Abstract
Genetic mapping on fully sequenced individuals is transforming our understanding of the relationship between molecular variation and variation in complex traits. Here we report a combined sequence and genetic mapping analysis in outbred rats that maps 355 quantitative trait loci for 122 phenotypes. We identify 35 causal genes involved in 31 phenotypes, implicating novel genes in models of anxiety, heart disease and multiple sclerosis. The relation between sequence and genetic variation is unexpectedly complex: at approximately 40% of quantitative trait loci a single sequence variant cannot account for the phenotypic effect. Using comparable sequence and mapping data from mice, we show the extent and spatial pattern of variation in inbred rats differ significantly from those of inbred mice, and that the genetic variants in orthologous genes rarely contribute to the same phenotype in both species.
Introduction
Unraveling the complex relationship between phenotype and genotype poses a formidable challenge for biomedical science. Despite considerable success in identifying genetic loci that contribute to quantitative variation and disease susceptibility in humans1, in most organisms the causal genetic variants at loci that contribute to complex phenotypes remain unclear2. Finding the responsible molecular changes would allow us to understand how phenotypic variation arises and confirm the identity of relevant genes.
In this report we present results from an outbred rat heterogeneous stock (hereafter NIH-HS) in a combined sequence-based and genetic mapping analysis of 160 phenotypes. The NIH-HS, established in the 1980s in NIH, is descended from eight inbred progenitors, BN/SsN, MR/N, BUF/N, M520/N, WN/N, ACI/N, WKY/N, and F344/N3, containing segregating variation representative of commonly used laboratory rats.
Heterogeneous stocks (HS) have three characteristics suited to genetic mapping: (i) quantitative trait loci (QTLs) can be resolved to megabase resolution; (ii) the complete sequence of genotyped HS animals can be imputed with high accuracy from the progenitor genomes; (iii) the population has a well-defined haplotype space that can be exploited to determine whether genetic association is caused by single sequence variants or by haplotypes4-6. This distinction is fundamental to understanding the signals from genome wide association studies, where it is unknown how often causality can be attributed to a single variant. In natural populations it is rarely feasible to test for haplotypic effects because of the difficulty of estimating the large number of unknown rare haplotypes7.
Here we describe the sequence of the eight progenitors, the development of a rat SNP array, the genotyping and phenotyping of 1,407 outbred NIH-HS rats, and the mapping of hundreds of quantitative trait loci (QTLs). We use the haplotypic properties of the NIH-HS to investigate the molecular basis of these QTLs.
Results
Sequence analysis
We generated sequence data equivalent to an average of 22X SOLiD coverage of the eight NIH-HS inbred founder strains. After mapping to the reference strain (BN/NHsdMcwi8) we report our results with respect to an accessible genome, which represents ~88% of the reference genome (Table 1). We identified 7.2M SNP sites (containing 19.8M genotypes differing from the reference in at least one strain), 633,000 indels (<10bp with the majority consisting of one (79.3%) or two (12.3%) basepair changes) and 44,000 structural variants.
Table 1. Sequence variation in the eight progenitor strains of the NIH-HS rats.
The amount of sequence mapped to the reference is shown in gigabases (Gb), the coverage, the percent of the genome deemed inaccessible, and the numbers of the three classes of variants compared to the reference strain. Note that ‘private variants’ refers to the number of variants that distinguish a specified strain from all the others; most of the alleles private to BN/SsN will be reference alleles.
| Strain | Gb of mapped data |
Coverage | % of genome inaccessible |
SNPs | Private SNPs |
Indels | Private indels |
Structural variants |
Private structural variants |
|---|---|---|---|---|---|---|---|---|---|
| ACI/N | 65.9 | 26.3 | 12.6 | 2,883,405 | 228,468 | 166,425 | 12,646 | 19,499 | 756 |
| BN/SsN | 54.4 | 21.7 | 9.4 | 71,038 | 563,308 | 0 | 14,839 | 27 | 4,203 |
| BUF/N | 62.3 | 24.9 | 12.7 | 2,748,633 | 125,202 | 172,934 | 7,195 | 22,176 | 1,002 |
| F344/N | 77.9 | 31.1 | 11.8 | 2,831,144 | 97,951 | 157,522 | 5,007 | 25,257 | 1,003 |
| M520/N | 72.5 | 28.9 | 12.3 | 2,836,898 | 89,277 | 170,031 | 5,008 | 24,090 | 915 |
| MR/N | 62.4 | 24.9 | 12.3 | 2,664,124 | 223,514 | 151,099 | 12,005 | 18,306 | 1,004 |
| WKY/N | 63.4 | 25.3 | 12.1 | 3,088,953 | 496,327 | 164,634 | 23,979 | 28,270 | 3,357 |
| WN/N | 62.3 | 24.9 | 12.2 | 2,698,493 | 249,563 | 154,769 | 13,541 | 18,563 | 700 |
We assessed the sensitivity and specificity of variant calls by comparison with 2.1 Mb of DNA from one non-reference strain, LE/Stm, finished to an estimated accuracy of one error per 100,000 bp9. Although LE/Stm is not an NIH-HS progenitor strain, it is one of the few non-reference rat strains cloned into a library of bacterial artificial chromosomes (BACs) (so suitable for highly accurate clone based sequencing)9 and one that is similarly diverged from the reference strain (BN/NHsdMcwi). Comparison of SOLiD and capillary variant calls showed 2.7% of SNPs, 2.2% of indels and 16.7% of structural variants were false positive calls. These error rates were independently confirmed in the NIH-HS strains by analysis of a randomly selected subset of variants using PCR-based resequencing, which confirmed all selected SNPs (84/84) and indels (80/80) and most of the called structural variants (53/54). In contrast, false-negative rates were much higher: 17.2% for SNPs, 41.4% for indels and 65% for structural variants. Most false-negative SNPs and indels are next to repeats (77.9 and 80.8% respectively).
Table 1 summarises the variation in each strain. Excluding BN/SsN (which is a sub-strain of the reference, with consequently far fewer differences than the other strains), the average number of SNPs per strain is 2.8M.
Nucleotide diversity in NIH-HS progenitors
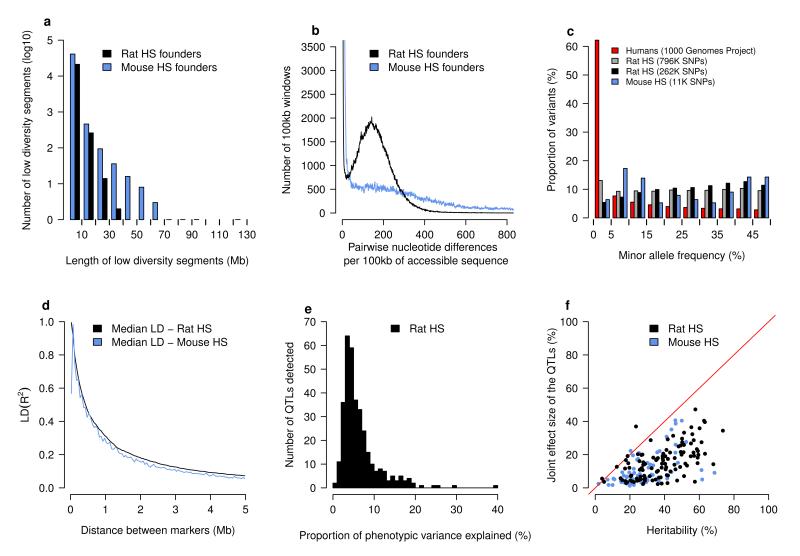
Sequence diversity in the NIH-HS progenitors has the following characteristics. First, diversity between all pairs of strains is similar, so that there are no extremely sequence divergent strains (Supplementary Figure 1). Second, in total 29% of 7.2M SNPs are private to a strain, hence unique haplotypes are relatively common in the NIH-HS. Third, regions of low diversity are small (median 400kb), with no blocks over 35 Mb (Figure 1a). Within divergent regions, there is a median of 151 differences per 100kb (Figure 1b).
