Summary
Two-component signal transduction systems typically involve a sensor histidine kinase that specifically phosphorylates a single, cognate response regulator. This protein-protein interaction relies on molecular recognition via a small set of residues in each protein. To better understand how these residues determine the specificity of kinase-substrate interactions, we rationally rewired the interaction interface of a Thermotoga maritima two-component system, HK853-RR468, to match that found in a different two-component system, E. coli PhoR-PhoB. The rewired proteins interacted robustly with each other, but no longer interacted with the parent proteins. Analysis of the crystal structures of the wild-type and mutant protein complexes, along with a systematic mutagenesis study, reveals how individual mutations contribute to the rewiring of interaction specificity. Our approach and conclusions have implications for studies of other protein-protein interactions, protein evolution, and the design of novel protein interfaces.
Introduction
Interacting protein partners must recognize each other while avoiding unproductive interactions within the crowded milieu of the cell. The residues important for a given protein-protein interface must therefore both promote interaction between cognate proteins and prevent, or at least minimize, all possible non-cognate pairings. The challenge of maintaining specificity is particularly acute for proteins that belong to large paralogous protein families, which often share significant similarity to one another at the sequence and structural levels (Gao and Stock, 2009; Keskin et al., 2008).
In bacteria, two-component signal transduction proteins are a prevalent mechanism for sensing and responding to the environment. These signaling pathways rely on a sensor histidine kinase that can autophosphorylate and transfer its phosphoryl group to a cognate response regulator (Stock et al., 2000). Many histidine kinases are bi-functional and can directly dephosphorylate their cognate response regulators (Huynh and Stewart, 2011; Igo et al., 1989). Histidine kinases and response regulators are two of the largest protein families in bacteria, with most organisms encoding tens to hundreds of each type of protein (Alm et al., 2006; Galperin, 2005). However, most histidine kinases phosphorylate only a single cognate response regulator and there is very little cross-talk observed between non-cognate partners (Capra et al., 2012; Laub and Goulian, 2007). Systematic studies of phosphotransfer have demonstrated that histidine kinases typically exhibit a strong kinetic preference for their cognate response regulators in vitro, suggesting that the interaction specificity of these signaling pathways is driven largely by molecular recognition rather than the cellular context (Fisher et al., 1996; Skerker et al., 2005).
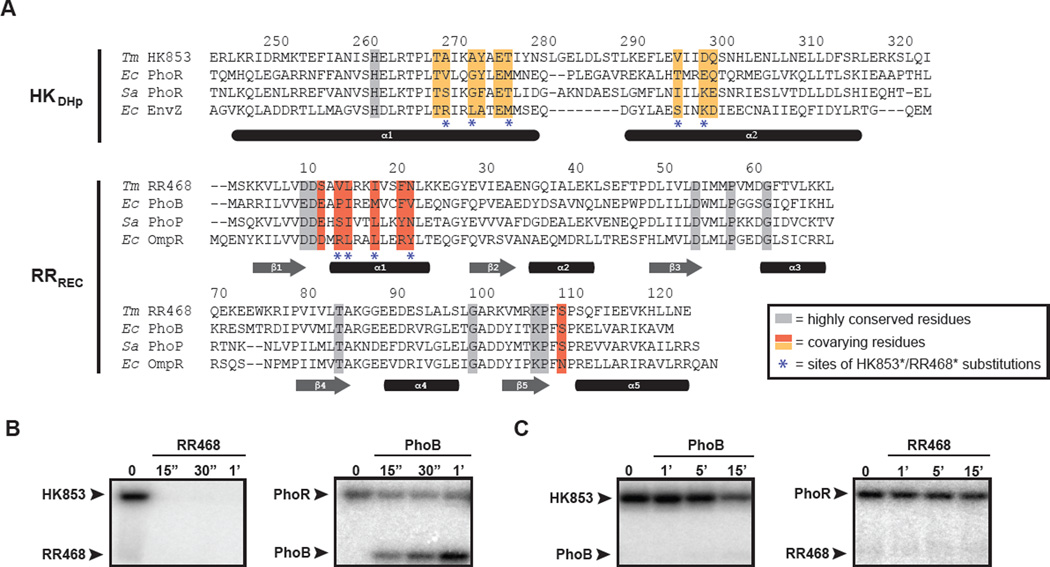
Previous studies have demonstrated that interaction specificity is dictated by a small subset of residues on each protein (Capra et al., 2010; Skerker et al., 2008). These studies relied on the identification of coevolving amino acids in large multiple sequence alignments of cognate kinase-regulator pairs from a diverse range of bacterial species (Codoner and Fares, 2008). The importance of these residues was validated through the rational rewiring of phosphotransfer specificity. Substituting the specificity residues in the E. coli histidine kinase EnvZ with those found in other kinases was sufficient to drive phosphotransfer toward previously non-cognate response regulators. A similar rewiring of the response regulator OmpR allowed it to receive phosphoryl groups from other histidine kinases. These coevolving specificity residues were confirmed as critical to molecular recognition when the first structure of a histidine kinase in complex with its cognate response regulator was solved (Casino et al., 2009; Casino et al., 2010). The complex of Thermotoga maritima kinase HK853 bound to a phosphorylated form of RR468 demonstrated that the primary basis of interaction involves the docking of helix α1 (α1) in the response regulator with both helices of the DHp (Dimerization and Histidine phosphotransfer) domain in the kinase. Nearly all of the specificity residues identified via coevolution studies are found within these helices (Figure 1A).
Figure 1. Specificity residues in two-component signaling proteins.
(A) Multiple sequence alignment of histidine kinases (DHp domain only) and response regulator (receiver domain only), with specificity residues and highly conserved residues highlighted. Species abbreviations: (Ec) Escherichia coli; (Sa) Staphylococcus aureus; (Tm) Thermotoga maritima. Interacting partners are arranged in the same order in both HK and RR alignments. Sequences are numbered according to the Tm proteins, with the last digit of each number positioned above the relevant amino acid residue. (B) HK853 and (C) PhoR phosphotransfer specificity. Each histidine kinase construct was autophosphorylated with [32P-γ]ATP and then incubated with the response regulator indicated at room temperature. Samples were taken at the time points indicated and phosphotransfer assessed by SDS-PAGE and phosphorimaging. Arrowheads indicate the position of autophosphorylated kinase or phosphorylated response regulator. Also see Figure S1.
Although two-component proteins have been successfully rewired, it remains unclear how a newly introduced set of specificity residues is accommodated at the molecular interface formed by a histidine kinase and a response regulator. How do individual residues contribute to the rewired specificity of a complex? How do the new residues pack together? Do changes at the interface affect other, distal regions of the proteins? To tackle these questions, we rationally rewired the interaction interface of Thermotoga maritima proteins HK853 and RR468 to harbor the specificity-determining residues of an unrelated two-component pathway, E. coli PhoR and PhoB. We solved crystal structures of complexes formed by the rewired proteins, as well as the structures of the rewired HK853 and RR468 alone. Comparison of these structures with the native HK853-RR468 complex, along with a systematic mutational analysis of the interface, helps reveal the structural basis of specificity in two-component signaling proteins. More generally, they provide insight into the rules of molecular recognition and coevolution in protein-protein interfaces.
