML [19] and other malignancies, and its inhibition may represent a novel approach to overcoming drug resistance mediated by ABCB1 and ABCG2.
The imidazo[1,2-b]pyridazine small molecule SGI-1776 [20-22] is the first Pim kinase inhibitor to have undergone clinical testing. SGI-1776 inhibits Pim-1, Pim-2 and Pim-3 with IC50 values of 7 nM, 363 nM, and 69 nM, respectively, in kinase inhibition assays [20], but is more than 95% bound to human plasma protein [20], and concentrations that inhibit Pim kinase activity are therefore approximately 100-fold higher in cell culture-based assays, in relation to kinase inhibition assays. We previously demonstrated that SGI-1776 inhibits Pim-1 at a concentration of 1 μM in a cell culture-based assay of inhibition of phosphorylation of Bcl-2-associated death promoter (BAD) protein at serine 112, and is not cytotoxic at this concentration, but is cytotoxic at higher concentrations [17]. SGI-1776 has been shown to sensitize ABCB1-overexpressing drug-resistant cells to ABCB1 substrate cancer chemotherapy drugs [17, 20], but was found to directly inhibit drug transport mediated by ABCB1 as a mechanism of chemosensitization [20]. While silencing of Pim-1 expression with siRNA was found to chemosensitize ABCG2-overexpressing drug-resistant cells to ABCG2 substrate chemotherapy drugs [18], the effects of SGI-1776 on resistance mediated by ABCG2 have not been studied.
In the work presented here, we studied the effect of SGI-1776 on chemosensitivity of cells overexpressing ABCG2, as well as ABCB1, and characterized the effects of SGI-1776 on ABCG2 and ABCB1 cell surface expression and on ABCG2- and ABCB1-mediated drug transport as mechanisms of chemosensitization.
2. Materials and Methods
2.1. Cell lines
HL60 and K562 leukemia cells were obtained from the American Type Culture Collection (ATCC), Manassas, VA, vincristine-selected HL60/VCR cells, overexpressing ABCB1, from Dr. Ahmad R. Safa, Indiana University, Indianapolis, IN [23], and 8226 and mitoxantrone-selected ABCG2-overexpressing 8226/MR20 myeloma cells [24] from Dr. William S. Dalton, Moffitt Cancer Center, Tampa, FL. HL60/VCR cells were maintained in drug-free RPMI 1640 medium with 10% fetal bovine serum (FBS) and 8226/MR20 cells in RPMI 1640 medium with 10% FBS and 20 nM mitoxantrone. K562/ABCB1 [25] and K562/ABCG2 [26] cells, transfected cell lines stably overexpressing ABCB1 and ABCG2, were gifts from Dr. Michael Gottesman, National Cancer Institute, Bethesda, MD and Dr. Yoshikazu Sugimoto, Kyoritsu University of Pharmacy, Tokyo, Japan, respectively, and flavopiridol-selected MCF-7/Flv1000 cells [27], overexpressing ABCG2, from Dr. Susan Bates, National Cancer Institute.
2.2. Materials
SGI-1776 was a gift from Tolero Pharmaceuticals, Inc. (Salt Lake City, UT). It was stocked at 50 mM in DMSO at −20°C, and was used at a concentration of 1 μM, as we previously demonstrated that it inhibits Pim-1 in cell culture-based assays and is not cytotoxic at this concentration [17]. DMSO was used in control treatments, with a maximum final concentration of 0.05%. The chemotherapy drugs daunorubicin, etoposide, paclitaxel, mitoxantrone and topotecan were purchased from Sigma-Aldrich (St Louis, MO), and flavopiridol from Enzo Life Sciences (Farmingdale, NY). Chemical structures of SGI-1776, the structurally similar kinase inhibitor ponatinib [28] and of the chemotherapy drugs studied are shown in Figure 1.
Figure 1. Chemical Structures.
Chemical structures of SGI-1776, ponatinib, mitoxantrone, topotecan, flavopiridol, daunorubicin, etoposide and paclitaxel are shown.
Fluorescein isothiocyanate (FITC)-conjugated annexin V and propidium iodide (PI) were purchased from Trevigen (Gaithersburg, MD), APC annexin V from BD Biosciences and LIVE/DEAD fixable near-IR dead cell stain from Invitrogen (Carlsbad, CA). MRK16 antibody to a cell surface epitope of ABCB1 was purchased from Alexis Biochemicals (San Diego, CA), allophycocyanin conjugate (APC)-tagged 5D3 antibody to a cell surface epitope of ABCG2 from BD Biosciences (San Jose, CA) and BXP-21 antibody to BCRP from Signet Laboratories (Dedham, MA). The fluorescent ABCB1 substrate 3,3′-diethyloxacarbocyanine iodide [DiOC2(3)] [29] was purchased from Sigma-Aldrich, and the fluorescent ABCG2 substrate pheophorbide A (PhA) [29] from Frontier Scientific (Logan, VT). The ABCB1 inhibitor PSC-833 was obtained from Novartis Pharmaceutical Corporation (East Hanover, NJ), and the ABCG2 inhibitor fumitremorgin C (FTC) was purchased from Sigma-Aldrich [30].
[125I]Iodoarylazidoprazosin (IAAP) (2200 Ci/mmol) was purchased from PerkinElmer Life and Analytical Sciences (Waltham, MA).
2.3. Immunoblotting
Cells were lysed in RIPA buffer (Sigma-Aldrich) supplemented with protease and phosphatase inhibitor cocktails (Roche Applied Science, Indianapolis, IN), protein concentrations were measured using the Bio-Rad Protein Assay KIT I (Bio-Rad Laboratories, Inc, Hercules, CA) according to kit instructions, and immunoblotting was performed as described previously [17]. Briefly, 20 μg or 100 μg of protein from each sample were electrophoresed and immunoblots were incubated with individual primary antibodies, including 1:400 dilution of rabbit anti-Pim-1, anti-Pim-2 and anti-Pim-3 (Cell Signaling Technology, Danvers, MA), 1:1,000 dilution of anti-ABCB1 (Santa Cruz), 1:500 dilution of anti-ABCG2 (Millipore, Billerica, MA) and 1:10,000 dilution of mouse anti-(GAPDH) glyceraldehyde-3-phosphate dehydrogenase (Millipore), for one hour at room temperature or overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated secondary antibody for one hour at room temperature.
2.4. Densitometric analysis
Band intensities of chemiluminescent protein immunocomplexes were measured by densitometry (Visionworks LS image acquisition and analysis software, UVP, Upland, CA). The relative intensity (RI) of ABCB1 or ABCG2 over GAPDH was calculated for SGI-1776 and DMSO control treatments as ABCB1 intensity/GAPDH intensity or ABCG2 intensity/GAPDH intensity. The relative expression (RE) of ABCB1 or ABCG2 intensity/GAPDH intensity in the presence of SGI-1776 (treatment) relative to DMSO (control) was then calculated as RI(treatment)/RI(control).
2.5. Reverse transcriptase polymerase chain reaction
Total RNA was extracted using the RNeasy kit (Qiagen) according to manufacturer instructions. cDNA was then reverse transcribed from the total RNA samples using the SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen). The cDNAs prepared were subsequently used as templates for real-time semi-quantitative PCR with the following primers:
Pim-1 sense: 5′-ACTTGCCGGTGGCCATCAAACA-3′
Pim-1 anti-sense: 5′-ACCCGAGCTCACCTTCTTCAGCA-3′
Pim-2 sense: 5′-AAGCCGCTGCTTCTTTGGCCAA-3′
Pim-2 anti-sense: 5′-AATGAGTTTGGCACAGCCACGG-3′
Pim-3 sense: 5′-TTCGACTTTATCACGGAGCGCG-3′
Pim-3 anti-sense: 5′-AGCGCAGGTCCACAAGCAGATT-3′
GAPDH sense: 5′-ACCATGGAGAAGGCTGGGGCTCAT-3′
GAPDH anti-sense: 5′-ACAGTTTCCCGGAGGGGCCAT-3′.
Expression of each Pim mRNA was quantified relative to control GAPDH mRNA and plotted graphically.