Figure 1. Sequence diversity among progenitor strains and genetic architecture of the rat NIH-HS.
a) Regions of low diversity in the rat (black) and mouse (blue) progenitors. The horizontal axis shows the length in megabases of genomic regions with little sequence divergence (less than 13 SNPs/100kb). The vertical axis shows the numbers of segments observed in the eight progenitors. b) Sequence divergence in the progenitors. The horizontal axis is a measure of pairwise sequence diversity, the number of sequence differences observed in windows of 100 kilobases, the vertical axis gives the number of observations. The horizontal axis is truncated at 800 sequence differences and the vertical axis at 3500 windows. c) Minor allele frequencies in rat (gray & black), mouse (blue) and human (red) populations. The rat analysis was performed with the set of autosomal markers used to reconstruct haplotypes (261,684) as well as the complete set of 796,187 autosomal variants on the RATDIV array. d) The extent of linkage disequilibrium (measured as R2) in the rat NIH-HS. Distances between pairs of autosomal markers were binned (horizontal axis). The vertical axis shows the median of the corresponding distribution of LD. e) The distribution of effect sizes for the 343 loci mapped by mixed models in the rat NIH-HS. The horizontal axis is the proportion of phenotypic variance attributable to each locus. f) The proportion of heritability that can be explained by the joint effect of the QTLs detected for each phenotype. Each dot represents a single phenotype, with the horizontal axis showing the heritability and the vertical axis the joint QTL effect for that phenotype.
In comparison with the eight inbred strains that founded the mouse HS4,10, the rat founders are less diverse (10.2M SNPs in the mouse founders), but that diversity is more homogeneous: in the mouse genomes, long tracts of identical haplotypes alternate with segments of much greater diversity (Figures 1a, 1b).
Phenotypes and genotypes
The NIH-HS rats were phenotyped with a protocol that includes six disease models (anxiety, diabetes, hypertension, aortic elastic lamina rupture, multiple sclerosis, osteoporosis) and measures of risk factors for common diseases (e.g. lipid and cholesterol levels and cardiac hypertrophy)11(Table 2). In total, 160 phenotypes were measured (Supplementary Table 1). We selected 1,407 animals for genotyping and 198 non-phenotyped parents, together with the HS founders.
Table 2. Summary of phenotypes collected.
| Phenotype | Disease model | Number of measures |
Age (weeks) |
|---|---|---|---|
|
| |||
| Coat colour | 4 | 7 | |
| Wound healing | 1 | 7 and 17 | |
| Fear related behaviours | Anxiety | 10 | 8 to 10 |
| Glucose tolerance | Type II diabetes | 6 | 11 |
| Cardiovascular function | Hypertension | 2 | 12 |
| Body weight | Obesity | 1 | 13 |
| Basal hematology | 26 | 13 | |
| Basal immunology | 34 | 13 | |
| Induced neuroinflammation | Multiple sclerosis | 11 | 13 to 17 |
| Bone mass and strength | Osteoporosis | 43 | 17 |
| Arterial elastic lamina ruptures | 6 | 17 | |
| Serum biochemistry | 15 | 17 | |
| Renal agenesis | 1 | 17 | |
We designed a high density Affymetrix SNP genotyping array (RATDIV), using sequences from 13 inbred strains, which interrogated 803,485 SNPs. The SOLiD and RATDIV calls agreed at 99.98% of the 560,000 SNPs segregating in the 8 NIH-HS founders. We genotyped the NIH-HS with the array, and reconstructed the mosaics of NIH-HS founder haplotypes from 265,551 polymorphic high quality SNPs. In the NIH-HS the mean minor allele frequency is 22% (Figure 1c) and linkage disequilibrium falls below 0.2 (median R2) within 1Mb on the autosomes (Figure 1d). Four pairs of loci show high interchromosomal LD, due to mis-assembly of the reference sequence used here (Rnor3.4); these loci were excluded from the analysis (Supplementary Table 2).
Quantitative Trait Loci
The NIH-HS contains individuals of varying relatedness that generate population structure and hence false positive genetic associations. We evaluated two strategies for dealing with relatedness; mixed models in which the genotypic similarity matrix between individuals modeled their phenotypic correlation12, and resampling methods to identify loci that replicate consistently across multiple QTL models fitted on subsamples of the mapping population13. In both strategies, QTLs were detected by haplotype association14.
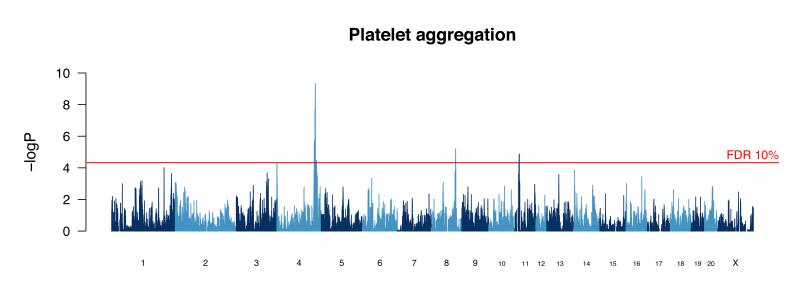
We compared the methods by simulation to find out which best controlled the false positive rate while retaining power. Mixed models performed better than resampling when phenotypes were simulated to be normally distributed, but the reverse was true for non-normally distributed phenotypes (i.e. binary phenotypes and those with a negative binomial distribution). Since these methods have different advantages, we mapped all traits with both methods, but only report those QTLs detected at false discovery rate (FDR) of 10% by that method which performed best for each trait (thresholds in Supplementary Table 1). Figure 2 shows a genome scan for one phenotype (platelet aggregation), revealing three loci at 10% FDR.
Figure 2. Genome scan for platelet aggregation.
The scan shows the results of a haplotype mixed model. The vertical scale is the negative logarithm of the P-value (logP) for association with variation in platelet aggregation. The peak on chromosome 4 harbors the von Willebrand factor gene that is identified through sequence analysis as the causative gene.
We identified 355 QTLs for 122 phenotypes with a mean of 2.9 QTLs per phenotype (Supplementary Table 3). The number of QTLs per phenotype and the QTL effect sizes (Figure 1e) have markedly skewed distributions, with a median effect size of 5% (mean effect size 6.5 %). Large effect QTLs are rare: only 22 QTLs explain more than 15% of the variance. We identified 28 QTLs that explained less than 2.5% of the phenotypic variance.
Figure 1f shows the correlation between the heritability and the total variance explained jointly by the detected QTLs. On average the QTLs explain 42% of the heritable phenotypic variance. In comparison with QTLs mapped in other rat crosses in the Rat Genome Database, there is significant overlap with NIH-HS QTLs for the number of arterial elastic lamina ruptures, total cholesterol levels and heart weight (at a nominal p-value of 0.05; Supplementary Table 4).
We estimated the QTLs’ confidence intervals by simulating a large number of QTLs throughout the genome with various effect sizes, and calculated the distribution of the confidence intervals’ widths as a function of their significance (Supplementary Figure 2). The median size of the 90% confidence interval is 4.5Mb, on average containing more than 40 genes.
Incorporation of sequence with mapping data
We investigated the extent to which our near complete catalogue of segregating sequence variants would identify genes and causative mutations. The HS permits a test, called merge analysis6, of whether a variant is responsible for phenotypic variation, under the assumption that a single imputed variant, or variants on a single progenitor haplotype, are causal. Because genetic variation segregates in the form of the progenitor haplotypes in the HS, QTLs can always be explained by variation in the haplotypes. When a QTL is due to a single variant though, genotypic variation at the variant will explain phenotypic variation better than progenitor haplotypes. To measure whether a single variant explains the QTL we calculated the difference d = logPmerge – logPhap where logPhap, is the maximum negative log 10 p-value of the haplotype test of no association and logPmerge is the maximum logP of all merge logP values of variants under the QTL. Any imputed variant that exceeds the maximum haplotype logP is termed a candidate variant. If d<0 then no candidate variants exist at the QTL. We investigated the characteristics of these candidate variants at 343 QTLs mapped using mixed models: at 131 QTLs (38%) we identified at least one candidate variant (Supplementary Table 3).