Results
HK853-RR468 and PhoR-PhoB have different phosphotransfer specificities
To investigate the structural consequences of rewiring a kinase-substrate interface, we rationally mutated the specificity residues of the T. maritima two-component pathway HK853-RR468 to match those of another two-component system. Previous work has shown that unlike the model kinase EnvZ, the HK853 homodimer autophosphorylates in cis, such that the histidine on a given chain is autophosphorylated by the ATP-binding domain of the same chain (Casino et al., 2009). We therefore aimed to reprogram the specificity of the HK853-RR468 system to match that of another system in which the kinase autophosphorylates in cis, the Escherichia coli system PhoR-PhoB (Ashenberg et al., 2013; Casino et al., 2009). HK853 and E. coli PhoR are ~32% identical at the amino acid level across their DHp and CA (Catalytic and ATP binding) domains and share four identities at the nine specificity positions. RR468 and E. coli PhoB are ~38% identical across their receiver domains and share two identities at the seven specificity positions (Figure 1A, Figure S1A).
To confirm that the different specificity residues in HK853-RR468 and PhoR-PhoB yield different phosphotransfer specificities, we purified His6-tagged versions of each protein. For HK853 and PhoR, we truncated the transmembrane domains, purifying only the soluble, cytoplasmic portions of each kinase (see Methods). RR468 has only a receiver domain, whereas PhoB has a receiver domain and a DNA-binding domain; we purified the receiver domain portion of each regulator. We first autophosphorylated each kinase in the presence of [γ-32P]ATP and then added cognate substrates to examine phosphotransfer. At room temperature, HK853 rapidly phosphorylated RR468 (Figure 1B). Because HK853 is bi-functional with strong phosphatase activity for RR468~P (Casino et al., 2009), the combination of phosphotransfer and subsequent dephosphorylation of RR468 led to a loss of radiolabeled HK853 and RR468 within 15 seconds (Figure 1B). The rapid disappearance of phosphorylated HK853 resulted from phosphotransfer to the regulator, not simply dephosphorylation of HK853 through hydrolysis. When HK853 was incubated with RR468(D53A), which cannot be phosphorylated, the kinase remained phosphorylated for extended periods of time (Casino et al., 2009). Like HK853, the histidine kinase PhoR rapidly phosphorylated its cognate partner, PhoB (Figure 1B). As the PhoR construct harbors only modest phosphatase activity for PhoB~P, continuous phosphotransfer resulted in the accumulation of phosphorylated PhoB over a 1-minute time course. Finally, we examined phosphotransfer from HK853 and PhoR to the non-cognate regulators PhoB and RR468, respectively. Neither HK853 nor PhoR phosphorylated the non-cognate substrate (Figure 1C, Figure S1B). The modest decrease in intensity of the HK853 band likely results from dephosphorylation, not phosphotransfer (Fig. S2B). These experiments demonstrate that HK853-RR468 and PhoR-PhoB have different phosphotransfer specificities, consistent with their different specificity residues.
Rewiring the specificity of HK853-RR468 to match that of PhoR-PhoB
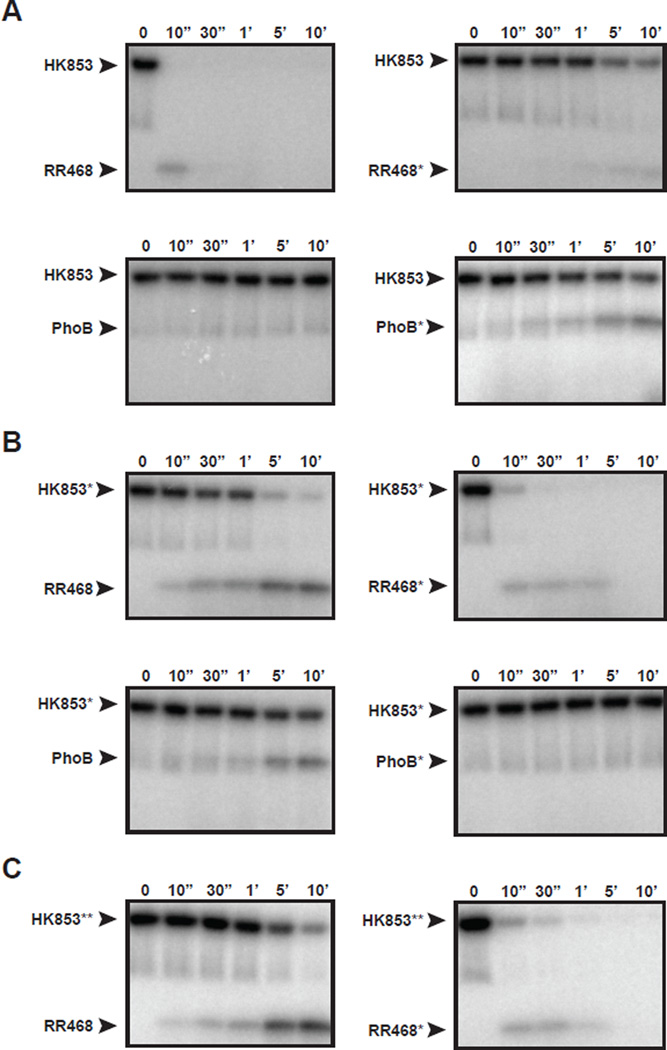
To rewire HK853-RR468, we substituted the specificity residues of HK853 and RR468 with those found in PhoR and PhoB, respectively, producing HK853* (A268V, A271G, T275M, V294T, D297E) and RR468* (V13P, L14I, I17M, N21V) (Figure 1A). We also substituted the specificity residues of PhoB with those found in RR468, producing PhoB* (P13V, I14L, M17I, V21N). The substitutions introduced into HK853 did not significantly affect kinase autophosphorylation (Figure S2B). We then examined phosphotransfer from HK853 and HK853* to the response regulators RR468, RR468*, PhoB, and PhoB* (Figure 2, Figure S3A). Because phosphotransfer from HK853 to RR468 is so rapid at room temperature (Figure 1B), we performed these assays at 4°C to facilitate the comparison of relative phosphotransfer rates. As before, HK853 rapidly phosphorylated and dephosphorylated RR468, with a complete loss of radiolabel within 30 seconds (Figure 2A). HK853 phosphorylated RR468* at an extremely slow rate, indicating that the substitutions introduced into RR468* disrupted the cognate interaction. As expected, wild-type HK853 did not transfer to the non-cognate regulator PhoB under these reaction conditions, even after 10 minutes. However, HK853 was capable of phosphorylating PhoB*, indicating that the introduction of RR468-like specificity residues into PhoB was sufficient to promote phosphorylation by HK853 (Figure 2A).
Figure 2. Rational rewiring of phosphotransfer specificity.