2.6. Cell survival assay
Cells treated with SGI-1776 at a range of concentrations or chemotherapy drugs at a range of concentrations with and without SGI-1776 were cultured in 96-well microculture plates using 96-hour continuous exposure conditions, and viability was then quantified by the WST-1 colorimetric assay, which assesses cell survival by metabolic activity. In this assay, water-soluble tetrazolium salt-1 or 2-(4-Iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium is cleaved to a soluble formazan by a complex cellular mechanism that is largely dependent on the glycolytic production of NAD(P)H in viable cells, so that the amount of formazan dye formed correlates directly with the number of metabolically active cells per well. Briefly, 10 μL WST-1 (Roche Diagnostics (Indianapolis, IN) was added to each well, incubation was continued at 37°C for 2 to 4 additional hours and the color developed was quantified according to the manufacturer’s instructions. Each experiment was performed in triplicate at least three times.
2.7. Curve shift assay
The effect of SGI-1776 on the sensitivity of drug-resistant and parental cells to chemotherapy drugs was determined by plating cells with chemotherapy drugs at a range of concentrations in a cell viability assay, as described above, in the presence and absence of 1 μM SGI-1776, with analysis by the WST-1 colorimetric assay, as described above. The resistance-modifying factor (RMF) was calculated as the ratio of IC50 values in the absence and presence of SGI-1776.
2.8. Measurement of apoptosis
8226/MR20 and K562/ABCG2 cells, overexpressing ABCG2, were incubated with mitoxantrone, topotecan or flavopiridol and HL60/VCR and K562/ABCB1 cells, overexpressing ABCB1, were incubated with etoposide or paclitaxel at serial concentrations for 48 hours in the presence and absence of SGI-1776, and apoptosis and necrosis were measured by staining with annexin V-FITC and PI, as previously described [28]. HL60/VCR and K562/ABCB1 cells were also incubated with daunorubicin at serial concentrations for 48 hours in the presence and absence of SGI-1776, and apoptosis and necrosis were measured using APC-conjugated annexin V and LIVE/DEAD fixable near-IR dead cell stain, to avoid spectral overlap with daunorubicin, as previously described [28].
2.9. Colony formation assay
Briefly, 500 cells were cultured in RPMI 1640 medium supplemented with 10% FBS for 10 days in methylcellulose-based medium (R&D Systems, Inc, Minneapolis, MN) with chemotherapy drugs at a range of concentrations in the presence and absence of 1μM SGI-1776, and colonies were counted on a colony counter (Microbiology International, Frederick, MD). Each treatment was performed in triplicate.
2.10. Cell surface ABCB1 and ABCG2 expression
HL60/VCR, 8226/MR20, K562/ABCG2 and K562/ABCB1 cells were incubated for 96 hours in medium with 1 μM SGI-1776 or DMSO control, with addition of fresh medium with SGl-1776 or DMSO at 48 hours. Cell surface expression of ABCB1 or ABCG2 was then measured by labeling with MRK16 or 5D3 antibody, respectively, as previously described [28].
2.11. Pim-1 shRNA knockdown
K562/ABCB1 cells were transduced with Control or Pim-1 MISSION shRNA lentiviral transduction particles (Sigma-Alrdich Co. LLC) by spinoculation using lipofectamine 2000 (Invitrogen) according to an established viral transduction protocol [31] with minor modifications, and cultured for three days in complete growth medium. shRNA-transduced K562/ABCB1 cells were then selected by culture for an additional three days in growth medium supplemented with 100 μg/ml puromycin hydrochloride (Sigma-Aldrich). ABCB1 cell surface expression was then measured by flow cytometry and total ABCB1, Pim-1 and GAPDH expression by immunoblotting.
2.12. Effect of SGI-1776 on ABCB1 serine phosphorylation
100 μg crude membrane extracts of High-Five insect cells overexpressing ABCB1 were diluted with RIPA buffer supplemented with 1% aprotinin (Sigma-Aldrich), and 100 μL protein A sepharose beads (Sigma-Aldrich) were added, followed by overnight incubation at 4°C with 10 μg ABCB1 antibody (C219, Fujirebio Diagnostics, Inc., Malvern, PA). The immunoprecipitates were washed twice with RIPA buffer, then with 50 mM Tris-Hcl, and 0.5 μg GST (Sigma-Aldrich) or GST-Pim-1 (Millipore) pre-incubated in 40μL 1× kinase buffer [32] with or without 1μM SGI-1776 for 5 minutes at 30°C was added, followed by additional incubation at 30°C for 25 minutes, addition of 2× sample loading buffer, and incubation at 42°C for 1 hour. The ABCB1 immunoprecipitates were electrophoresed on an 8% tris-gycine gel, probed overnight at 4°C with anti-rabbit phosphoserine primary antibody (Millipore), and then with HRP-labeled secondary antibody for 1 hour at room temperature. Control immunoprecipitation with mouse IgG was also performed. The blot was reprobed with ABCB1 antibody to confirm uniform loading.
2.13. Accumulation of fluorescent ABCB1 and ABCG2 substrates
To measure the effect of SGI-1776 on steady-state accumulation of fluorescent ABC protein substrates as an assessment of transport function, HL60/VCR and K562/ABCB1 cells (1 × 106) were incubated for 30 minutes at 37°C with DiOC2(3) (0.6 ng/mL) and SGI-1776 (0-5 μM) or PSC-883 (2.5 μM) as a control, and 8226/MR20 and K562/ABCG2 with PhA (1 μM) and SGI-1776 (0-5 μM) or FTC (10 μM) as a control. Cells were then washed twice with PBS, resuspended in PBS, then analyzed as previously described [28]. Substrate content after uptake with and without modulator was compared using the Kolmogorov-Smirnov statistic, expressed as a D-value ranging from 0 (no difference) to 1 (no overlap) [33], with D-values ≥0.2 indicating significant modulation, based on previous work [34].
2.14. Photoaffinity labeling of ABCG2 and ABCB1 with [125I]IAAP
High-Five insect cell membrane vesicles expressing ABCB1 and crude membranes from MCF-7/Flv1000 cells (30 μg) expressing ABCG2 were incubated with SGI-1776 at the indicated concentrations for 5 minutes at 21-23°C in 50 mM Tris-HCl, pH 7.5. [125I]-IAAP (2200 Ci/mmole), 3-6 nM, was added and photoaffinity labeling of ABCB1 and ABCG2 by [125I]IAAP was measured as previously described [35, 36].
2.15. ABCG2 and ABCB1 ATPase assay
The ATPase activity of vanadate (Vi)-sensitive ABCB1 and beryllium fluoride (BeFx)-sensitive ABCG2 expressed in the membrane vesicles of High-Five insect cells, in the presence of SGI-1776 at serial concentrations, was measured as previously described [36, 37].
2.16. Statistical analyses
All statistical analyses were performed using Graphpad Prism V software (GraphPad Software, Inc., La Jolla, CA). IC50 values were calculated by the least square fit of dose-response inhibition non-linear regression model. Comparisons for apoptosis assays and colony assays were performed using two-way ANOVA with post hoc Bonferroni testing [38]. ABCG2 and ABCB1 protein expression data were compared using the unpaired Student T-test. Statistical significance was defined by p<0.05.