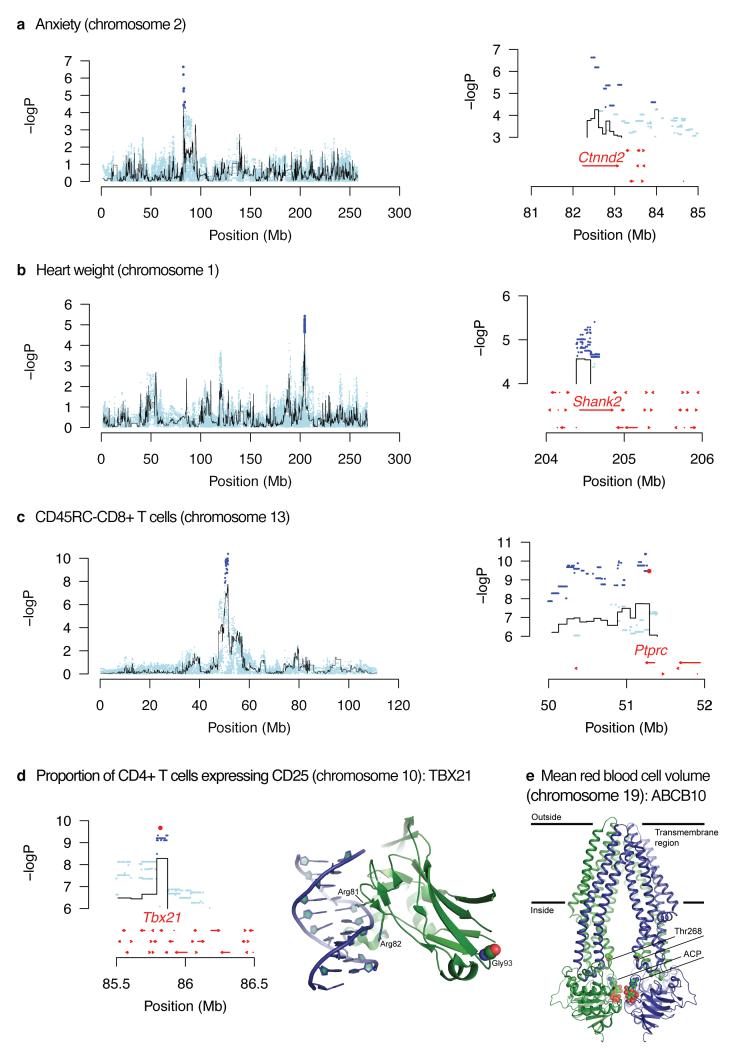
There are three ways in which focusing on these candidate variants helps identify genes at a QTL. First, we increase resolution by ruling out the great majority (usually over 90%) of sequence variants under most QTLs as being causal. We found 28 QTLs at which only a single gene contained candidate variants (Table 3). An example is Catenin-delta 2 (Ctnnd2) at a QTL for an anxiety-related phenotype (Figure 3a). CTNND2 is a protein found in complexes with cadherin cell adhesion molecules at neuronal synapses15. Figure 3b shows another example for a locus influencing heart weight, where out of 82 coding genes under the QTL, only Shank2 contained candidate SNPs. Shank2 encodes a synaptic protein 16 not previously associated with cardiovascular physiology.
Table 3. Summary of genes identified at QTLs and potential functional variants.
Shown are the phenotype measured, the chromosome (chr.), the start and stop coordinates of the QTL, gene symbol and description, whether the gene is the only one at a QTL with candidate variants, whether a variant alters an amino acid and, if so, the residue changed and the potential consequences.
| Measure | Chr. | QTL location (Mb) |
Gene | Gene description | Only gene with candidate variants in QTL |
Amino acid change with potential effect |
Location of the residue, potential effect |
|---|---|---|---|---|---|---|---|
|
Mean response
latency |
2 | 80.23– 84.83 |
Ctnnd2 | Catenin δ2 | + | None | − |
| Femur neck width | 1 | 156.27– 160.9 |
Fchsd2 | FCH and double SH3 domains protein 2 | + | None | − |
|
Distal femur total
density |
2 | 152.74– 157.22 |
Kcnab1 | Voltage-gated potassium channel subunit β1 |
+ | None | − |
|
Femoral neck
total density |
5 | 4.03– 8.22 |
Eya1 | Eyes absent homolog 1 | + | None | − |
|
Femur midshaft
cortical density |
6 | 38.24– 41.52 |
Lpin1 | Phosphatidate phosphatase LPIN1 | + | None | − |
|
Femur midshaft
total area |
2 | 43.96– 48.57 |
Ndufs4 | NADH dehydrogenase (ubiquinone) iron- sulfur protein 4, mitochondrial |
+ | None | − |
|
Femur work to
failure |
8 | 21.57– 26.17 |
Dpy19l1 | Protein dpy-19 homolog 1 | + | None | − |
|
Lumbar
trabecular area |
20 | 21.1– 25.75 |
F1LW02_RAT | Uncharacterized protein | + | None | − |
| Heart weight | 1 | 202.15– 206.63 |
Shank2 | SH3 and multiple ankyrin repeat domains protein 2 |
+ | None | − |
|
Area under
glycemia curve over baseline |
2 | 80.5– 85.11 |
Ctnnd2 | Catenin δ2 | + | None | − |
|
Hemoglobin
concentration |
12 | 1.62– 5.77 |
Insr | Insulin receptor subunit α, insulin receptor subunit β |
+ | None | − |
|
Mean platelet
mass |
1 | 193.98– 197.88 |
Dock1 | Dedicator of cytokinesis protein 1 | + | None | − |
|
Mean platelet
mass |
9 | 52.53– 88.11 |
ErbB4 | Receptor tyrosine protein kinase erbB– 4ERBB4 intracellular domain |
+ | None | − |
| Platelet clumps | 8 | 100.57– 104.81 |
Clstn2 | Calsyntenin-2 | + | None | − |
| Platelet count | 11 | 14.47– 18.54 |
Hspa8 | Heat shock 70-kDa protein 8 | + | None | − |
|
Absolute
CD25+CD4+ cells |
19 | 50.71– 54.96 |
Galnt2 | Polypeptide N- acetylgalactosaminyltransferase 2 |
+ | None | − |
|
Absolute CD8+ T
cells |
20 | 1.00– 8.90 |
RT1-Db2 | RT1 class II, locus Db2 | + | None | − |
|
Proportion of B
cells in white blood cells |
10 | 27.1– 31.59 |
D3ZTU5_RAT | Uncharacterized protein | + | None | − |
|
Proportion of B
cells in white blood cells |
20 | 1.00– 2.66 |
Olr1687 | Olfactory receptor Olr1687 | + | None | − |
|
Proportion of
CD4+ cells expressing CD45RC |
13 | 36.86– 62.54 |
Ptprc | Receptor-type tyrosine protein phosphatase C |
+ | None | − |
|
Proportion of
CD4+ cells in T cells |
20 | 14.83– 19.43 |
RGD1559903 | Uncharacterized protein | + | None | − |
|
Proportion of
CD8+ cells expressing CD45RC |
13 | 50.49– 55.97 |
Ptprc | Receptor-type tyrosine protein phosphatase C |
+ | None | − |
|
Proportion of
CD8+ cells with high expression of CD25 |
19 | 52.29– 56.8 |
Sipa1l2 | Signal-induced proliferation-associated 1– like protein 2 |
+ | None | − |
| Lowest weight | 3 | 121.45– 126.25 |
Pak7 | Serine/threonine protein kinase PAK 7 | + | None | − |
|
Weight loss
compared to day 0 |
2 | 169.79– 174.4 |
Fam198b | Protein FAM198B | + | None | − |
|
Serum alkaline
phosphatase |
3 | 18.49– 23.11 |
Lrp1b | Low-density lipoprotein–related protein 1B (deleted in tumors) |
+ | None | − |
|
Serum chloride
concentration |
9 | 32.72– 36.5 |
Uggt1 | UDP- glucose:glycoproteinglucosyltransferase 1 |
+ | None | − |
|
Serum
triglycerides |
4 | 74.8– 79.28 |
Dfna5 | Deafness, autosomal dominant 5 | + | None | − |
|
Weight loss
compared to day 0 |
20 | 2.48– 7.07 |
RT1-Da | RT1 class II histocompatibility antigen Da chain |
− | p.Thr182Ala | Surface exposed, disturbed intermolecular interactions |
|
Weight loss
compared to day 0 |
20 | 2.48– 7.07 |
RT1-Da | RT1 class II histocompatibility antigen Da chain |
− | p.Thr182Met | Surface exposed, disturbed intermolecular interactions |
|
Weight loss
compared to day 0 |
20 | 2.48– 7.07 |
RT1-Bb | RT1 class II histocompatibility antigen, B- 1β chain |
− | p.His200Arg | Surface exposed, disturbed intermolecular interactions |
|
Weight loss
compared to day 0 |
20 | 2.48– 7.07 |
RT1-Bb | RT1 class II histocompatibility antigen, B- 1β chain |
− | p.Thr165Met | Surface exposed, disturbed intermolecular interactions |
|
Weight loss
compared to day 0 |
20 | 2.48– 7.07 |
RT1-Bb | RT1 class II histocompatibility antigen, B- 1β chain |
− | p.Gln162Arg | Surface exposed, disturbed intermolecular interactions |
|
Expression on
RT1B on B cells |
17 | 26.63– 27.55 |
Tbc1d7 | TBC1-domain family member 7 | − | p.Ser116Leu | Surface exposed, disturbed intermolecular interactions |
|
Proportion of B cells in white
blood cells |
1 |
182.36–
186.67 |
Itgal | Integrin αL | − | p.Asn890Ser | Abolished glycosylation |
|
Proportion of CD4+ cells with
high expression of CD25 |
10 | 84.27– 87.32 |
Tbx21 | T-box transcription factor TBX21 | − | p.Gly175Arg | Surface exposed, additional interactions with DNA |
| Ratio of T cells to B cells | 1 | 183.58– 187.41 |
Rabep2 | Rab GTPase–binding effector protein 2 |
− | p.Ile336Thr | Partially buried, disturbed oligomerization |
| Ratio of T cells to B cells | 1 | 183.58– 187.41 |
Itgal | Integrin αL | − | p.Leu806Ser | Surface exposed, disturbed intermolecular interactions |
|
Mean corpuscular red blood cell
volume |
19 | 53.11– 55.80 |
Abcb10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
− | p.Thr233Met | Transport channel exposed, altered transport |
| Platelet count | 12 | 1.00–7.47 | Rfc3 | Replication factor C (Activator 1) | − | p.Pro173Ala | Surface exposed, alteration of the α helix |
|
Proportion of monocytes in
white blood cells |
1 | 250.37– 254.00 |
Pdcd11 | Protein RRP5 homolog | − | p.Glu160Gly | Surface exposed |
Figure 3. Merge analysis to identify causative genes and sequence variants.