Phosphotransfer assays for wild-type and mutant two-component proteins. In each panel, the histidine kinase indicated was autophosphorylated with [32P-γ]ATP and then incubated with the response regulator indicated at 4 °C. Samples were taken at the time points indicated and phosphotransfer assessed by SDS-PAGE and phosphorimaging. (A) Wild-type HK853 and (B) HK853* (which harbors the substitutions A268V, A271G, T275M, V294T, D297E) were tested for phosphotransfer to RR468, RR468*, PhoB, and PhoB*. RR468* contains the substitutions V13P, L14I, I17M, and N21V. PhoB* contains the substitutions P13V, I14L, M17I, and V21N. (C) HK853**, which harbors the substitutions A268V, A271G, and T275M was tested for phosphotransfer to RR468 and RR468*. Also see Figures S2–S3.
For HK853*, we observed significantly reduced rates of phosphotransfer to RR468, indicating that changes to the specificity residues of HK853 had diminished the cognate pairing of HK853 and RR468 (Figure 2B). Strikingly, however, HK853* rapidly phosphorylated and dephosphorylated RR468*, indicating that the substitutions in RR468* restored a robust interaction with HK853*. Consistent with its new specificity residues, we found that HK853* could phosphorylate PhoB, but not PhoB* (Figure 2B).
We confirmed that HK853* has phosphatase activity toward RR468*, but not RR468, by using [32P] acetyl-phosphate to radiolabel RR468*; subsequent addition of HK853* led to a rapid loss in radiolabel compared to buffer alone (Figure S2C). This result indicates that HK853* stimulates the dephosphorylation of RR468*.
A mutant of HK853, designated HK853**, harboring only three of the five specificity mutations (A268V, A271G, T275M) behaved similar to HK853*, suggesting that these three residues in the middle of α1 were sufficient to rewire interaction specificity (Figure 2C). Collectively, these findings demonstrate that introduction of PhoR-like and PhoB-like substitutions into HK853 and RR468 was sufficient to reprogram their phosphotransfer specificity while maintaining phosphotransfer and dephosphorylation rates comparable to those seen in the wild-type proteins.
Structural characterization of the rewired complex reveals changes in relative orientation between interacting proteins
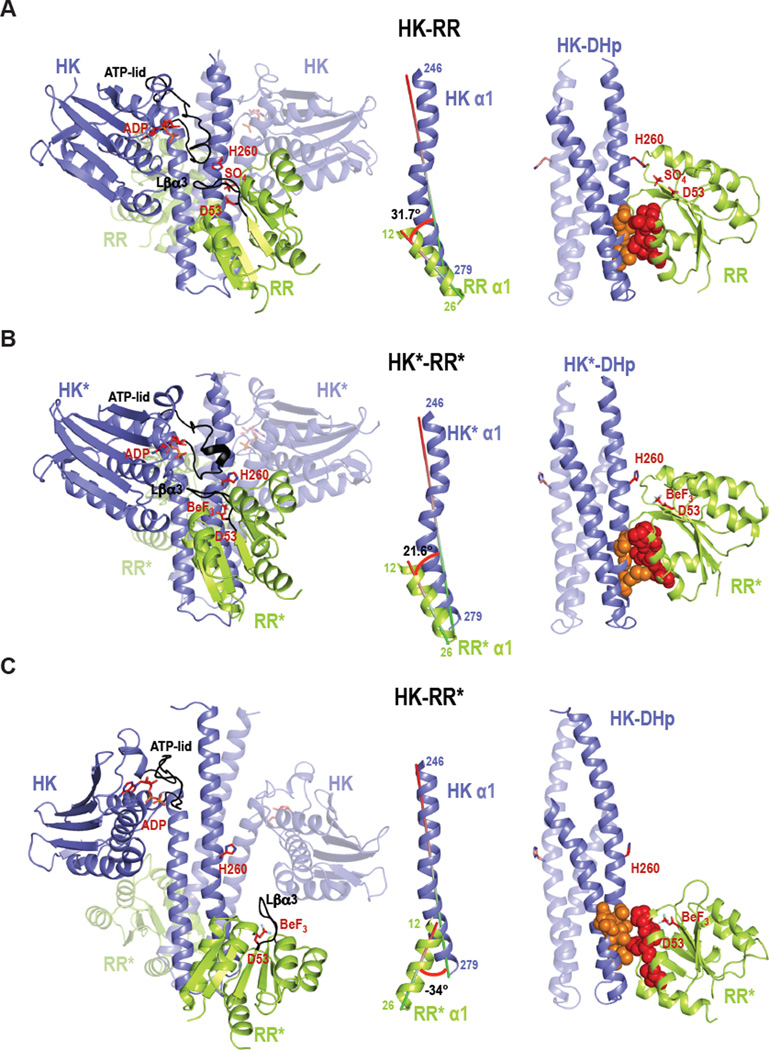
To investigate the effects of the new specificity residues on partner recognition, we solved an X-ray crystal structure of the rewired complex formed by HK853* and RR468* (HK*-RR*) (Table 1), which can engage in both phosphotransfer and phosphatase reactions (Figure 2B, Figure S2C). The rewired complex preserves the stoichiometry of the original complex (HK-RR; PDB:3DGE) (Casino et al., 2009): HK853* forms a homodimer that interacts with two molecules of RR468* (HK2-2RR) (Figure 3). In the rewired complex, the HK2-2RR is generated by crystallographic two-fold symmetry while in the HK-RR complex the asymmetric unit contained HK2-2RR (Table 1). Further, the structures of the individual components are highly similar. The kinases superimpose with an rmsd value of 1.82 Å (Table 2, Figure S4A). The superposition of individual domains shows even more similarity, with rmsd values of 1.0 Å and 0.87 Å for the DHp (residues 245–317) and CA (residues 320–480) domains, respectively. The slightly greater rmsd value for the kinases relative to individual domains results from a 19.7° rigid body rotation of the HK853* CA domain toward α1 of the DHp domain, which agrees with the structural plasticity and dynamic nature reported for the CA domain (Albanesi et al., 2009). RR468* has an almost identical structure to RR468 in the two functional complexes, HK-RR and HK*-RR*, with an rmsd of 0.6 Å (Table 3).
Table 1.