3. Results
3.1. Expression of Pim kinase isoforms in resistant and parental cells
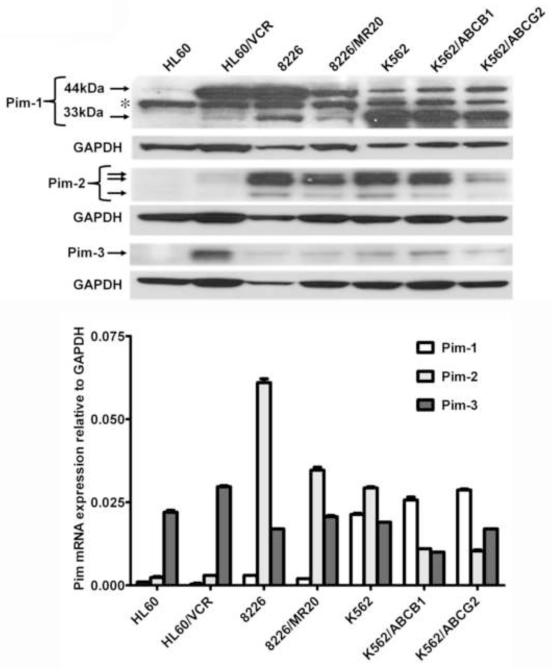
Prior to studying the effects of the Pim kinase inhibitor SGI-1776 on drug resistance of cells overexpressing ABCB1 or ABCG2, we characterized resistant and parental cells for Pim-1, Pim-2 and Pim-3 kinase expression by immunoblotting of 20 μg protein from K562, K562/ABCB1 and K562/ABCG2 and 100 μg protein from HL60, HL60/VCR, 8226 and 8226/MR20 cells, due to differences in Pim-1 expression levels (Figure 2A). 33 kDa Pim-1 was strongly expressed in K562, K562/ABCB1 and K562/ABCG2 and expressed at lower level in HL60/VCR, 8226 and 8226/MR20 cells, but not in parental HL60. Expression of 33 kDa Pim-1 generally correlated with total Pim-1 mRNA expression measured by RT-PCR (Figure 2B). A band was also seen at 44 kDa, but did not correlate with the 33 kDa band, nor, importantly, with Pim-1 mRNA expression measured by RT-PCR (Figure 2B). This finding is consistent with previously reported expression of Pim-1 exclusively as a 33 or 34 kDa protein in human cells, in contrast to its expression as both 33 or 34 kDa and 44 kDa proteins in murine cells [39], though expression of 44 kDa Pim-1 has been reported in human prostate cancer cell lines [40]. The exact identify of the 44 kDa band is the human leukemia and myeloma cells that we studied is not clear; 44 kDa Pim-1 has been reported to originate from an alternative translation initiation site [39].
Figure 2. Pim kinase expression in ABCB1- and ABCG2- overexpressing and parental cell lines.
A. Pim-1, Pim-2 and Pim-3 kinase expression in cell lines expressing ABCG2 or ABCB1 and parental cells was studied by immunoblotting, and GAPDH expression was measured as a loading control. A representative blot from three separate experiments is shown. * indicates a non-specific band. In the Pim-1 immunoblot a five-fold higher quantity of protein was loaded in the HL60, HL60/VCR, 8226 and 8226/MR20 lanes. B. Pim-1, Pim-2 and Pim-3 mRNA expression, measured by RT-PCR and normalized to GAPDH expression.
Pim-2 and Pim-3 expressed was variable (Figure 2, A and B). In the cell lines studied, there was no apparent systematic difference in Pim-1, Pim-2 or Pim-3 kinase expression levels between cells with ABCB1 or ABCG2 expression and parental cells.
Also noted was dissociation between mRNA and protein levels, e.g. low Pim-1 mRNA level but presence of 33 kDa Pim-1 protein in 8226 cells, and Pim-3 mRNA in HL60 cells, but no protein. This apparent post-transcriptional regulation of expression may be due to regulation by microRNA (miR) species. Pim-1 expression has been found to be regulated by miR-1 [41-43], miR-16 [44], miR-33a [45, 46] and miR-328 [47] in different cell types. miR regulation of Pim-2 and Pim-3 expression has not yet been characterized.
3.2. SGI-1776 has similar cytotoxic effects in resistant and parental cells
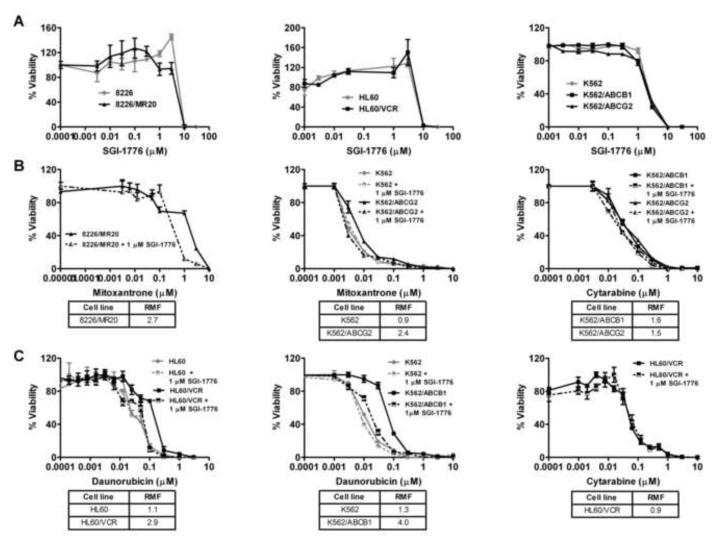
We next measured the cytotoxic effects of SGI-1776 on parental and multidrug resistant 8226 and 8226/MR20, HL60 and HL60/VCR and K562, K562/ABCG2 and K562/ABCB1 cells in cell viability assays (Figure 3A). 8226 and 8226/MR20, HL60 and HL60/VCR and K562, K562/ABCG2 and K562/ABCB1 cells were incubated with SGI-1776 at a range of concentrations, and IC50 values were derived. Cytotoxicity was similar in ABCG2- and ABCB1-overexpressing cells and parental cells, with steep slopes and estimated IC50 values of approximately 5 μM in all cell lines. Of note, consistent with our previous findings, SGI-1776 was not cytotoxic at 1 μM in any of the cell lines studied.
Figure 3.
A. SGI-1776 is similarly cytotoxic to cells expressing ABCG2 or ABCB1 and parental cells. 8226/MR20, HL60/VCR, K562/ABCG2 and K562/ABCB1 cells, expressing ABCG2 or ABCB1, and HL60, 8226 and K562 parental cells in log growth phase were cultured in 96-well plates at a density of 1× 103 cells per well in the presence of SGI-1776 in increasing concentrations for 96 hours. Viable cells were subsequently quantified using the WST-1 assay as detailed in Materials and Methods. Results are shown as mean ± SEM (n=6). B SGI-1776 (1μM) sensitizes ABCG2-overexpressing cells to ABCG2 substrate, but not non-substrate, drugs. 8226/MR20 and K562/ABCG2 cells, expressing ABCG2, and K562 parental cells were plated with mitoxantrone or cytarabine at increasing concentrations in the presence and absence of 1 μM SGI-1776. Resistance modifying factors (RMF) were calculated and are shown below the graphs. C. SGI-1776 sensitizes ABCB1-overexpressing, but not parental, cells to ABCB1-substrate, but not non-substrate, drugs. HL60/VCR and K562/ABCB1, expressing ABCB1, and their respective parental cell lines were plated as above with daunorubicin or cytarabine in increasing concentrations in the presence and absence of 1 μM SGI-1776, and viable cells were subsequently quantified using the WST-1 assay. The experiment was repeated three times and means ± SEM are shown. IC50s in the presence and absence of SGI-1776 were derived, and RMFs for 1 μM SGI-1776 were then calculated for each cell line as detailed in Materials and Methods. RMF values are shown below the graphs.
3.3. SGI-1776 sensitizes ABCG2-, as well as ABCB1-overexpressing, cells to substrate chemotherapy drugs in cell survival assays
We then determined whether SGI-1776 sensitized multidrug resistant cells with ABCG2 expression to ABCG2 substrate chemotherapy drugs, and confirmed previously demonstrated sensitization of cells with ABCB1 expression to ABCB1 substrate chemotherapy drugs. The cytotoxic effects of the ABCG2 or ABCB1 substrate chemotherapy drugs daunorubicin or mitoxantrone, respectively, were measured in the absence and presence of SGI-1776 in ABCG2- or ABCB1-overexpressing cell lines. SGI-1776 at 1 μM sensitized ABCG2-overexpressing 8226/MR20 and K562/ABCG2 cells to mitoxantrone, with RMFs of 2.7 and 2.4 (Figure 3B), respectively and sensitized ABCB1-overexpressing HL60/VCR and K562/ABCB1 cells to daunorubicin, with RMFs of 2.9 and 4.0 (Figure 3C), respectively. In contrast, SGI-1776 did not sensitize parental HL60 or K562 cells to daunorubicin (Figure 3C) or mitoxantrone (Figure 3B, middle panel) and did not sensitize ABCG2- or ABCB1-overexpressing cells to the ABCG2 and ABCB1 non-substrate drug cytarabine (Figure 3, B and C, right panels). Thus SGI-1776 enhances the cytotoxic effects of ABCG2, as well as ABCB1, substrate, but not non-substrate, chemotherapy drugs in cells with ABCG2 or ABCB1 expression, respectively, but not in parental cells.