The top three panels (a – c) show, on the left, the scans for a whole chromosome, with the name of the phenotype. The black lines represent the haplotype analysis and the blue dots are the merge analysis results of testing for association with all sequence variants identified in the progenitor strains. On the right is an enlargement of the highest peak showing the location of candidate variants and genes. Candidate variants are those whose significance exceeds that of the haplotype analysis (i.e. blue dots are above the highest value of the black line). Genes are shown by red arrows. Panel (d) shows candidate variants on chromosome 10 for the proportion of CD4+ cells with high expression of CD25. The highest variant lies within the TBX21 protein. The crystal structure of human TBX5-DNA complex (PDB code 2X6V) maps the location of the rat TBX21 mutation Gly175Arg to the DNA binding domain. The structure of TBX5 (green) complexed with DNA (blue) is shown in ribbon representation. Gly93 is shown as spheres (C atoms in green, O atoms in red N atoms in blue). Gly93 and corresponding Gly175 (rat) are conserved. Side chains of two arginines that mediate interactions with DNA are shown as sticks. Panel (e) shows a candidate variant in the Abcb10 gene on chromosome 19 for a locus influencing mean red cell volume. The structure of the homodimeric ABCB10 (PDB code 4AYT) is shown in ribbon representation, with the monomers in blue and green. Two ATP analogues (ACP) and side chains of Thr268 are shown as spheres (C atoms in green, O atoms in red N atoms in blue and P in orange). Thr268 in the human protein corresponds to the conserved Thr233 residue in the rat protein. The rat ABCB10 mutation Thr233Met lies in the central cavity of the translocation pathway. Amino acid sequence identity between rat and human ABCB10 is 84% (587 aligned residues).
Second, merge analysis identifies some candidate variants lying within coding regions. Those predicted to affect protein structure are more likely to be causal. Thus we identified a potential causal nucleotide in a QTL for antibody recognition of CD45RC on CD4+ and CD8+ T cells (Figure 3c). The antibody used binds to the CD45RC isoform, which expresses a C-domain, encoded by the sixth exon, in which we found a candidate variant changing an amino acid (p.Arg114His).
At 43 out of 91 non-synonymous candidate variants, where similar protein structures were available17 we predicted the structural consequences of mutations (for a further 48 candidate variants there were no homologies with known protein structures). Nine genes, listed in Table 3, contained candidate variants for which structural evidence suggests protein structure or interactions might be altered.
An example is shown in Figure 3d, for the protein TBX21, encoded by a gene under a QTL influencing the proportion of CD4+ cells with high expression of CD25.) Here the candidate variant changes glycine to arginine (p.Gly175Arg). The additional arginine could alter the DNA-binding characteristics of this protein.
Figure 3e shows the crystal structure of human ABCB10, a mitochondrial transporter induced by GATA1 during erythroid differentiation18,19. The candidate variant (p.Thr233Met), predicted to influence mean red cell volume, maps to a position in the protein structure where the residue side chain points to the centre of the transporter channel (Figure 3e). Threonine has a polar uncharged side chain while methionine has a hydrophobic side chain, a difference likely altering transporter function.
Third, merge analysis eliminates candidate genes at a QTL that are distant from any candidate variant. This approach confirmed a well-established relationship between a cluster of apolipoprotein genes at a QTL on chromosome 1 and cholesterol biosynthesis (HDL, LDL and total cholesterol). Similarly, merge analysis identified a locus influencing platelet aggregation, on chromosome 4, that harbors the von Willebrand factor gene, encoding a key glycoprotein involved in blood coagulation.
Merge analysis also contributes to an understanding of the pathogenesis of experimental autoimmune encephalomyelitis (EAE), an autoimmune neuroinflammatory disease with clinical and pathological similarities to multiple sclerosis (MS)20. The MHC class II region on chromosome 20 (Eae1) is known to influence EAE susceptibility. However, attempts to identify the responsible gene have had limited success. In this study, the two variants most likely to be causative for the QTL on chromosome 20 (i.e. highest logP) are a variant in an intron of Btnl2 and a variant 274 bp upstream of RT1-Db1, both in the class II region. The human orthologue of RT1-Db1, HLA-DRB1, is associated with multiple sclerosis with risk allele HLA-DRB1*15:0121.
Single variants rarely account for NIH-HS QTL genetic effects
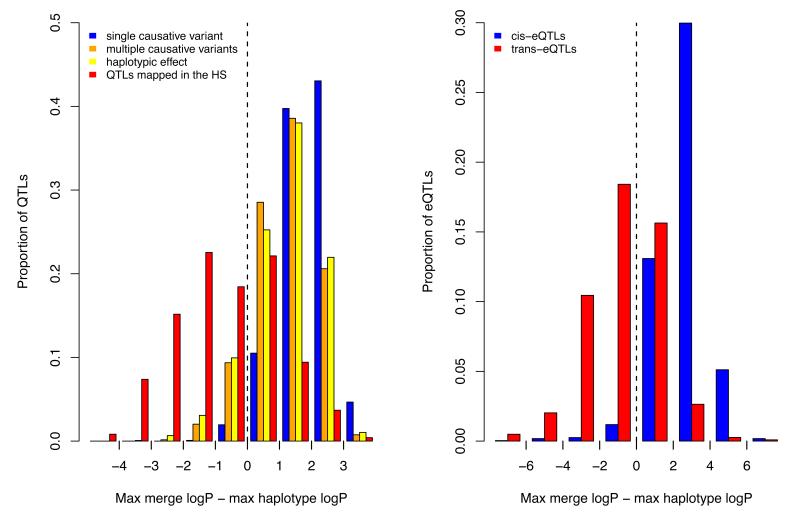
Unexpectedly 212 QTLs (62%) had no candidate variant (Figure 4a). We considered four explanations for this observation: (i) causative variants were missing from the sequence catalogue; (ii) haplotype mapping is biased towards QTLs without candidate variants; (iii) the merge analysis underestimated statistical significance compared to single marker association; (iv) the presence of multiple causal variants.
Figure 4. Merge analysis and simulations.
The figure plots the difference between the negative logarithm p-value of association (logP) of imputed variants and haplotype-based logP for the rat QTLs (4a) and a set of 1,386 cis-acting and 7,464 trans-acting expression QTLs mapped in a mouse HS (4b). In cases where there is a single causal variant at a QTL, the logP of some imputed variants will exceed that of the haplotypes, so that the mean of the distribution of the difference between these two logP values will be greater than zero. This is shown as a blue histogram on the plot 4a. The distribution observed for the phenotypic QTLs, shown in red in 4a, has a mean less than zero. The results of simulating haplotypic effects are shown in yellow, and in orange the consequence of simulating multiple causative variants. The distribution of the difference in logP for the cis-eQTLs is shown in blue in 4b to highlight the resemblance with the results of simulating single causative variants. The distribution for the trans-eQTLs is shown in green in 4b and is most similar to that for the phenotypic QTLs.
First, causal variants may have been missed because our sequence data are incomplete. Despite linkage disequilibrium extending over a few megabases, not all variants are tagged by a nearby variant with identical strain distribution pattern (SDP) in the founders. For example, only 50% of the structural variants are tagged by a SNP lying within 1Mb.