Crystallographic data and refinement statistics
| Processed data | HKf* | RRf* | HK*-RR* | HK-RR* |
|---|---|---|---|---|
| Wavelength (Å) | 0.92 | 0.87 | 0.98 | 0.87 |
| Resolution (Å) | 72.98-2.70 (2.85-2.70) | 35.74-1.80 (1.88-1.79) | 48.34-3.00 (3.16-3.00) | 46.32-3.10 (3.27-3.10) |
| Rmerge (%) | 0.057 (0.337) | 0.078 (0.279) | 0.060 (0.389) | 0.071 (0.401) |
| Rpim (%) | 0.023 (0.13) | 0.030 (0.107) | 0.036 (0.226) | 0.035 (0.198) |
| Mean I/δ(I) | 21.0 (5.6) | 19.7 (7.6) | 15.0 (3.7) | 17.9 (4.1) |
| N° reflections (observed/unique) | 60367/8277 (8550/1162) | 84433/10950 (11971/1566) | 43627/12140 (6406/1750) | 108863/22038 (15794/3160) |
| Completeness (%) | 99.5 (99.2) | 100.0 (100.0) | 98.3 (98.9) | 99.9 (100.0) |
| Redundancy | 7.3 (7.4) | 7.7 (7.6) | 3.6 (3.7) | 4.9 (5.0) |
| Space group | C2221 | I222 | I222 | C2221 |
| a=81.96 | a=53.98 | a=75.71 | a=119.32 | |
| Cell dimensions (Å) | b=160.38 | b=58.06 | b=85.31 | b=143.93 |
| c=43.89 | c=71.47 | c=185.59 | c=138.97 | |
| Refined data | ||||
| Rfactor (%) | 0.236 | 0.186 | 0.209 | 0.202 |
| Rfree (%) | 0.279 | 0.223 | 0.252 | 0.253 |
| Asymmetric unit composition | 1 HK | 1RR | 1HK:1RR | 2HK:2RR |
| N° protein atoms | 1890 | 976 | 2856 | 5842 |
| N° water molecules | 44 | 91 | 13 | 13 |
| N° ligand/ion | 1 | 5 | 4 | 14 |
| RMSD | ||||
| Bond deviation (Å) | 0.009 | 0.008 | 0.005 | 0.004 |
| Angle deviation (°) | 1.3 | 1.3 | 1.0 | 0.9 |
| Media B-factor (Å2) | ||||
| Main chain | 70.3 | 11.4 | 72.7 | 71.3 |
| Side chain | 71.2 | 13.5 | 75.2 | 73.2 |
| All atoms | 70.7 | 12.4 | 73.9 | 72.2 |
| Ramachandran Map (%) | ||||
| Favoured | 96.88 | 99.15 | 94.89 | 96.81 |
| Allowed | 3.12 | 0.85 | 4.83 | 3.06 |
| Disallowed region | 0 | 0 | 0.28 | 0.14 |
| PDB accession code | 4JAU | 4JA2 | 4JAS | 4JAV |
Values in parentheses correspond to data for the highest resolution shell
Rmerge = ΣhklΣi | I(hkl)i - <I(hkl)> | / ΣhklΣi<I(hkl)i>
Rpim = Σhkl√(1/(n-1)) Σi | I(hkl)i - <I(hkl)> | /ΣhklΣi I(hkl)i
Rfactor= Σ∥Fo|−|Fc∥/Σ|Fo|
Rfree is the Rfactor calculated with 5 to 7% of the total unique reflections chosen randomly and omitted from refinement.
Figure 3. Crystal structures of the wild-type complex HK-RR, the rewired and functional HK*-RR* complex, and the impaired HK-RR* complex.
Cartoon representation of (A) HK-RR complex with HK853 bound to ADP and RR468 D53 bound to SO4, (B) HK*-RR* complex with HK853* bound to ADP and RR468* D53 bound to BeF3− and (C) HK-RR* complex with HK853 bound to ADP and RR468* D53 bound to BeF3−. Left; cartoon representations of the overall structure of the three complexes formed by a homodimeric HK (blue colored with one subunit transparent) bound to two molecules of RR (yellow-green colored with one molecule transparent). In each complex, the ATP-lid in the HK and the β3-α3 linker in the RR are colored in black; the phosphorylatable residues H260 and D53 as well as bound ligands ADP, sulfate (SO4) and beryllium trifluoride (BeF3) are shown as sticks. Middle; the angle formed between the interacting helices HK α1 (246–279) and RR α1 (12–26) for each complex is shown. Right; HK-RR interface shown by the DHp domain (with one subunit transparent) bound to one RR with the critical specificity residues (13, 14, 17 and 21 in red for RR468 and 268, 271, 275, 294 and 297 in orange for HK853) highlighted in space-filling spheres. Also see Figures S4–S6.
Table 2.
RMSD differences in the HK component (residues 245–480)
| Rmsd (Å) | HK-RR (3DGE)1 |
HK*-RR* | HK-RR* | HKf (2C2A)1 |
HKf* |
|---|---|---|---|---|---|
| HK-RR | - | 1.82 | 5.6 | 5.2 | 4.95 |
| HK*-RR* | - | - | 4.8 | 4.35 | 4.1 |
| HK-RR* | - | - | - | 2.16 | 1.92 |
| HKf | - | - | - | - | 0.89 |
PDB codes for the previously published structures.
Table 3.
RMSD differences in the RR component (residues 2–121)
| Rmsd (Å) | HK-RR | HK*-RR* | HK-RR* | RRf* |
|---|---|---|---|---|
| HK-RR | - | 0.61 | 0.69 | 0.67 |
| HK*-RR* | - | - | 0.6 | 0.54 |
| HK-RR* | - | - | - | 0.47 |
Despite high similarity at the domain level, the arrangement and relative orientation of the kinase with respect to the regulator differs in the rewired complex. In particular, RR468* is rotated 17.5° and translated 0.5 Å along the DHp domain, such that the angle between α1 of HK853* (residues 246–279) and α1 of RR468* (residues 12–26), the two key structural elements for complex formation, is 21.6° (Figure 3B). In the HK-RR complex this same angle is 31.7° (Figure 3B). Because of this rotation, the HK853*-RR468* complex has a more parallel orientation between α1 in the regulator and the kinase helical bundle. In this orientation RR468* loses some interactions with the DHp α-helix 2 and increases interactions with the CA domain. In particular, the β2-α2 and β3-α3 loops of RR468* now interact directly with the α3-β1 loop and the ATP-lid of the HK853* CA domain. These new contacts lead to a larger interaction surface area in the HK*-RR* complex relative to HK-RR (2690 Å2 vs 1866 Å2) (Figure S5 and Table S1). However, the new contacts specific to the HK*-RR* complex appear largely dispensable for phosphotransfer and dephosphorylation (see Supplemental Text). The differences in orientation are likely due to the mutations introduced and not to differences in crystal packing since RR468* in the HK*-RR* complex could acquire the disposition of RR468 in HK-RR without steric crystallographic clashes.
Although HK853 phosphorylates and dephosphorylates the mutant RR468* very slowly (Figure 2A, Figure S2C), we were able to solve a structure of these proteins in complex (HK-RR*). The asymmetric unit showed HK2-2RR stoichiometry with almost two-fold symmetry (Table 1), broken due to a slightly different relative disposition of the CA domain with respect to the DHp domain (~18.8°), confirming the previously mentioned plasticity of the CA domain (Figure S4C). Both HK subunits in the HK-RR* complex adopt a conformation that is more similar to the free form of HK853 (HKf; PDB:2C2A; rmsd = 2.2 Å) than to the kinase in the HK-RR complex (rmsd=5.6 Å) (Table 2, Figure S4). The structure of RR468* in the HK-RR* complex is similar to RR468* in the HK*-RR* complex (rmsd=0.6 Å, Table 3), but the RR468* molecule in HK-RR* adopts a totally different position relative to the kinase. In the HK-RR* complex, RR468* α1 is rotated ~55° and slightly displaced (~1.0 Å) relative to RR468* and RR468 in the rewired HK*-RR* and native HK-RR complexes, respectively (Figure 3C). Many of the intermolecular contacts seen in the productive HK-RR and HK*-RR* complexes are lost in the HK-RR* complex, consistent with its greatly diminished phosphotransfer rate (Table S1). Finally, the phosphorylatable residues in RR468* (Asp53) and HK853 (His260) are extremely far apart (19.0 Å) and improperly oriented for any catalytic reaction (Figure 3C).