3.4. SGI-1776 sensitizes ABCG2-, as well as ABCB1-overexpressing, cells to apoptosis induced by substrate chemotherapy drugs
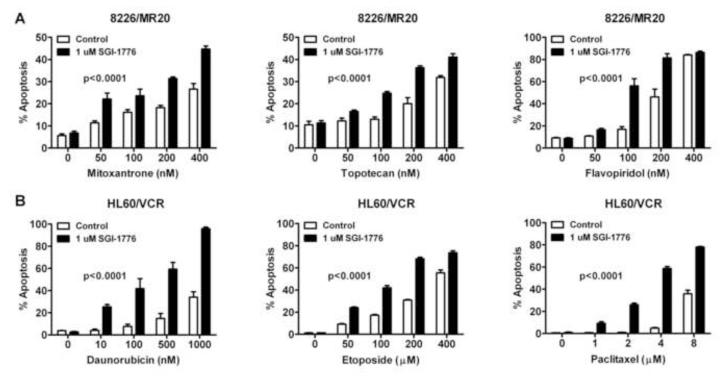
Since SGI-1776 sensitized cells with ABCG2 or ABCB1 overexpression to substrate chemotherapy drugs in cell survival assays, we studied its effect on apoptosis induction by ABCG2 or ABCB1 substrate chemotherapy drugs in cells with ABCG2 or ABCB1 expression. Co-incubation with 1 μM SGI-1776 enhanced apoptosis induction by mitoxantrone, topotecan or flavopiridol (p<0.0001 for each) at a range of concentrations in 8226/MR20 cells, with ABCG2 overexpression (Figure 4A) and by daunorubicin, etoposide or paclitaxel (p<0.0001 for each) at a range of concentrations in HL60/VCR cells, expressing ABCB1 (Figure 4B).
Figure 4. Concomitant treatment with SGI-1776 significantly enhances apoptosis induction by ABCG2 and ABCB1 substrate chemotherapy drugs.
1×105 cells were plated per well in 6-well plates with chemotherapy drugs in the presence (closed bars) and absence (open bars) of 1 μM SGI-1776.and apoptosis was measured by flow cytometry, as described in Materials and Methods, 48 hours after plating. Data shown are means ± SEM of triplicate values A. Percentage of apoptotic cells in 8226/MR20 cells, expressing ABCG2, treated with the ABCG2 substrate drugs mitoxantrone, topotecan and flavopiridol at increasing concentrations B. Percentage of apoptotic cells in HL60/VCR cells, expressing ABCB1, treated with the ABCB1 substrate drugs daunorubicin, etoposide and paclitaxel at increasing concentrations.
3.5. SGI-1776 enhances ABCG2, as well as ABCB1, substrate chemotherapy drug inhibition of colony formation by cells expressing ABCG2 or ABCB1
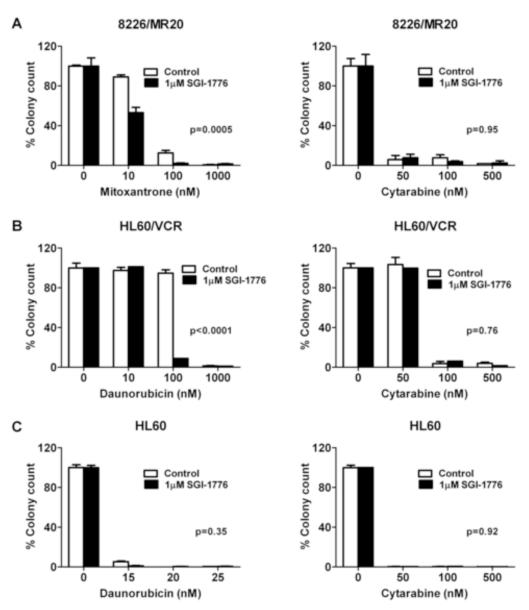
Further, we investigated the effect of SGI-1776 on ABCG2 or ABCB1 substrate chemotherapy drug inhibition of colony formation of cells with ABCG2 or ABCB1 expression, respectively. Co-incubation with 1 μM SGI-1776 decreased colony formation of 8226/MR20 cells, with ABCG2 overexpression, in the presence of mitoxantrone, but not cytarabine (Figure 5A) and of HL60/VCR cells, with ABCB1 expression, in the presence of daunorubicin, but not cytarabine (Figure 5B). Additionally, there was no effect on HL60 colony formation in the presence of daunorubicin or cytarabine (Figure 5C).
Figure 5. SGI-1776 decreases colony formation by ABCG2- or ABCB1-expressing, but not parental, cells in the presence of substrate, but not non-substrate, drugs.
A. 8226/MR20 cells were plated in methylcellulose-based medium in 24-well plates at a density of 500 cells per well with mitoxantrone or cytarabine at increasing concentrations with and without 1 μM SGI-1776 for 10 days, and colonies were counted as described in Materials and Methods. B. HL60/VCR cells were plated as above with daunorubicin or cytarabine at increasing concentrations in the presence and absence of 1 μM SGI-1776 for 10 days, and colonies were counted C. Parental HL60 cells were cultured as above with daunorubicin or cytarabine at increasing concentrations in the presence and absence of 1 μM SGI-1776 and colonies formed after 10 days were counted. Data are expressed as means ± SEM of triplicate treatments.
3.6. SGI-1776 decreases ABCB1 and ABCG2 surface expression on resistant cells with strong Pim-1 expression
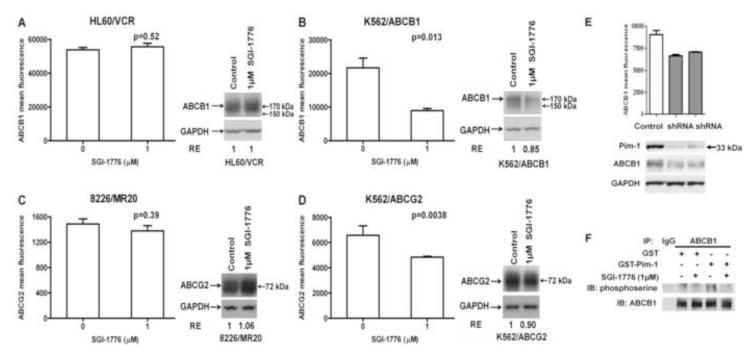
Given that SGI-1776 sensitized ABCB1- or ABCG2-expressing cells to ABCB1 or ABCG2 substrate chemotherapy drugs in cell survival, apoptosis and colony formation assays, and that inhibition or downregulation of Pim-1 decreases ABCG2 and ABCB1 cell surface translocation [17, 18], we studied the effects of SGI-1776 on cell surface expression of ABCB1 and ABCG2 (Figure 6). Incubation with 1 μM SGI-1776 significantly decreased ABCB1 and ABCG2 surface expression on K562/ABCB1 (Figure 6B) and K562/ABCG2 (Figure 6D) cells, but did not decrease ABCB1 on HL60/VCR (Figure 6A) or ABCG2 on 8226/MR20 (Figure 6C) cells. Moreover, ABCB1 and ABCG2 surface expression decreased on both of the resistant K562 cell lines (Figure 6B and D) without a significant decrease in total ABCB1 and ABCG2 expression, as measured by Western blot analysis (Figure 6). Additionally, incubation with SGI-1776 decreased expression of 150 kDa ABCB1, seen in Western blot analysis (Figure 6B), consistent with our previous demonstration of Pim-1 stabilization of 150 kDa ABCB1 [17]. Differential effects on K562/ABCB1 and K562/ABCG2, in relation to HL60/VCR and 8226/MR20, may be attributable to the fact that both K562/ABCB1 and K562/ABCG2 have markedly elevated levels of Pim-1 expression, while HL60/VCR and 8226/MR20 both have lower levels of Pim-1 expression (Figure 2).
Figure 6. SGI-1776 treatment decreases surface expression of ABCB1 and ABCG2 on cells with high, but not low, Pim-1 expression.