However, because only a limited set of possible SDPs exist in the HS, we can test whether missing genotypes are responsible for failure to detect candidate variants. We generated SDPs for all diallelic and tri-allelic variants at every locus within the 212 QTLs and tested each by merge analysis, to see how many would have been candidates. Only 44 QTLs had candidate diallelic variants and 165 had diallelic or triallelic variants. Thus if the effects are attributable to a single diallelic variant that we had failed to sequence, then there are still 168 QTLs (49%) without a candidate variant. If the effects are attributable to a di-allelic or tri-allelic variant, the fraction reduces to 14%. However, triallelic SNPs are very uncommon and therefore unlikely to explain the large number of QTLs without candidate variants.
Second, haplotype mapping might simply not be powerful enough to detect candidate variants, or be biased towards QTLs without candidate variants. We addressed the first possibility by simulation and show the results in Figure 4a. In each case we report the distribution of the difference d between maximum merge and haplotype logPs, so that if candidate variants exist then d > 0 (where d = logPmerge – logPhap as defined above). When simulated QTLs arise from single causal variants, merge analysis does indeed identify candidate variants at almost all QTLs placed at random regions of the genome as well as at QTLs simulated in the same locations as the detected QTLs.
We also considered the performance of the method at QTLs where a single variant is highly likely to be the causal variant, namely at cis-acting expression QTLs 22,23. We tested 1,398 eQTLs detected in the hippocampus of HS mice24. We found that the merge analysis identified variants that exceed the haplotype-based test at 97% of QTLs (Figure 4b). Interestingly, when we carried out the same analysis on trans eQTLs, the distribution of d values was similar to that seen for the rat phenotypic QTLs (Figure 4b). This difference between cis and trans-eQTLs is true across all logP values, indicating that the difference is not due to lower power to detect trans eQTLs.
Since mapping QTLs using haplotype analysis might bias results towards finding loci without candidates (a winner’s curse is likely to operate), we used merge analysis to map QTLs genome-wide. The two methods do not identify the same QTLs (152 are unique to the merge method) but the merge method identified 16% fewer than the haplotype method. Importantly, only 9% of the merge-identified QTLs had no candidate variants (Supplemental figure 3). Consequently haplotype mapping will overestimate the number of QTLs without a candidate variant while merge analysis underestimates it. Therefore our best estimate of the proportion of QTLs without candidate variants is obtained from combining both methods. From the set of QTLs found by either merge or haplotype mapping we find that 44% of QTLs cannot be explained by single causal variants (instead of 62% when only the haplotype-based QTLs were considered). Thus while a winner’s curse does operate in favour of the haplotype analysis, it cannot account for all QTLs without a candidate variant.
The third explanation was that the merge analysis under-estimates statistical significance. We compared the performance of the merge analysis with single marker association at genotyped SNPs. Across all phenotypes, the R2 between the logP values was 0.9; agreement was strongest for the most highly associated SNPs. This result indicates that merge analysis performs as well as SNP analysis.
Finally, we investigated the extent to which multiple variants at QTLs would account for our findings. We investigated the consequences of a variety of complex QTL architectures by simulation and show the results in Figure 4a. Simulating multiple causal variants, on different haplotypes, reduced the frequency that any single variant exceeded the maximum haplotype logP, although this was still insufficient to mimic the observed distribution (Figure 4a). Simulating irreducible haplotypic effects arising from the reconstructed haplotype mosaics in the HS (rather than from a selection of sequence variants) also led to fewer QTLs with candidate variants (Figure 4a), although again it did not match the proportion observed with the real QTLs. Our simulations suggest that the presence of multiple causal variants at a locus accounts in part for the failure to find candidate causal variants.
Concordance between species
It is often assumed that genetic loci underlying a phenotype identified in one species are homologous to those underlying the same phenotype in another, and that natural variation within these loci will pinpoint the same genes 25-27. However, there have been no genome-wide tests of the hypothesis for natural variation. Our data allowed us to examine whether genes and QTLs identified in the NIH-HS overlapped those found for the same phenotypes in a mouse HS10.
In total 38 measures were common to both studies, and were mapped using the same mixed model method. Only one measure, the ratio of CD4+ to CD8+ T-cells, showed overlap (using a FDR of 10% and looking in the 90% QTL confidence interval) but this was not significant (empirical p-value of 0.1). We repeated the analysis using QTLs called at a lower significance threshold (20th percentile of the extreme value distribution for each measure) and expanding the width of each QTL to 8Mb. Table 4 shows overlap for eight phenotypes, only two of which were significant at an empirical p-value of 0.05: serum urea concentration and the ratio of CD4+ to CD8+ T-cells. Overall, genetic variants in orthologous genes rarely contribute to the same phenotype in the two populations.
Table 4. Syntenic QTLs mapped in the rat and mouse HS for the same measure.
The table shows the 8 measures (out of 38) that have syntenic QTLs, the QTL coordinates (chromosome, start and stop) and the p-value of the overlap (one p-value per measure).
| Phenotype | Rat chr. | Rat QTL (Mb) | Mouse chr. |
Mouse QTL (Mb) | P-value of overlap |
|---|---|---|---|---|---|
|
| |||||
| CD4/CD8 ratio | 2 | 80.51 - 88.51 | 8 | 71.7 - 79.7 | |
| CD4/CD8 ratio | 20 | 1.00 - 21.13 | 17 | 29.77 - 37.77 | |
| CD4/CD8 ratio | 9 | 0.16 - 8.16 | 17 | 50.77 - 58.77 | 0.009 |
|
| |||||
| Serum urea | 3 | 42.22 - 50.22 | 2 | 62.25 - 70.25 | 0.017 |
|
| |||||
| Serum calcium | 12 | 32.82 - 40.82 | 5 | 122.62 - 130.62 | 0.082 |
|
| |||||
| White blood cells | 10 | 57.69 - 71.77 | 11 | 64.92 - 72.92 | |
| White blood cells | 20 | 47.41 - 55.24 | 10 | 40.74 - 48.74 | 0.115 |
|
| |||||
| T/B cells ratio | 13 | 76.73 - 84.73 | 1 | 169.63 - 177.63 | |
| T/B cells ratio | 20 | 37.59 - 45.59 | 10 | 36.25 - 48.68 | 0.149 |
|
| |||||
| Serum chloride | 9 | 30.61 - 38.61 | 13 | 2.91 - 15.19 | 0.22 |
|
| |||||
| Monocytes | 20 | 0.17 - 8.17 | 17 | 21.00 - 29.00 | 0.301 |
|
| |||||
| Serum total cholesterol | 4 | 17.09 - 25.09 | 5 | 12.52 - 20.52 | 0.598 |
To test whether QTL overlap existed within similar pathways we compared the enrichment of KEGG pathways28. Only one measure, the proportion of B cells in the white blood cells, showed significant enrichment of a pathway (corrected p-value < 0.05). Even at a more relaxed significance threshold of 0.05 (non-corrected for multiple testing), only three measures show significant enrichment in the same KEGG pathways.
Discussion
Using 1,407 outbred rats we have mapped 122 phenotypes and identified 355 QTLs at high resolution. We have shown how combining sequence with high resolution mapping data can lead to the immediate identification of candidate genes, and in some cases to the identification of candidate causal variants at many QTL. We highlight two examples here.
The locus on chromosome 10 regulating frequency of CD25+ CD4 T cells, and the frequencies of CD4 and CD8 T cells, has previously been shown to control CD4 and CD8 frequencies in a cross between ACI and F34429, both represented in the NIH-HS rats. The amino acid substitution at position 175 (p.Gly175Arg) of the TBX21 protein is a very strong causal candidate at this QTL since this domain is important for DNA interactions. Tbx21 has been implicated in the genetic control of regulatory T cells30, a subset of T cells with high surface expression of CD25, and might indirectly regulate the frequency of CD4+ and CD8+ T cells via the transcriptional repressor Sin3a31,32.
We implicated Abcb10 in red blood cell differentiation. Evidence from mouse knockouts indicates that this gene is essential for erythropoiesis18,19,33. The p.Thr233Met mutation positions a larger, bulkier residue into a region that is tightly packed in the open-outwards conformation of ABC transporters, potentially interfering with conformational changes which are essential for transport of the substrate.
Two noteworthy features of the genetic architecture of complex traits in the rat emerge from this study: (i) the contrast with human GWAS findings; (ii) about half of QTLs cannot be attributed to a single causal variant. We discuss these points below.