Given the structural data for the functional complexes, HK-RR and HK*-RR*, we conclude that there is a permissible range of rotational motion of the kinase relative to the regulator that allows proper positioning of the active site while accommodating new interfacial residues.
The active center of the rewired complex is a snapshot of a new intermediate state in phosphotransfer
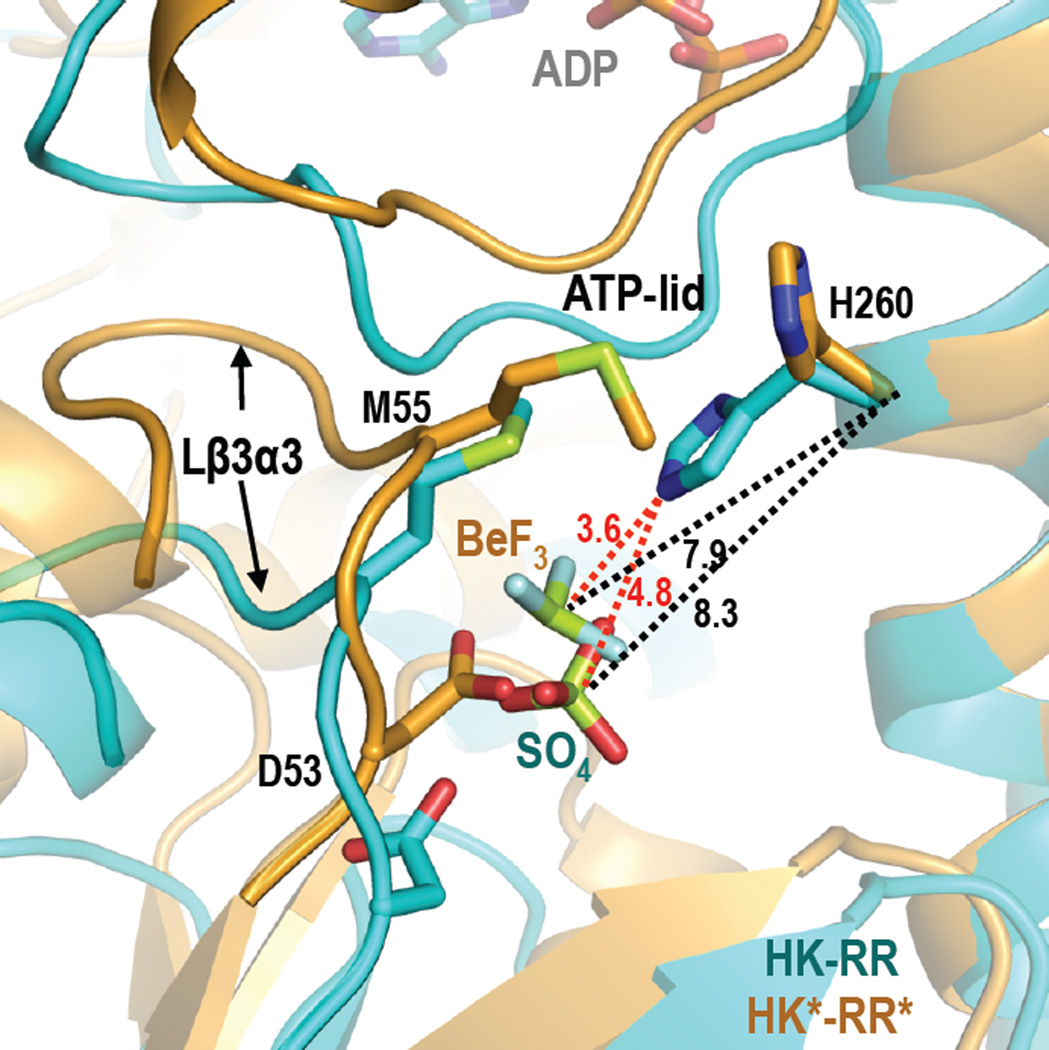
A closer view of the active sites in the HK*-RR* and HK-RR* complexes shows the phosphomimetic beryllium trifluoride (BeF3−) bound to the catalytic Asp53 of RR* in both cases. In HK*-RR* the BeF3− is placed similarly to the sulfate found in the active site of the HK-RR complex, but the Be atom is slightly closer to the phosphorylatable His260 (Be-His260 Cα=7.85 Å) than the sulphur atom of the sulfate in the HK-RR complex (S-His260 Cα=8.30 Å) (Figure 4). However, Met55 of RR468* is interposed between the His and the BeF3− in the HK*-RR* structure, forcing an alternative rotamer for His260 that points away from the active site. Thus, the structure of the HK*-RR* mutant complex may represent an earlier phase of the phosphatase reaction, when the phosphoryl group is still bound to the response regulator. Alternatively, if His260 acquired the rotamer conformation observed in the HK-RR complex, the distance between the phosphoacceptor nitrogen of this residue and the Be atom in the HK*-RR* complex would be 3.6 Å, a distance compatible with phosphotransfer (Figure 4). This observation suggests that the conformation captured in the crystal could, instead, correspond to the end of the phosphotransfer reaction just prior to complex dissociation. Because the phosphatase reaction is not the reverse of a phosphotransfer reaction (Hsing and Silhavy, 1997), the active center observed in the crystal could correspond to either of these reactions.
Figure 4. Comparison of the active center in the HK-RR and HK*-RR* complexes.
Close-up view of the active center with a superposition of HK-RR (in cyan) and HK*-RR* (in orange) in cartoon representation. The phosphorylatable residues H260 and D53, residue M55, and the bound ligands (sulfate (SO4) in the HK-RR complex and beryllium trifluoride (BeF3) in the HK*-RR* complex) are shown as sticks. Distances are shown by dashed lines; in black color for Cα of H260 in HK-RR with the sulfur atom of SO4 (8.3 Å) and for Cα of H260 in HK*-RR* with the Be atom of BeF3 (7.9 Å); in red color for εN of H260 in HK-RR with the sulfur atom of SO4 (4.8 Å) and with the Be atom of BeF3− (3.6 Å). Also see Figures S4–S6.