A-D. HL60/VCR and K562/ABCB1 cells, expressing ABCB1, and 8226/MR20 and K562/ABCG2 cells, expressing ABCG2, were plated at a density of 1 × 104 cells/ml with and without SGI-1776 1 μM for 48 hours, re-fed with fresh medium with and without the drug and cultured for 48 additional hours. ABCB1 surface expression was measured on 1 × 105 HL60/VCR (A) and K562/ABCB1 (B) cells and ABCB1 surface expression on 1 × 105 8226/MR20 (C) and K562/ABCG2 (D) cells by flow cytometry, as detailed in Materials and Methods. Data are mean ± SEM of triplicate values per treatment. Protein lysates were prepared from the remaining cells and total ABCB1 or ABCG2 expression was quantified by densitometric analysis following immunoblotting. Immunoblots of ABCB1 or ABCG2 and GAPDH loading controls are shown. Relative expression (RE), calculated as detailed in Materials and Methods, is shown. E. Pim-1 knockdown was performed in K562/ABCB1 cells as described in Materials and Methods, resulting in decreased cell surface expression of ABCB1. Cell surface expression, measured by flow cytometry, is shown graphically in the upper panel, with the lower panel showing immunoblots measuring Pim-1, ABCB1 and GAPDH expression. F. ABCB1 and control IgG were immunoprecipitated from crude membrane extracts of High-Five insect cells overexpressing ABCB1 and serine phosphorylation of the immunoprecipitated ABCB1 by recombinant GST-tagged Pim-1 kinase in the presence and absence of 1 μM SGI-1776 was studied using an in vitro kinase assay as described in Materials and Methods. The resultant immunoblot probed with antibodies against phosphoserine and ABCB1 is shown. .
To further demonstrate that effects on ABC protein cell surface expression are Pim-1-dependent, Pim-1 knockdown was performed in K562/ABCB1 cells, as described in Materials and Methods. Pim-1 knockdown resulted in decrease in ABCB1 cell surface expression (Figure 6E). Of note, decreased cellular expression of ABCB1 was also seen, consistent with Pim-1 stabilizing 150 kDa ABCB1 [17] and Pim-1 knockdown and, to a lesser degree, inhibition destabilizing 150 kDa ABCB1, and therefore resulting in decreased levels of glycosylated 170 kDa ABCB1. Additionally, effects of SGI-1776 on ABCB1 serine phosphorylation in membrane extracts of High-Five insect cells overexpressing ABCB1 were studied in vitro (Figure 6F); decreased serine phosphorylation of ABCB1 was seen when membrane extracts were incubated with, compared to without, 1 μM SGI-1776 in the presence of GST-Pim-1.
3.7. SGI-1776 inhibits substrate transport mediated by ABCG2, as well as ABCB1
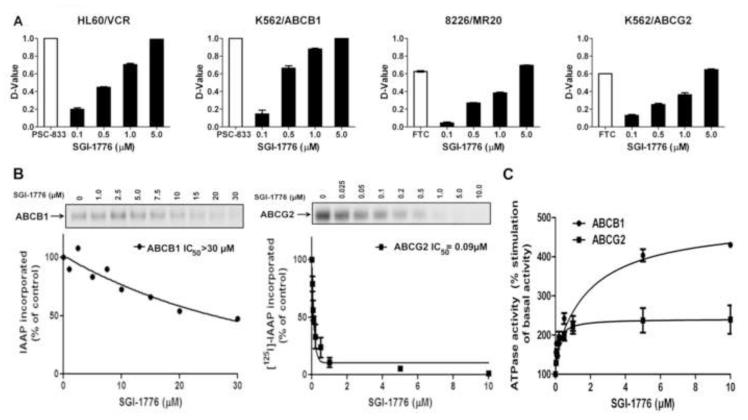
Since SGI-1776 sensitized ABCG2- and ABCB1-expressing cells to ABCG2 and ABCB1 substrate chemotherapy drugs in cell survival, apoptosis and colony formation assays, but only decreased ABCG2 and ABCB1 cell surface expression on cells with strong Pim-1 expression, we postulated that SGI-1776 might inhibit substrate transport mediated by ABCG2, as well as ABCB1, independently from its effect on Pim-1. To test this, cells expressing ABCG2 or ABCB1 were incubated with the fluorescent ABCG2 and ABCB1 substrates PhA and DiOC2(3), respectively, in the presence of SGI-1776 at a range of concentrations. SGI-1776 enhanced accumulation of PhA in ABCG2-overexpressing 8226/MR20 and K562/ABCG2 cells, as well as DiOC2(3) in ABCB1-overexpressing HL60/VCR and K562/ABCB1 cells, in a concentration-dependent manner (Figure 7A), consistent with inhibition of ABCG2-, as well as ABCB1-, mediated transport by SGI-1776.
Figure 7.
A. SGI-1776 increases substrate uptake in cells expressing ABCB1 or ABCG2. 1 × 106 HL60/VCR and K562/ABCB1 cells, expressing ABCB1, and 8226/MR20 and K562/ABCG2 cells, expressing ABCG2, were exposed to their respective fluorescent substrates DiOC2(3) and pheophorbide A (PhA) in the presence of SGI-1776 in increasing concentrations or the established ABCB1 modulator PSC-833 or ABCG2 modulator fumitremorgin C (FTC) for 30 minutes at 37°C. Cellular fluorescence was measured by flow cytometry, and compared by D-values. Means ± SEM. of triplicate treatments are shown. B. SGI-1776 decreases [125I]-IAAP photolabeling of ABCB1 and ABCG2. Crude membranes containing ABCB1 or ABCG2 were incubated with 0-30 μM SGI-1776 for 5 minutes, then 3-6 nM [125I]-IAAP (2200 Ci/mmole) for five additional minutes. IAAP labeling of ABCB1 or ABCG2 was then measured as described in Materials and Methods and plotted against SGI-1776 concentration. Means ± SD from three independent experiments are shown. C. SGI-1776 increases ABCB1 and ABCG2 ATPase activity. Crude membrane preparations containing ABCB1 or ABCG2 were incubated with SGI-1776 in increasing concentrations in the presence or absence of beryllium sulfate and sodium fluoride (BeFx) or sodium orthovanadate, respectively. The amount of inorganic phosphate released and the vanadate or BeFx-sensitive ATPase activity were measured as described in Materials and Methods. The average from independent duplicate experiments is shown, with error bars representing standard error.
3.8. SGI-1776 binds to ABCB1 and ABCG2 drug-binding sites and stimulates ATPase activity
To study the mechanism of SGI-1776 inhibition of ABCB1- and ABCG2-mediated transport, we measured the effects of SGI-1776 on [125I]IAAP photoaffinity labeling of ABCB1 and ABCG2 and on ABCB1 and ABCG2 ATPase activity. SGI-1776 weakly inhibited [125I]IAAP binding to ABCB1 and strongly inhibited [125I]IAAP binding to ABCG2, with IC50 values of >30 μM and 0.09 μM, respectively (Figure 7B). SGI-1776 stimulated both ABCB1 and ABCG2 ATPase activity in a concentration-dependent manner, with similar stimulation of ABCB1 and ABCG2 ATPase activity at 1 μM, but stronger stimulation of ABCB1 ATPase activity at higher concentrations (Figure 7C). The discrepancy between the effects of SGI-1776 on ABCB1 [125I]IAAP photoaffinity labeling and ATPase activity may be explained by binding of SGI-1776 to an ABCB1 drug-binding site different from the IAAP binding site, as it is generally accepted that the drug-binding pocket of ABCB1 contains multiple overlapping sites [48]. Taken together, the findings were consistent with SGI-1776 binding to ABCB1 and ABCG2 drug-binding sites and inhibiting substrate transport.