Rat and mouse HS experiments differ from human GWAS in two ways. In the rodent GWAS, far fewer subjects are required to detect a significant effect and fewer loci of larger effect explain more of the variance. In rats the median proportion of heritability explained by joint QTLs is 39.1% (mean 42.3%), in mice 32.2% (mean 42.0%). In humans the equivalent figure is often less than 10%.
One explanation for these differences is the markedly different allele frequencies: human populations are characterized by a preponderance of rare alleles (minor allele frequency less than 1%); HS populations have a relatively uniform distribution of minor allele frequencies (Figure 1c). However, it is important to realize that the mouse and rat differ in the degree of segregating variation (in the rat NIH-HS there are 7.2M SNP sites, compared to 10.2M in the mouse HS). In the rat there are 2.8M SNPs per HS strain, the corresponding number in the mouse HS is 4.4M. In other words, total sequence variation per se is not a critical determinant of the explanatory power of the QTLs. Furthermore, the heritabilities of the homologous phenotypes in rat NIH-HS and in HS mice are highly correlated (0.6, p-value 0.0002) (Supplemental Figure 4), implying that the additional sequence variation in the mouse does not give rise to an increase in heritability.
The failure to detect a single candidate variant at half of rat QTLs was surprising. We showed that while reliance on haplotype mapping can underestimate the number of QTLs without candidate variants, after taking this bias into account (by detecting QTLs with merge and haplotype analysis) there is still a large fraction (44%) of QTLs without candidate variants. The contrast between the 44% figure and the 97% that emerged from an analysis of variants at cis-eQTLs is striking. It is also notable that the findings from trans-eQTLs are so similar to those of the rat phenotypes (Figure 4) suggesting that cis-eQTLs are atypical. Our simulations indicate, but have not proven, that multiple causal variants are in part to blame. At present, we can only conclude that single causal variants are not always responsible for the genetic signal. Whether the lack of single causal variants at many loci is a general feature of loci influencing complex traits or not remains to be determined.
One simple interpretation of human GWAS is that each locus represents the presence of a single, relatively common, functional variant. Our results indicate that more complex models are required. Such alternative hypotheses exist, in which for example multiple alleles of varying frequency at the same or closely linked loci, contribute to the signal. Identifying the correct model of genetic action is critical for finding causative variants, since incorrect assumptions about the number and mode of action of genetic variants reduce power and can lead to false positive results34. The extent and nature of sequence diversity may be partly responsible for the complex way sequence variation acts at a QTL.
It is sometimes hoped that loci found in the rat could be typed and identified in humans, thus providing a cost-efficient way to find medically relevant genes. We observe some examples where the same loci act in different species, the most notable example being for variation in the ratio of CD4+ to CD8+ T-cells: the locus lies within the MHC in rats, humans35 and mice36 and its molecular nature in mouse has been identified as a deletion in the promoter of the Class II H2-Ea gene36. However formal tests for overlap between rat and mouse at the level of the gene or of a pathway yielded little that was statistically significant. Since the amount of sequence variation segregating within the two HS populations is relatively limited, failure to detect shared loci may be due to sampling. Also, the relatively small number of genes found for each phenotype reduces our power to detect pathways. We suspect that currently it is not possible to accurately assess overlap between the two species.
This study strengthens the rat’s role as a model organism in physiology and disease. Our mapping and sequencing data provide an important resource for addressing many biomedical questions.
Methods
Sequencing of HS founder genomes
Genome sequencing
DNA libraries for SOLiD sequencing were generated from genomic DNA from samples of the original rats that were used to create the HS population. The libraries were generated using standard protocols (Life Technologies) and had a median insert size of between 109 and 196 bp. All libraries were sequenced with fragment (50 bp) and paired-end (50+35 bp) runs using SOLiD 4 and SOLiD 5500 sequencers to a depth of at least 22x base coverage for each of the eight HS progenitors and for the strain LE/Stm, which was used to estimate error rates in comparison with hand-finished BAC sequence.
Sequence alignment
Sequence reads were mapped against contigs of the Rnor3.4 rat reference genome assembly (reference strain BN) using BWA v0.5.937 with parameters -c –l 25 –k 2 –n 10. Alignments from different libraries of the same HS progenitor were combined into a single BAM file.
Variant calling
Variant calling was performed independently on each strain. SNPs and short indels (<10bp) were called using a modified Samtools38 pipeline: Only unambiguously mapped reads were used. Sites with coverage below 4 or over 2000 were not used for SNP calling. Read bases with base-quality below 30 were ignored. Duplicate reads starting at the same position and mapped to the same strand as another read were discarded as likely PCR artifacts. Each of the called alleles had to be supported by at least one read where the variant mapped within the seed part of the read (first 25 bases). Non-reference alleles called with fewer than 3 reads were set to missing. Variable sites with more than 2 alleles within one founder were set to missing. The remaining variants were considered to be homozygous non-reference alleles (frequency of non-reference call > 2/3) or heterozygous alleles (frequency between 1/3 and 2/3) – however, we set to missing the small number of heterozygote calls as these were likely to be artefacts, for example due to unknown duplications. We later attempted to call all the missing genotypes by imputation (see below).
Copy number variants were called using depth-of-coverage approach implemented in DWAC-Seq v. 0.56 (https://github.com/Vityay/DWAC-Seq) using default parameters. Structural variants (SVs) were called using discordant pair mapping implemented in 1-2-3-SV v. 1.0 (https://github.com/Vityay/1-2-3-SV), requiring unambiguous mapping of both paired tags and at least 4 tag pairs per SV event. SV calls from these tools were merged. Prediction of the functional effect of each variant was performed by Variant Effect Predictor tool VEP 2.1 tool39.
We defined inaccessible regions of the HS rat genomes in a similar way as was done for mouse genomes4. A base was considered as accessible if it did not overlap simple, tandem repeats or low complexity sequence (defined by Dust, source: Ensembl release 66; http://www.ensembl.org), was not covered by more than 150 reads, and average mapping quality was at least 40. Nucleotide positions within 15bp of indels were also considered as inaccessible for SNP calling.
False positive and false negative rates
Thirteen BACs from the strain LE/Stm were sequenced using capillary methods, assembled and manually edited, producing a total of 2.1 Mb finished sequence. BAC sequences were aligned using BLAT40 For each BAC a single contiguous alignment was obtained, which was used to extract single base changes (SNPs) short indels (1-10 bp) and structural variants (100 bp and above). False positive and false negative rates were estimated from 1.9 Mb of genome sequence syntenic between BACs and genome assembly, excluding low quality BAC sequence (as defined by the BAC finishing team) and inaccessible regions (as defined above). False positive and false negative rates within this 1.9Mb were estimated from the discordance between our allele calls and those in the BACs.
Low false positive rates were independently confirmed by analysis of a randomly selected subset of 96 SNPs and 96 indels using PCR-based resequencing. Oligonucleotide primers were selected to amplify 300 bp fragments around the candidate polymorphism. When amplification was successful (SNPs: 84, indels: 80), amplicons were sequenced on an Applied Biosystems ABI 3730XL sequencer using Big-Dye terminator and analyzed with Polyphred software manually.
For CNVs and SVs 184 variants were selected and PCR primers were designed in such way that the presence or absence (depending on the variation type) of a PCR product could confirm the presence of the variation. After PCR, samples were run on agarose gel and analyzed manually. Of the 184 amplicons, 93 gave a PCR product. Of these 93, a group of 39 variants that were predicted SVs in the NIH-HS founders, were also confirmed by PCR in BN/NHsdMcwi indicating that these are probably assembly errors in the current reference genome (Rn3.4). Of the remaining 54 variants, 53 gave a banding pattern according to our expectance and in one case the predicted variation type was not correctly predicted.
Sequence divergence
Genotypes and genome accessibility data for HS rats (this study) and HS mice4 were used to characterize patterns of nucleotide diversity in these two panels. We partitioned each genome into non-overlapping windows such that each window contained 100 kb of accessible sequence (defined relative to the rat BN strain or mouse C57BL6 strain). The number of sequence differences per window was calculated for all windows and for all possible pairs of strains.
Low diversity regions
We found the spatial distribution of pairwise differences in the rat progenitors was bimodal with modes at 0 and 150 SNPs per 100 kb window (Figure 1b). Based on this distribution we defined a region of low nucleotide diversity between two strains as consecutive windows with nucleotide diversity below 13 SNPs per 100kb window.
Phenotyping
Animals
The rat NIH-HS originates from a colony established in the 1980s in NIH3. Since its creation, the stock has been bred using a rotational outbreeding regime in order to minimize the extent of inbreeding, drift, and fixation.