Introduction of new specificity residues does not affect global structural integrity
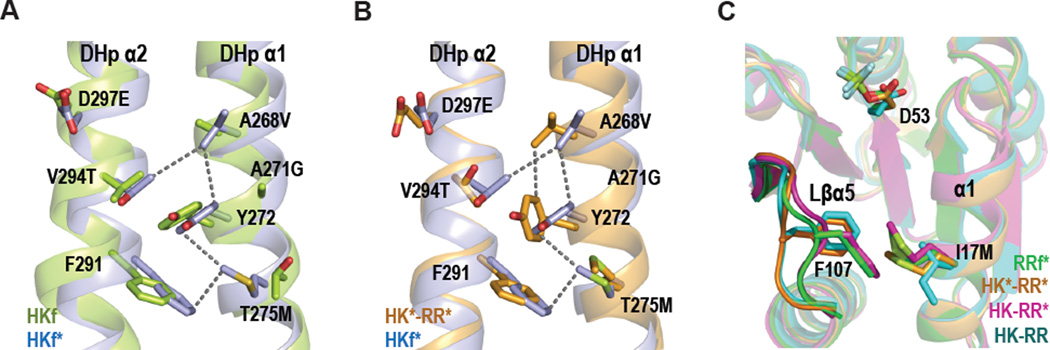
Structural differences between the different HK-RR complexes could be imposed by specific requirements for partner recognition, or could reflect intrinsic changes in the individual proteins resulting from the point mutations. To address this issue, we solved the structures of the free forms of HK853* (HKf*) bound to ADP and RR468* (RRf*) bound to BeF3− (Table 1). The conformations of HKf* and HKf are almost identical (rmsd = 0.9 Å for the superposition of the structures; Table 2, Figure S4B). However, there are local changes around the mutated residues that can be attributed to interactions between DHp α-helices 1 and 2 mediated by the new side chains. In HKf* V268 and M275, together with T294 and F291 from α2 and Y272 from α1, generate a hydrophobic network (Figure 5A). These interactions induce changes in the exposed recognition surface and could impair interaction with the regulator. The flexibility of the new Met side chain seems to play a key role in this network, since it is sandwiched between the Y272 and F291 aromatic rings (Figure 5A). The interaction of V268 and M275 with the complementary surface provided by the mutated residues of RR468* promotes the disappearance of this hydrophobic network in the productive complex (Figure 5B).
Figure 5. Differential interactions in free and complex structures.
(A) DHp domains with a superposition of HKf (in green) and HKf* (in blue) to show new interactions resulting from the mutations introduced into HKf*: A268V, A271G, T275M, V294T and D297E. (B) DHp domains with a superposition of HKf* and HK853* from the HK*-RR* complex (in orange) to show changes in the interactions for M275. (C) Superposition of RR468 in RRf* (green), HK-RR (cyan), HK*-RR* (orange) and HK-RR* (magenta) structures shows the interaction M17-F107 and the different conformations of F107 and the β5-α5 linker in the RR alone or in complex. All the structures are shown in cartoon representation with the selected residues labeled in black and drawn as sticks. Also see Figures S4–S6.
As with HK853*, comparison of RR468* in isolation and in the HK*-RR* complex shows minimal structural differences (Table 3) that are localized around the mutated residues. The I17M mutation seems to have the biggest structural impact on RR468* as the new Met side chain now mediates a hydrophobic interaction with F107 that was not present in RR468 (Figure 5C). This interaction involves a 2.4 Å displacement of F107 toward M17, a movement that brings the β5-α5 linker (Lβα5) closer to α1. In the HK-RR complex, these two structural elements of RR468 clamp α1 of the kinase's DHp domain. Although the M17-F107 interaction occurs in both RRf* and HK-RR*, F107 is positioned perpendicular to the M17 side chain in the HK*-RR* complex, suggesting that mutations in HK* may compensate for the presence of M17 (Figure 5C). The positioning of F107 in HK*-RR* is similar to that seen in HK-RR, suggesting this conformation may be important for the formation of a productive complex.
Systematic mutational characterization of the rewired interface
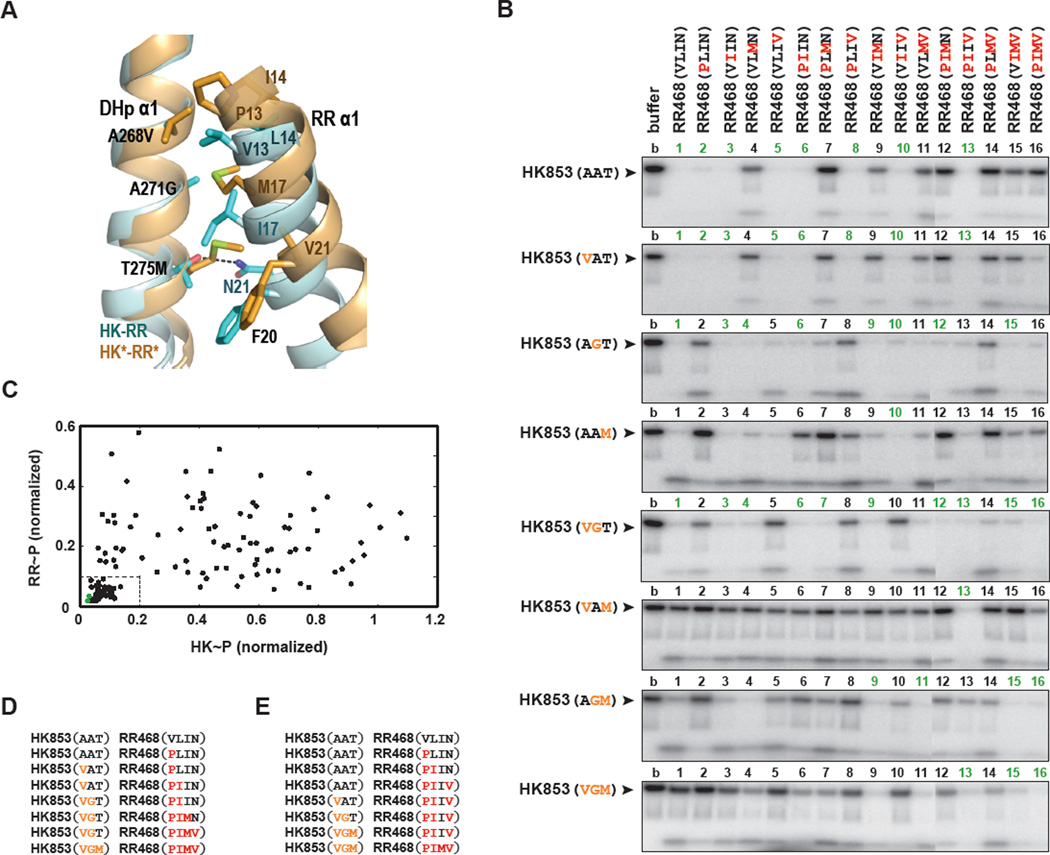
To further analyze the effects of individual substitutions at the HK853-RR468 interface (Figure 6A), we constructed all possible mutational intermediates separating the HK-RR and HK*-RR* pairings. Consistent with our previous finding that only three of the five mutations (A268V, A271G, T275M) in HK853 are necessary to rewire the interface, residues V294 and D297 do not make inter-protein contacts in the HK*-RR* structure (Table 2). We therefore constructed three single and three double mutants in HK853; along with the wild type and triple mutant, there were eight different residue combinations for the kinase. For RR468 we made four single, six double, and four triple mutants; along with the wild type and quadruple mutant (V13P, L14I, I17M, N21V), there was a total of 16 specificity residue combinations for the regulator. For simplicity, we refer to each of the mutants according to the identities of their specificity residues.