4. Discussion
Our group and our collaborators previously demonstrated that Pim-1 phosphorylates ABCG2 and ABCB1 and thereby enables their translocation to the cell surface, where they function as drug efflux pumps [17, 18]. Here we have studied the Pim kinase inhibitor SGI-1776, and demonstrated that it sensitizes ABCG2-, as well as ABCB1-, overexpressing multidrug resistant cells to ABCB1 or ABCG2 substrate, but not non-substrate, chemotherapy drugs. We further demonstrated that SGI-1776 has distinct effects on expression and on function of both ABCB1 and ABCG2. First, it decreases surface expression of ABCB1 and ABCG2 on ABCB1- and ABCG2-overexpressing K562 cells, which have a high level of Pim-1 expression, but not on HL60/VCR and 8226/MR20 cells, which have lower-level Pim-1 expression, and this effect is therefore likely due to decreased cell surface translocation of ABCB1 and ABCG2 resulting from inhibition of Pim-1. Secondly, SGI-1776 directly inhibits ABCG2-, as well as ABCB1-, mediated transport of their substrates in ABCG2- or ABCB1-overexpressing cells, and this effect is unrelated to level of Pim-1 expression and is likely due to binding of SGI-1776 to the drug-binding sites of ABCG2 and ABCB1 and competitive inhibition of transport of their substrates.
The observation that SGI-1776 decreases ABCB1 and ABCG2 surface expression on K562/ABCB1 and K562/ABCG2 cells, which express Pim-1 at high levels, but not on HL60/VCR and 8226/MR20 cells, which express it at lower levels, is consistent with the previous demonstration that Pim-1 phosphorylates ABCB1 and ABCG2 and promotes their cell surface translocation [17, 18]. Effects of Pim-2 and Pim-3 on drug resistance proteins have not been studied. Of note, SGI-1776 IC50s for Pim-2 and Pim-3 inhibition in kinase inhibition assays are 363 nM and 69 nM, respectively [20], and SGI-1776 is more than 95% bound to human plasma proteins [21]; therefore SGI-1776 may not inhibit Pim-2 or Pim-3 at 1 μM in cell culture-based assays.
In addition to effects of SGI-1776 on ABCG2 and ABCB1 cell surface expression, we found that SGI-1776 inhibited substrate transport by ABCG2, as well as ABCB1, in ABCG2- or ABCB1-expressing cells independently from level of Pim-1 expression. SGI-1776 inhibition of substrate transport by ABCB1 was indeed consistent with previously published data [17, 18], while the effect on substrate transport by ABCG2 had not been previously studied. The data on SGI-1776 effects on [125I]IAAP photoaffinity labeling of ABCB1 and ABCG2 and on ABCB1 and ABCG2 ATPase activity were consistent with SGI-1776 binding to ABCB1 and ABCG2 drug-binding sites and thereby inhibiting substrate transport. In this regard, it is a priori surprising that the IC50 values of SGI-1776 were identical in ABCG2- and ABCB1-overexpressing and parental cells (Figure 3A), but these data can be explained by SGI-1776 inhibiting its own transport, as evidenced by its effects on [125I]-IAAP binding and ATPase activity.
A number of therapeutically active kinase inhibitors have been demonstrated to be inhibitors and/or substrates of ABCB1 and/or ABCG2. These include the BCR-ABL inhibitors imatinib mesylate, nilotinib and dasatinib [49-55] and the fms-like tyrosine kinase 3 (FLT3) inhibitors PKC412 or midostaurin [56], tandutinib [57], sorafenib [58] and sunitinib [59], as well as inhibitors used in therapy of solid tumors. Additionally, we recently demonstrated that the novel BCR-ABL and FLT3 inhibitor ponatinib is a potent inhibitor of ABCG2, a less potent inhibitor of ABCB1, and a substrate of both proteins, consistent with competitive inhibition [28]. Activity of therapeutically active kinase inhibitors as inhibitors and/or substrates of ABCB1 and/or ABCG2 has implications not only for chemosensitization, but also for clinically important drug interactions.
We have demonstrated that SGI-1776, which is an imidazo[1,2-b]pyridazine small molecule, is an ABCG2 and ABCB1 inhibitor. The ABCG2 and ABCB1 drug binding cavity pharmacophore requirements are satisfied by SGI-1776, e.g., aromatic ring centers, hydrogen bond acceptors and a basic tertiary amine of the piperidine ring. Interestingly, ponatinib, which we recently found to inihibit ABCG2 and ABCB1, albeit with more potent effects on ABCG2 [28], is also an imidazo[1,2-b]pyridazine [60]. In addition to SGI-1776 and ponatinib, other described imidazo[1,2-b]pyridazine small molecules have activity as inhibitors of cyclin-dependent kinase (CDK) [61], IkappaB kinase (IKK) beta [62] and vascular endothelial growth factor (VEGF) [63]. If therapeutically applicable, these molecules should be characterized with regard to interactions with ABCG2 and ABCB1.
Pim-1 is thought to be a clinically promising therapeutic target in leukemias and other malignancies [16, 21, 22], and other Pim kinase inhibitors are in preclinical and clinical development. It will be of interest to characterize subsequent clinically applicable Pim kinase inhibitors with regard to interactions with ABCB1 and ABCG2. In particular, while therapeutic strategies based on inhibition of drug transport mediated by ABCB1 with competitive inhibitors including PSC-833, zosuquidar and cyclosporine A have largely been clinically unsuccessful, inhibition of ABCG2 and ABCB1 cell surface translocation by Pim-1 kinase inhibitors may have therapeutic implications. Additional clinically applicable Pim kinase inhibitors are in development and should be characterized with regard to effects on ABCG2 and ABCB1 cell surface expression and drug transport.
Acknowledgements
Grant support: Leukemia and Lymphoma Society Translational Research Award (M.R. Baer), University of Maryland, Baltimore UMMG Cancer Research Grant #CH 649 CRF, State of Maryland Department of Health and Mental Hygiene (DHMH) under the Cigarette Restitution Fund Program (M.R. Baer), NCI Cancer Center Support Grant P30 CA134274 (UMGCC), and NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research (S. Shukla, S.V. Ambudkar).
The authors thank James Steinhardt, University of Maryland School of Medicine, for providing the modified shRNA transduction protocol.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to disclose.