Phenotyping
A full description of the phenotyping protocol is given in Supplementary Note (“Phenotyping”).
All procedures were carried out in accordance with the Spanish legislation on “Protection of Animals Used for Experimental and Other Scientific Purposes” and the European Communities Council Directive (86/609/EEC) on this subject. The experimental protocol was approved by the Autonomous University of Barcelona Ethics committee (permit CEEAH 697).
Quality control, covariate analysis and normalisation of phenotypes
The phenotype data were uploaded to a database (Integrated Genotyping System41) in batches over the three years of data collection. All relevant covariates were evaluated for their effect on each measure. The final set of covariates and transformations applied to each phenotype, as well as the number of data points for each measure, are given in Supplementary Table 1.
Genotyping
The RATDIV array was developed as a general SNP genotyping array, applicable both to the rat HS project and other populations of laboratory rats. Full descriptions of the development of the rat array and of the selection of the 265,551 SNPs used in this study are given in the Supplementary Note (sections “Development of the rat array” and “Selection of SNPs for this study” respectively).
Linkage disequilibrium analysis
Linkage disequilibrium (LD) between SNPs in the rat and mouse HS was calculated using PLINK42 from the genotypes called for the 261K autosomal rat SNPs and 12K autosomal mouse SNPs10. In the rat HS, eight regions with very high interchromosomal LD were identified, and excluded from subsequent analyses (Supplemental Table 2). Using UCSC liftover tool43, these regions mapped in the new rat reference genome assembly (RGSC 5.0) to the regions with which they were in high LD in the current assembly (Rnor3.4).
QTL Mapping
Reconstruction of HS rat genomes as mosaics of founder haplotypes
All genetic analysis was performed using R44. We used the R HAPPY package14 to calculate the descent probabilities from the eight HS founders for each animal at each of 265,551 inter-markers intervals, and then averaged these probabilities over 90kb windows, so that we eventually worked with 24,196 probability matrices. The density of the 265k SNPs was much greater than the density of recombinants in the HS, so the averaging did not cause any reduction in mapping resolution (most QTLs are mapped to intervals over a Mb wide, containing over ten 90kb intervals).
Accounting for confounding in the HS
HS rats with different levels of relatedness were used in this study, including siblings, half sibs, cousins, uncles, great-uncles, etc. This unequal genome-wide genetic similarity means that correlations exist in the HS between distant markers. These long-range correlations (as opposed to short-range correlations due to physical linkage) can be responsible for false associations if not accounted for. We used two methods to control for unequal relatedness: Resample Model Averaging (as implemented in BAGPHENOTYPE 13) for non normally distributed phenotypes, and Mixed Models for normally distributed phenotypes. Information on the scope and the performance of the methods is given in the Supplementary Note (section “Comparison between mixed models and resample model averaging”). Because most of the phenotypes were normally distributed and the merge analysis was run in the mixed model framework, we present the mixed models briefly here. They were implemented in R so that haplotype mapping could be carried out using the descent probabilities output by HAPPY14. The model used to test for association between the ancestral haplotypes segregating at a locus L and phenotypic variation was:
| (1) |
where yi is the phenotypic value of the rat i, βc the regression coefficient of covariate c and xic the value of the covariate c in rat i. Importantly, the covariates include a dummy intercept term. TLs is the deviation in phenotypic value that results from carrying one copy of a haplotype from strain s at locus L and PLi(s) the expected number of haplotypes of type s carried by rat i at locus L output by HAPPY14. ui and εi are random effects, with cov(ui,uj) = σg2Ki,j and cov(εi,εj) = σe2Ii,j where σg2 and σe2 are estimated in the null model (no locus effect, TLs=0) using the R package EMMA12. K is the genetic covariance matrix, and is estimated from the genome-wide genotypic data using identity by state (IBS, the proportion of shared alleles between any two animals). The IBS matrix was calculated using the R package EMMA12. I is the identity matrix. The total covariance matrix V= σg2K + σe2I can be factorized as V=A2. Writing the equation (1) in matrix form,
| (2) |
Pre-multiplying (2) by A−1 gives a transformed equation
in which the variance-covariance structure of the random term A−1(u + ε) is now proportional to a diagonal matrix and so can be fitted as a standard linear model.
Thresholds and confidence intervals
Calculations of the significance thresholds (when the phenotype was analysed with mixed models), inclusion probability thresholds (when it was analysed by resample model averaging), and confidence intervals are described in the Supplementary Note (section “QTL calling”).
Incorporation of sequence into QTL mapping
Implementation of the merge analysis in the mixed model framework
Merge analysis is a form of imputation appropriate to HS-type populations whose genomes are mosaics of known haplotypes. Merge analysis asks two questions at each imputed variant: is the variant associated with the phenotype? (a standard test of association), and is it as significant as the test of haplotype association in the locality of the variant? We implemented merge analysis6 in a mixed models framework by comparing the models:
| (2 above) |
| (3) |
where V is a sequence variant in interval L, and MV is the merge matrix for the variant, formed by summing those columns of PL that carry the same allele at V (each column of PL represents one founder strain). This can be computed efficiently by defining a matrix BV that encodes the columns to be merged such that MV = PVBV. This test is applied at every variable site in the catalogue of single nucleotide variants that segregate between the 8 HS founders. From a statistical point of view, there is no difference between two variants with the same strain distribution pattern at a locus; they will give the same merge analysis result.
Because the two models (2), (3) are nested, the best possible fit (in terms of variance explained) is obtained with the haplotype model (2). If the QTL arises from variation at a single variant V, the fit of the merge model (3) for variant V will be as good as the fit of (2) and its significance will be greater due to the fewer number of degrees of freedom (for a diallelic variant, the degrees of freedom is 1 rather than 7 for the haplotype model). The merge model is fitted by multiplying by A−1 as before.
Simulating all possible strain distribution patterns at a QTL
For each QTL lacking variants with a merge logP exceeding the haplotype logP, we looked for unobserved causal variants that might not have been sequenced. We simulated candidate variants with every possible strain distribution pattern (SDPs; 127 possible SDPs for diallelic variants, 1,094 possible SDPs when allowing for 3 alleles). The simulated variants were repeated within each QTL interval.
Simulating different QTL architectures
To investigate the hypothesis that failure to detect candidate variants by merge analysis reflected a complex architecture of the QTLs, we simulated QTLs arising from a single causal variant, QTLs arising from multiple causal variants within the same locus and/or multiple causal variants at linked loci, and QTLs arising from haplotypic effects not reducible to individual variants. In all cases the phenotypes were simulated from three components: a genetic random effect explaining 20% of phenotypic variation, uncorrelated errors explaining 75% of phenotypic variation, and a single QTL explaining 5% of phenotypic variation. When multiple causal variants were simulated, each explained the same proportion of phenotypic variation (5% divided by the number of causal variants). The effect sizes calculated a posteriori could be quite different from their target values due to correlations between the different components of the simulated phenotypes. For the simulations reported in Figure 4a, either a single causal variant was simulated, or nine causal variants in three linked loci (each locus within 2Mb of the central locus and distant by at least 200kb from each other locus). Alternatively, the probabilities PL were used to simulate irreducible QTLs. We analysed each simulation by merge analysis and when logPhaplotype was between 4 and 6 (to have a similar distribution of logP values to that of the rat QTLs) we calculated d = max logPmerge – max logPhaplotype. We compared the distribution of d from the different simulation sets to determine the likely genetic architecture of the QTLs.
eQTL mapping and merge analysis in the mouse Heterogeneous Stock
Hippocampus expression levels in 460 HS mice measured using 12 thousand probes of Illumina Mouse WG-6 v1 BeadArrays 24 were mapped to the mouse ancestral haplotypes in the mixed model framework. QTLs were called in the same way as for the rat QTLs but using a confidence interval of 8Mb and a significance threshold of 4. cis-eQTLs were defined as within 2Mb of the beginning of the probe, and trans-eQTLs as those QTLs on a different chromosome from that of the probe or more than 10Mb away from it. Merge analysis was carried out at each eQTL and the difference between the maximum merge logP and the maximum haplotype logP was calculated.
Homology modeling
To assess the potential effect of mutations on protein structure, homology models of target proteins were constructed and analysed. Amino acid sequences of target proteins were retrieved from Ensembl or UniProt databases45 and analysed using HHPred46 web server to identify structures with similar amino acid sequences in the Protein Data Bank17 for homology modelling with MODELLER47. Potential location of the mutation-bearing side chains (buried or surface exposed) and effect on the structure-function (e.g. disturbed hydrophobic core) was evaluated manually in PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.).