Figure 6. Phosphotransfer between all possible mutational intermediates separating HK-RR and HK*-RR*.
(A) Superposition of HK-RR (in cyan) and HK*-RR* (in orange) complexes highlighting how mutations in DHp α1 (A268V, A271G and T275M) affect interactions with mutations in RR α1 (V13P, L14I, I17M and N21V) and F20. A dashed line represents a polar interaction between N21 and T275 in the HK-RR complex. Also see Figures S4–S6. (B) Phosphotransfer assays for wild-type HK853 and HK853 harboring all possible combinations of one, two, or three PhoR-like specificity substitutions present in HK853** (A268V, A271G and T275M). Each lane represents the incubation of the indicated autophosphorylated kinase with the indicated response regulator for 15 seconds at room temperature. Reactions 1–11 and 12–16 were run on separate SDS-PAGE gels; the resulting phosphorimages were contrasted identically and stitched together. (C) The histidine kinase (HK) and response regulator (RR) bands from the phosphotransfer experiments in panel B were quantified and plotted. For each mutational pairing, the x-axis value indicates the intensity of the autophosphorylated HK band (HK~P) normalized to the intensity of the autophosphorylated kinase band and the y-axis value indicates the intensity of the phosphorylated response regulator band (RR~P). In each case, band intensities were normalized to the intensity of the autophosphorylated kinase incubated without response regulator (lane 1 of each gel in panel B). Green points indicate the pairs HK853-RR468 and HK853*-RR468*. The box in the lower left indicates pairings deemed functional; a low level of both the kinase and regulator bands reflects efficient phosphotransfer and dephosphorylation. The 43 functional pairings are underlined in panel B. (D) One example of a mutational path from the wild-type to the rewired complex in which each intermediate state is functional. (E) An example of a mutational path in which all mutations to the kinase occur in three successive steps.
We systematically tested all 128 pairwise combinations of the 8 kinases and 16 regulators for activity by incubating each autophosphorylated kinase with each regulator for 15 seconds (Figure 6B). Residue combinations that supported both phosphotransfer and dephosphorylation, such as the wild-type residues, led to a depletion of radiolabeled kinase with minimal accumulation of radiolabeled regulator. Combinations that supported only phosphotransfer led to an accumulation of phosphorylated regulator, and unproductive combinations retained phosphorylated kinase. Although the in vivo role of HK853/RR468 is unknown, studies with other two-component systems have demonstrated that both phosphotransfer and phosphatase activity are important for proper signal transduction (Huynh and Stewart, 2011). Therefore we identified functional kinase-regulator pairs for which (i) the intensity of the phosphorylated kinase band in the presence of regulator was less than 20% the intensity of the kinase band in the presence of buffer alone and (ii) the intensity of the phosphorylated regulator band was less than 10% the intensity of the autophosphorylated kinase band in the presence of buffer. Of the 128 combinations, 43 pairs satisfied these criteria (Figure 6B–C).
Our systematic mutagenesis shed light on the amino acid combinations permissible at this protein interface. For example, the profile of HK853 against the 16 regulator mutants demonstrates that robust phosphotransfer and dephosphorylation are retained for all regulators except those harboring the I17M mutation (Figure 6B). For most of these pairings, the defect resulting from an I17M substitution can be rescued by the substitution A271G in the kinase. Given the similarity in size and nature between isoleucine and methionine, it is difficult to rationalize the strong impact of this mutation. However, our structural data indicated that the interaction of M17 with F107 could compromise proper packing of RR468* with HK853 DHp α1, thereby precluding a productive interaction (Figure 5C). Similarly, substituting a methionine residue into the kinase disrupted phosphatase activity, as evidenced by the inability of HK853(AAM), HK853(VAM), and HK853(AGM) to dephosphorylate the majority of regulator mutants (Figure 6B). In the HK*-RR* structure, the side chain of M275 lies within the hydrophobic pocket generated by RR468* residues M17, F20 and V21 (Figure 6A). A comparison with HK-RR shows that the residues M275 and M17 would have clashed if the displacement of RR468* α1 had not generated the hydrophobic pocket in HK*-RR*. In the new interface, the polar bond between T275 and N21 is replaced by a hydrophobic interaction between M275 and M17. This arrangement is further facilitated by the A271G mutation in HK*, as the original alanine side chain would have clashed with M17 in RR*. Together, these findings illuminate the conformational restrictions imposed by individual residues that underlie amino acid coevolution, which in turn enabled the identification of specificity residues by statistical covariation analyses (Skerker et al., 2008).
Some combinations of residues yielded more promiscuous proteins. For instance, HK853(VGT) phosphorylated and dephosphorylated 10 of the 16 regulators, including both the wild-type regulator and the quadruple mutant (Figure 6B). By comparison, wild-type HK853 phosphorylates and dephosphorylates 8 of the regulators, including its partner RR468 but not the quadruple mutant. The flexibility introduced by glycine (A271G) may lead to productive interactions with many regulators. Conversely, some residue combinations interact with a very limited number of regulators. For instance, RR468(PLMV) interacted poorly with almost all kinase partners, and HK853(VAM) could phosphorylate and dephosphorylate only a single regulator.
Analysis of these mutational intermediates also demonstrated the interdependence of interface residues. The double mutant HK853(VAM) can phosphorylate all of the regulators to some extent, but its phosphatase activity is limited to RR468(PIIV) (Figure 6B). However, either the A268V or the T275M mutation in HK853 by itself allows the kinase to both phosphorylate and dephosphorylate eight or six of the regulators, respectively. As another example, the mutation A268V has little effect on the wild-type kinase and its phosphotransfer profile looks essentially identical to the starting protein (Figure 6B). However, when introduced in the context of HK853(AGT), this mutation affects interactions with several of the mutant regulators. Thus, the effect of individual substitutions on specificity is highly context-dependent and difficult to predict from the behavior of the individual mutations.
Mutational trajectories between the wild-type and mutant interfaces
Having a complete characterized set of mutational intermediates also shed light on the evolution of two-component signaling protein specificity following duplication and divergence. There are 5,040 possible mutational paths between two sequences that are 7 letters in length, assuming that only one amino acid is mutated at each step. Given our criteria for productive phosphotransfer and dephosphorylation (Figure 6C), we calculated that only 200 of these 5,040 possible paths, or 4%, retain functional kinase-regulator pairs along the entire path. Interestingly, two of the 43 kinase-substrate combinations that we deemed functional do not appear in any of the 200 mutational paths. For example, the pair HK853(AAM) and RR468(VILV) can be reached from a wild-type starting point, but all possible subsequent mutations produce a non-functional pair.