References
- [1].Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- [2].Benderra Z, Faussat AM, Sayada L, Perrot JY, Chaoui D, Marie JP, et al. Breast cancer resistance protein and P-glycoprotein in 149 adult acute myeloid leukemias. Clin Cancer Res. 2004;10:7896–902. doi: 10.1158/1078-0432.CCR-04-0795. [DOI] [PubMed] [Google Scholar]
- [3].Damiani D, Tiribelli M, Calistri E, Geromin A, Chiarvesio A, Michelutti A, et al. The prognostic value of P-glycoprotein (ABCB) and breast cancer resistance protein (ABCG2) in adults with de novo acute myeloid leukemia with normal karyotype. Haematologica. 2006;91:825–8. [PubMed] [Google Scholar]
- [4].Baer MR, George SL, Dodge RK, O’Loughlin KL, Minderman H, Caligiuri MA, et al. Phase 3 study of the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age and older with acute myeloid leukemia: Cancer and Leukemia Group B Study 9720. Blood. 2002;100:1224–32. [PubMed] [Google Scholar]
- [5].Greenberg PL, Lee SJ, Advani R, Tallman MS, Sikic BI, Letendre L, et al. Mitoxantrone, etoposide, and cytarabine with or without valspodar in patients with relapsed or refractory acute myeloid leukemia and high-risk myelodysplastic syndrome: a phase III trial (E2995) J Clin Oncol. 2004;22:1078–86. doi: 10.1200/JCO.2004.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van der Holt B, Löwenberg B, Burnett AK, Knauf WU, Shepherd J, Piccaluga PP, et al. The value of the MDR1 reversal agent PSC-833 in addition to daunorubicin and cytarabine in the treatment of elderly patients with previously untreated acute myeloid leukemia (AML), in relation to MDR1 status at diagnosis. Blood. 2005;106:2646–54. doi: 10.1182/blood-2005-04-1395. [DOI] [PubMed] [Google Scholar]
- [7].Kolitz JE, George SL, Marcucci G, Vij R, Powell BL, Allen SL, et al. P-glycoprotein inhibition using valspodar (PSC-833) does not improve outcomes for patients younger than age 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia Group B study 19808. Blood. 2010;116:1413–21. doi: 10.1182/blood-2009-07-229492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cripe LD, Uno H, Paietta EM, Litzow MR, Ketterling RP, Bennett JM, et al. Zosuquidar, a novel modulator of P-glycoprotein, does not improve the outcome of older patients with newly diagnosed acute myeloid leukemia: a randomized, placebo-controlled trial of the Eastern Cooperative Oncology Group 3999. Blood. 2010;116:4077–85. doi: 10.1182/blood-2010-04-277269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Raaijmakers M, de Grouw E, Heuver L, van der Reijden B, Jansen J, Scheper R, et al. Breast cancer resistance protein in drug resistance of primitive CD34+38− cells in acute myeloid leukemia. Clin Cancer Res. 2005;11:2436–44. doi: 10.1158/1078-0432.CCR-04-0212. [DOI] [PubMed] [Google Scholar]
- [10].Raaijmakers MH, de Grouw EP, van der Reijden BA, de Witte TJ, Jansen JH, Raymakers RA. ABCB1 modulation does not circumvent drug extrusion from primitive leukemic progenitor cells and may preferentially target residual normal cells in acute myelogenous leukemia. Clin Cancer Res. 2006;12:3452–8. doi: 10.1158/1078-0432.CCR-05-1945. [DOI] [PubMed] [Google Scholar]
- [11].Qadir M, O’Loughlin KL, Fricke SM, Williamson NA, Greco WR, Minderman H, et al. Cyclosporin A is a broad-spectrum multidrug resistance modulator. Clin Cancer Res. 2005;11:2320–6. doi: 10.1158/1078-0432.CCR-04-1725. [DOI] [PubMed] [Google Scholar]
- [12].List AF, Kopecky KJ, Willman CL, Head DR, Persons DL, Slovak ML, et al. Benefit of cyclosporine modulation of drug resistance in patients with poor-risk acute myeloid leukemia: a Southwest Oncology Group study. Blood. 2001;98:3212–20. doi: 10.1182/blood.v98.12.3212. [DOI] [PubMed] [Google Scholar]
- [13].Matsouka P, Pagoni M, Zikos P, Giannakoulas N, Apostolidis I, Asprogeraka T, et al. Addition of cyclosporin-A to chemotherapy in secondary (post-MDS) AML in the elderly. A multicenter randomized trial of the Leukemia Working Group of the Hellenic Society of Hematology. Ann Hematol. 2006;85:250–6. doi: 10.1007/s00277-005-0066-0. [DOI] [PubMed] [Google Scholar]
- [14].Chauncey TR, Gundacker H, Shadman M, List AF, Dakhil SR, Erba HP, et al. Sequential phase II Southwest Oncology Group studies (S0112 and S0301) of daunorubicin and cytarabine by continuous infusion, without and with ciclosporin, in older patients with previously untreated acute myeloid leukaemia. Br J Haematol. 2010;148:48–58. doi: 10.1111/j.1365-2141.2009.07919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Becton D, Dahl GV, Ravindranath Y, Chang MN, Behm FG, Raimondi SC, et al. Randomized use of cyclosporin A (CsA) to modulate P-glycoprotein in children with AML in remission: Pediatric Oncology Group Study 9421. Blood. 2006;107:1315–24. doi: 10.1182/blood-2004-08-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95:1004–15. doi: 10.3324/haematol.2009.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xie Y, Burcu M, Linn DE, Qiu Y, Baer MR. Pim-1 kinase protects P-glycoprotein from degradation and enables its glycosylation and cell surface expression. Mol Pharmacol. 78:310–8. doi: 10.1124/mol.109.061713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xie Y, Xu K, Linn DE, Yang X, Guo Z, Shimelis H, et al. The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. J Biol Chem. 2008;283:3349–56. doi: 10.1074/jbc.M707773200. [DOI] [PubMed] [Google Scholar]
- [19].Amson R, Sigaux F, Przedborski S, Flandrin G, Givol D, Telerman A. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci U S A. 1989;86:8857–61. doi: 10.1073/pnas.86.22.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mumenthaler SM, Ng PY, Hodge A, Bearss D, Berk G, Kanekal S, et al. Pharmacologic inhibition of Pim kinases alters prostate cancer cell growth and resensitizes chemoresistant cells to taxanes. Mol Cancer Ther. 2009;8:2882–93. doi: 10.1158/1535-7163.MCT-09-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen LS, Redkar S, Bearss D, Wierda WG, Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009;114:4150–7. doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen LS, Redkar S, Taverna P, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to Pim kinase inhibitor, SGI-1776, in acute myeloid leukemia. Blood. 2011;118:693–702. doi: 10.1182/blood-2010-12-323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ogretmen B, Safa AR. Identification and characterization of the MDR1 promoter-enhancing factor 1 (MEF1) in the multidrug resistant HL60/VCR human acute myeloid leukemia cell line. Biochemistry. 2000;39:194–204. doi: 10.1021/bi991943f. [DOI] [PubMed] [Google Scholar]
- [24].Hazlehurst LA, Foley NE, Gleason-Guzman MC, Hacker MP, Cress AE, Greenberger LW, et al. Multiple mechanisms confer drug resistance to mitoxantrone in the human 8226 myeloma cell line. Cancer Res. 1999;59:1021–8. [PubMed] [Google Scholar]
- [25].Hafkemeyer P, Licht T, Pastan I, Gottesman MM. Chemoprotection of hematopoietic cells by a mutant P-glycoprotein resistant to a potent chemosensitizer of multidrug-resistant cancers. Hum Gene Ther. 2000;11:555–65. doi: 10.1089/10430340050015743. [DOI] [PubMed] [Google Scholar]
- [26].Yanase K, Tsukahara S, Asada S, Ishikawa E, Imai Y, Sugimoto Y. Gefitinib reverses breast cancer resistance protein-mediated drug resistance. Mol Cancer Ther. 2004;3:1119–25. [PubMed] [Google Scholar]
- [27].Robey RW, Medina-Perez WY, Nishiyama K, Lahusen T, Miyake K, Litman T, et al. Overexpression of the ATP-binding cassette half-transporter, ABCG2 (Mxr/BCrp/ABCP1), in flavopiridol-resistant human breast cancer cells. Clin Cancer Res. 2001;7:145–52. [PubMed] [Google Scholar]
- [28].Sen R, Natarajan K, Bhullar J, Shukla S, Fang HB, Cai L, et al. The novel BCR ABL and FLT3 inhibitor ponatinib is a potent inhibitor of the multidrug resistance-associated ATP-binding cassette transporter ABCG2. Mol Cancer Ther. 2012 doi: 10.1158/1535-7163.MCT-12-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Minderman H, Suvannasankha A, O’Loughlin KL, Scheffer GL, Scheper RJ, Robey RW, et al. Flow cytometric analysis of breast cancer resistance protein expression and function. Cytometry. 2002;48:59–65. doi: 10.1002/cyto.10111. [DOI] [PubMed] [Google Scholar]