Genetic architecture
Heritability
Heritability is defined as the ratio of the genetic variance component by the sum of the variance components estimated in the null mixed model (covariates but no QTL).
QTL Effect sizes and joint effect sizes
Effect sizes are defined as the ratio between the fitted sum of squares and the total sum of squares in a model with covariates and without genetic random component. Joint effect sizes are defined as the ratio between the fitted sum of squares and the total sum of squares in a model without genetic random component including covariates and all the QTLs called for a given phenotype. Including the genetic random component would underestimate most of the effect sizes because part of the variance would have been attributed to it. Thus the QTL effect sizes reported are probably overestimates.
Number of genes under a QTL
The number of genes under each QTL confidence interval was calculated using Ensembl protein coding genes and genes coding for micro RNAs (downloaded from BioMart48).
Overlap with RGD QTLs and with QTLs detected in the mouse HS
The calculation of the overlap between RGD and rat HS QTLs, and between mouse and rat HS QTLs is given in the Supplementary Note (section “Overlap between sets of QTLs”).
Pathway analysis for the QTLs detected in the rat and mouse heterogeneous stocks
Kyoto Encyclopedia of Genes and Genomes pathways were retrieved using R KEGG.db package. We used INRICH 49 to find enrichment of pathways in the mouse and rat phenotypic QTLs (as defined by the 90% confidence interval) called at a low significance threshold (20th percentile of the extreme value distribution). We report the empirical and corrected p-values reported by INRICH.
Supplementary Material
Acknowledgements
We are grateful to T. Serikawa (Kyoto University) for the LE/Stm BAC clones. The Human Genome Sequencing Center sequence production teams at the Baylor College of Medicine produced the Sanger sequencing data for the eight sequenced strains used to define the RATDIV SNP genotyping array (see ref. 8 for a list of Baylor College of Medicine HGSC sequencing contributors). We thank E. Redei for providing the NIH-HS rat colony. The funders we would like to acknowledge are as follows: the European Union's Seventh Framework Programme (FP7/2007- 2013) under grant agreement HEALTH-F4-2010-241504 (EURATRANS); The Wellcome Trust (090532/Z/09/Z, 083573/Z/07/Z, 089269/Z/09/Z); The Swedish Research Council (grant K2008-66X-20776-01-4); the Harald and Greta Jeanssons Foundation; The Swedish Association for Persons with Neurological Disabilities; the Åke Wibergs Foundation; the Åke Löwnertz Foundation; Karolinska Institutet funds; the European Union's Sixth Framework Programme EURATools (grant LSHG-CT-2005-019015); the Bibbi and Nils Jensens Foundation; the Söderbergs Foundation; and the Knut and Alice Wallenbergs Foundation. We also thank the Ministerio de Ciencia e Innovación (reference PSI2009-10532 and the Formación de Personal Investigador fellowship to C.M.-C.); the Fundació La Maratò TV3 (reference 092630); the Direcció General de la Recerca (reference 2009SGR-0051); and the British Heart Foundation (BHFRG/07/005/23633). T.J.A. and S.S.A. acknowledge funding from the Imperial BHF Centre of Research Excellence. M. Johannesson acknowledges support from Prof. Nanna Svartz Foundation, The Swedish Rheumatism Association and The King Gustaf V 80th Anniversary Foundation. D. Gauguier acknowledges support from the Institute of Cardiometabolism and Nutrition (ICAN; ANR-10-IAHU-05). T.M. and E.Y.J. acknowledge support from Cancer Research UK (A10976) and the UK Medical Research Council (G9900061). T.F., D.L.K. and I.A. acknowledge support from the U.S. National Institutes of Health (R01 AR047822).
Footnotes
Mapping data are available at http://mus.well.ox.ac.uk/gscandb/rat (see Supplementary Note (“Guidelines to explore the genome scans and integrated sequence data”) for directions on how to explore the sequence data at each QTL).
Variant calls and inaccessible regions are available at http://www.hubrecht.eu/research/cuppen/suppl_data.html.
Accession numbers: Sequence data for the eight HS founders are available from EBI SRA archive under accession ERP001923. The LE/Stm BAC sequences are available in the NCBI Trace Archive (accessions FO181540, FO181541, FO117626, FO181542, FO117624, FO181543, FO117625, FO117627, FO117628, FO117629, FO117630, FO117631, and FO117632)
References
- 1.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–31. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 2.Flint J, Mackay TF. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–33. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen C, Spuhler K. Development of the National Institues of Health genetically heterogeneous rat stock. Alcoholism, Clinical and Experimental Research. 1984;8:477–479. doi: 10.1111/j.1530-0277.1984.tb05706.x. [DOI] [PubMed] [Google Scholar]
- 4.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talbot CJ, et al. High-resolution mapping of quantitative trait loci in outbred mice. Nature Genetics. 1999;21:305–308. doi: 10.1038/6825. [DOI] [PubMed] [Google Scholar]
- 6.Yalcin B, Flint J, Mott R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics. 2005;171:673–81. doi: 10.1534/genetics.104.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayosi BM, Keavney B, Watkins H, Farrall M. Measured haplotype analysis of the aldosterone synthase gene and heart size. Eur J Hum Genet. 2003;11:395–401. doi: 10.1038/sj.ejhg.5200967. [DOI] [PubMed] [Google Scholar]
- 8.Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 9.Serikawa T, et al. National BioResource Project-Rat and related activities. Exp Anim. 2009;58:333–41. doi: 10.1538/expanim.58.333. [DOI] [PubMed] [Google Scholar]
- 10.Valdar W, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–87. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 11.Johannesson M, et al. A resource for the simultaneous high-resolution mapping of multiple quantitative trait loci in rats: the NIH heterogeneous stock. Genome Res. 2009;19:150–8. doi: 10.1101/gr.081497.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–23. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdar W, Holmes CC, Mott R, Flint J. Mapping in structured populations by resample model averaging. Genetics. 2009;182:1263–77. doi: 10.1534/genetics.109.100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A. 2000;97:12649–54. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Israely I, et al. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol. 2004;14:1657–63. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 16.Berkel S, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–91. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 17.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirihai OS, Gregory T, Yu C, Orkin SH, Weiss MJ. ABC-me: a novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 2000;19:2492–502. doi: 10.1093/emboj/19.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyde BB, et al. The mitochondrial transporter ABC-me (ABCB10), a downstream target of GATA-1, is essential for erythropoiesis in vivo. Cell Death Differ. 2012;19:1117–26. doi: 10.1038/cdd.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallström EOT. Rat Models of Experimental Autoimmune Encephalomyelitis. In: Conn PJ, editor. Sourcebook of Models for biomedical research. Humana Press Inc; Otowa: 2007. [Google Scholar]
- 21.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang GJ, et al. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res. 2009;19:1133–40. doi: 10.1101/gr.088120.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagodic M, et al. A role for VAV1 in experimental autoimmune encephalomyelitis and multiple sclerosis. Sci Transl Med. 2009;1:10ra21. doi: 10.1126/scitranslmed.3000278. [DOI] [PubMed] [Google Scholar]
- 26.Swanberg M, et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat Genet. 2005;37:486–94. doi: 10.1038/ng1544. [DOI] [PubMed] [Google Scholar]
- 27.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner M, Laragione T, Yarlett NC, Gulko PS. Genetic regulation of T regulatory, CD4, and CD8 cell numbers by the arthritis severity loci Cia5a, Cia5d, and the MHC/Cia1 in the rat. Mol Med. 2007;13:277–87. doi: 10.2119/2007-00003.Brenner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang S, Collins PL, Aune TM. T-bet dependent removal of Sin3A-histone deacetylase complexes at the Ifng locus drives Th1 differentiation. J Immunol. 2008;181:8372–81. doi: 10.4049/jimmunol.181.12.8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowley SM, et al. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liesa M, et al. Mitochondrial transporter ATP binding cassette mitochondrial erythroid is a novel gene required for cardiac recovery after ischemia/reperfusion. Circulation. 2011;124:806–13. doi: 10.1161/CIRCULATIONAHA.110.003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–31. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira MA, et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet. 2010;86:88–92. doi: 10.1016/j.ajhg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yalcin B, et al. Commercially available outbred mice for genome-wide association studies. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiddy S, Cattermole D, Xie D, Duan XY, Mott R. An integrated system for genetic analysis. BMC Bioinformatics. 2006;7:210. doi: 10.1186/1471-2105-7-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–8. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R-Development-Core-Team . A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2004. [Google Scholar]
- 45.Magrane M, UniProt Consortium UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–59. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 48.Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database (Oxford) 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.