- [30].Robey RW, Steadman K, Polgar O, Morisaki K, Blayney M, Mistry P, et al. Pheophorbide a is a specific probe for ABCG2 function and inhibition. Cancer Res. 2004;64:1242–6. doi: 10.1158/0008-5472.can-03-3298. [DOI] [PubMed] [Google Scholar]
- [31].Horikawa K, Takatsu K. Interleukin-5 regulates genes involved in B-cell terminal maturation. Immunology. 2006;118:497–508. doi: 10.1111/j.1365-2567.2006.02382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim KT, Levis M, Small D. Constitutively activated FLT3 phosphorylates BAD partially through pim-1. Br J Haematol. 2006;134:500–9. doi: 10.1111/j.1365-2141.2006.06225.x. [DOI] [PubMed] [Google Scholar]
- [33].Young IT. Proof without prejudice: use of the Kolmogorov-Smirnov test for the analysis of histograms from flow systems and other sources. J Histochem Cytochem. 1977;25:935–41. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- [34].Minderman H, O’Loughlin KL, Pendyala L, Baer MR. VX-710 (biricodar) increases drug retention and enhances chemosensitivity in resistant cells overexpressing P-glycoprotein, multidrug resistance protein, and breast cancer resistance protein. Clin Cancer Res. 2004;10:1826–34. doi: 10.1158/1078-0432.ccr-0914-3. [DOI] [PubMed] [Google Scholar]
- [35].Sauna ZE, Ambudkar SV. Evidence for a requirement for ATP hydrolysis at two distinct steps during a single turnover of the catalytic cycle of human P-glycoprotein. Proc Natl Acad Sci U S A. 2000;97:2515–20. doi: 10.1073/pnas.97.6.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shukla S, Robey RW, Bates SE, Ambudkar SV. The calcium channel blockers, 1,4-dihydropyridines, are substrates of the multidrug resistance-linked ABC drug transporter, ABCG2. Biochemistry. 2006;45:8940–51. doi: 10.1021/bi060552f. [DOI] [PubMed] [Google Scholar]
- [37].Ambudkar SV. Drug-stimulatable ATPase activity in crude membranes of human MDR1-transfected mammalian cells. Methods Enzymol. 1998;292:504–14. doi: 10.1016/s0076-6879(98)92039-0. [DOI] [PubMed] [Google Scholar]
- [38].Miller RG., Jr. Simultaneous Statistical Inference. Springer-Verlag; New York: 1991. [Google Scholar]
- [39].Saris CJ, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–64. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xie Y, Xu K, Dai B, Guo Z, Jiang T, Chen H, et al. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene. 2006;25:70–8. doi: 10.1038/sj.onc.1209058. [DOI] [PubMed] [Google Scholar]
- [41].Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [42].Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang ZP, Li J, et al. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol. 2011;31:368–75. doi: 10.1161/ATVBAHA.110.218149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Katare R, Caporali A, Zentilin L, Avolio E, Sala-Newby G, Oikawa A, et al. Intravenous gene therapy with PIM-1 via a cardiotropic viral vector halts the progression of diabetic cardiomyopathy through promotion of prosurvival signaling. Circ Res. 2011;108:1238–51. doi: 10.1161/CIRCRESAHA.110.239111. [DOI] [PubMed] [Google Scholar]
- [44].Kim KT, Carroll AP, Mashkani B, Cairns MJ, Small D, Scott RJ. MicroRNA-16 is down-regulated in mutated FLT3 expressing murine myeloid FDC-P1 cells and interacts with Pim-1. PLoS One. 2012;7:e44546. doi: 10.1371/journal.pone.0044546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–24. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- [46].Thomas M, Lange-Grünweller K, Weirauch U, Gutsch D, Aigner A, Grünweller A, et al. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 2012;31:918–28. doi: 10.1038/onc.2011.278. [DOI] [PubMed] [Google Scholar]
- [47].Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–65. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gutmann DA, Ward A, Urbatsch IL, Chang G, van Veen HW. Understanding polyspecificity of multidrug ABC transporters: closing in on the gaps in ABCB1. Trends Biochem Sci. 2010;35:36–42. doi: 10.1016/j.tibs.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Illmer T, Schaich M, Platzbecker U, Freiberg-Richter J, Oelschlägel U, von Bonin M, et al. P-glycoprotein-mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia. 2004;18:401–8. doi: 10.1038/sj.leu.2403257. [DOI] [PubMed] [Google Scholar]
- [50].Burger H, van Tol H, Boersma AW, Brok M, Wiemer EA, Stoter G, et al. Imatinib mesylate (STI571) is a substrate for the breast cancer resistance protein (BCRP)/ABCG2 drug pump. Blood. 2004;104:2940–2. doi: 10.1182/blood-2004-04-1398. [DOI] [PubMed] [Google Scholar]
- [51].Houghton PJ, Germain GS, Harwood FC, Schuetz JD, Stewart CF, Buchdunger E, et al. Imatinib mesylate is a potent inhibitor of the ABCG2 (BCRP) transporter and reverses resistance to topotecan and SN-38 in vitro. Cancer Res. 2004;64:2333–7. doi: 10.1158/0008-5472.can-03-3344. [DOI] [PubMed] [Google Scholar]
- [52].Shukla S, Sauna Z, Ambudkar S. Evidence for the interaction of imatinib at the transport-substrate site(s) of the multidrug-resistance-linked ABC drug transporters ABCB1 (P-glycoprotein) and ABCG2. Leukemia. 2008;22:445–7. doi: 10.1038/sj.leu.2404897. [DOI] [PubMed] [Google Scholar]
- [53].Tiwari AK, Sodani K, Wang SR, Kuang YH, Ashby CR, Jr., Chen X, et al. Nilotinib (AMN107, Tasigna) reverses multidrug resistance by inhibiting the activity of the ABCB1/Pgp and ABCG2/BCRP/MXR transporters. Biochem Pharmacol. 2009;78:153–61. doi: 10.1016/j.bcp.2009.04.002. [DOI] [PubMed] [Google Scholar]
- [54].Lagas JS, van Waterschoot RA, van Tilburg VA, Hillebrand MJ, Lankheet N, Rosing H, et al. Brain accumulation of dasatinib is restricted by P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) and can be enhanced by elacridar treatment. Clin Cancer Res. 2009;15:2344–51. doi: 10.1158/1078-0432.CCR-08-2253. [DOI] [PubMed] [Google Scholar]
- [55].Dohse M, Scharenberg C, Shukla S, Robey RW, Volkmann T, Deeken JF, et al. Comparison of ATP-binding cassette transporter interactions with the tyrosine kinase inhibitors imatinib, nilotinib, and dasatinib. Drug Metab Dispos. 2010;38:1371–80. doi: 10.1124/dmd.109.031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hunter HM, Pallis M, Seedhouse CH, Grundy M, Gray C, Russell NH. The expression of P-glycoprotein in AML cells with FLT3 internal tandem duplications is associated with reduced apoptosis in response to FLT3 inhibitors. Br J Haematol. 2004;127:26–33. doi: 10.1111/j.1365-2141.2004.05145.x. [DOI] [PubMed] [Google Scholar]
- [57].Yang JJ, Milton MN, Yu S, Liao M, Liu N, Wu JT, et al. P-glycoprotein and breast cancer resistance protein affect disposition of tandutinib, a tyrosine kinase inhibitor. Drug Metab Lett. 2010;4:201–12. doi: 10.2174/187231210792928279. [DOI] [PubMed] [Google Scholar]
- [58].Lagas JS, van Waterschoot RA, Sparidans RW, Wagenaar E, Beijnen JH, Schinkel AH. Breast cancer resistance protein and P-glycoprotein limit sorafenib brain accumulation. Mol Cancer Ther. 2010;9:319–26. doi: 10.1158/1535-7163.MCT-09-0663. [DOI] [PubMed] [Google Scholar]
- [59].Shukla S, Robey RW, Bates SE, Ambudkar SV. Sunitinib (Sutent, SU11248), a small-molecule receptor tyrosine kinase inhibitor, blocks function of the ATP-binding cassette (ABC) transporters P-glycoprotein (ABCB1) and ABCG2. Drug Metab Dispos. 2009;37:359–65. doi: 10.1124/dmd.108.024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Huang WS, Metcalf CA, Sundaramoorthi R, Wang Y, Zou D, Thomas RM, et al. Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem. 2010;53:4701–19. doi: 10.1021/jm100395q. [DOI] [PubMed] [Google Scholar]
- [61].Byth KF, Cooper N, Culshaw JD, Heaton DW, Oakes SE, Minshull CA, et al. Imidazo[1,2-b]pyridazines: a potent and selective class of cyclin-dependent kinase inhibitors. Bioorg Med Chem Lett. 2004;14:2249–52. doi: 10.1016/j.bmcl.2004.02.008. [DOI] [PubMed] [Google Scholar]
- [62].Shimizu H, Tanaka S, Toki T, Yasumatsu I, Akimoto T, Morishita K, et al. Discovery of imidazo[1,2-b]pyridazine derivatives as IKKbeta inhibitors. Part 1: Hit-to-lead study and structure-activity relationship. Bioorg Med Chem Lett. 2010;20:5113–8. doi: 10.1016/j.bmcl.2010.07.026. [DOI] [PubMed] [Google Scholar]
- [63].Iwata H, Imamura S, Hori A, Hixon MS, Kimura H, Miki H. Biochemical characterization of TAK-593, a novel VEGFR/PDGFR inhibitor with a two-step slow binding mechanism. Biochemistry. 2011;50:738–51. doi: 10.1021/bi101777f. [DOI] [PubMed] [Google Scholar]