As noted above, the mutations T275M in the kinase and I17M in the regulator are often deleterious (Figure 6B). Consequently, these substitutions are often found along dead-end mutational paths. Of the 200 paths that maintain a productive interaction, 97 introduce T275M into the kinase as the final mutation, 64 paths introduce I17M into the regulator as the last mutation, and 29 paths end with the regulator mutation V13P. These patterns suggest that other mutations must be introduced first to effectively prime the interface for introduction of residues that restrict conformational freedom, such as methionine or proline. More generally, we infer that these residues contribute to specificity by eliminating interactions with non-cognate regulators, rather than promoting interaction with the cognate regulator. For instance, HK853(VGT) phosphorylates and dephosphorylates 10 of the 16 regulators; subsequent introduction of the T275M mutation eliminates interaction with 7 of these 10 regulators, but does not substantially improve interaction with the fully rewired, quadruple RR468 mutant (Figure 6B).
Although the majority of paths connecting the HK-RR and HK*-RR* pairs that maintain a functional interaction involve alternating mutations in the two proteins (Figure 6D), there are paths in which either the kinase or the regulator is completely changed through successive mutations while the other protein remains fixed. For instance, if the first three mutations produce RR468(PIIV) in any order, HK853 can then tolerate three successive mutations A268V followed by A271G/T275M in either order (Figure 6E). There are a total of 12 mutational paths in which the kinase accumulates all of its mutations in succession. Similarly, if the first two mutations in a path yield the double mutant HK853(VGT), then the regulator can accumulate four successive mutations, in seven different orders; there are 14 such paths, in which the regulator accumulates all mutations in succession. In both of these examples, the protein that remains fixed while the other protein accumulates substitutions is highly tolerant of mutations in its partner: RR468(PIIV) fully interacts with 5 out of 8 mutant kinases and HK853(VGT) fully interacts with 10 out of the 16 mutant regulators (Figure 6B). Such tolerant intermediates may play important roles in the rewiring of two-component signaling interfaces that occurs during evolutionary processes such as duplication and divergence.
Discussion
Despite the importance of protein-protein interaction specificity to the operation of cells, it remains relatively unclear how proteins use a finite set of amino acids to specifically recognize cognate partners. We addressed this question using two-component signaling proteins, which utilize a limited and known set of amino acids for partner recognition. The identification of these residues has guided the rational rewiring of two-component signaling pathways (Bell et al., 2010; Capra et al., 2010; Skerker et al., 2008), but a structural understanding of how rewiring is achieved was lacking. Here, we reprogrammed the structurally characterized complex T. maritima HK853-RR468 by introducing nine specificity residues from E. coli PhoR-PhoB. Although highly specific, phosphotransfer and dephoshorylation rates of PhoB-PhoR are slower than those of HK853-RR468. Strikingly, the introduction of PhoR-PhoB specificity residues did not impair the rapid reaction rates of HK853-RR468, despite the change in interaction specificity. This finding supports the notion that the specificity residues are required for recognition and proper positioning of the two partners, but other residues set the rates of the reactions (Pazy et al., 2009; Zapf et al., 1998).
The structure of the rewired complex HK*-RR* demonstrates that these functionally rewired two-component proteins preserve the overall structure of the wild-type complex along with catalytic activity. The kinase domains can reposition themselves slightly relative to each other and relative to the regulator to accommodate the foreign interfacial residues, but the overall complex retains wild-type character. Thus, rewiring leads primarily to local spatially restricted changes in the regions of each protein directly engaged in molecular docking.
Although the HK-RR and HK*-RR* complexes are similar, the HK-RR* complex harbors a completely different intermolecular orientation such that the phosphorylatable residues are no longer in close proximity. We do not yet have a structure of the HK*-RR complex, which can participate in slow phosphotransfer but not dephosphorylation. Because HK853* is competent as a phosphatase for other partners, the lack of activity with respect to RR is probably due to a mismatch at the interaction interface, which could force the regulator to dock in the wrong orientation. Alternatively, the regulator may bind at the correct position but too weakly to support rapid phosphotransfer and dephosphorylation.
How do individual residues contribute to specificity? In some cases, disruptive mutations that change the size or nature of a residue can be restored by balancing mutations at neighboring inter- or intra-molecular positions. This sort of intermolecular compensation is consistent with the extensive amino acid coevolution previously documented for two-component signaling proteins (Skerker et al., 2008). However, it is difficult to ascribe a role to individual specificity residues, as the effect of a given substitution can be highly context-dependent. In general, our systematic mutagenesis study indicated that individual residues do not typically contribute equally or additively to specificity. This interdependence of specificity residues resonates with other recent studies suggesting that amino acid epistasis in proteins is extensive and common (Breen et al., 2012; Levin et al., 2009; Ortlund et al., 2007).
Our work also has implications for understanding the evolutionary processes of duplication and divergence, which underlie the massive expansion of two-component signaling protein families in bacteria (Capra and Laub, 2012; Capra et al., 2012). Post duplication, paralogous signaling proteins must become insulated with respect to phosphotransfer while retaining an interaction with their cognate partners. Examples of the mutational trajectories that proteins follow, and any constraints they face, are largely unknown. Although T. maritima HK853-RR468 and E. coli PhoR-PhoB are not closely related, our systematic analysis of mutational trajectories between them provides insights into how specificity evolves. Our finding that only 4% of the theoretically possible evolutionary paths retain a functional interaction suggests that trajectories through sequence space may be severely constrained. Previous work on mutational trajectories that convert β-lactamase from a drug-sensitive to drug-resistant state also found strong constraints with only 8 of 120, or 6%, of paths permissible (Weinreich et al., 2006).
None of the mutational combinations we tested were fully insulated from both the starting HK-RR and the final HK*-RR* complex, i.e. none forms an orthogonal interacting pair. This finding highlights the potential evolutionary importance of promiscuous states of the binding interface (Aharoni et al., 2005; Matsumura and Ellington, 2001). We speculate that ancestral two-component proteins may have harbored such promiscuity; following duplication and divergence the paralogs could have gained specificity simply through the accumulation of mutations that disrupt a subset of interactions seen in the ancestral state. This process of sub-functionalization represents a rapid route to specificity and could help explain the apparent ease with which paralogous protein families have expanded.
Our results also have implications for protein design efforts. As noted, there are dependencies between neighboring residues on one molecule, and between residues on two different protein partners. Thus, the HK853-RR468 interface can tolerate certain substitutions only in combination – an effect not easily predicted based on a consideration of how the individual substitutions behave. Consequently, efforts to design or engineer novel protein-protein interfaces will have to tackle this combinatorial problem. Our findings also underline the importance of subtle backbone flexibility in protein design (Humphris and Kortemme, 2008; Smith and Kortemme, 2008), as a static complex would not properly accommodate the introduction of a new set of specificity residues. In sum, our studies have provided important new insights into the molecular and structural basis of two-component signaling specificity, but also highlight the significant challenges that remain in computationally predicting (Chen and Keating, 2012) the effects of mutations and in designing interfaces de novo.
Experimental Procedures
Cloning, mutagenesis, and protein purification
All site-directed mutagenesis (see Table S2 for primers) was done with Gateway (Invitrogen) cloning vectors as described previously (Skerker et al., 2008). Mutagenized and sequence-verified protein sequences were moved from pENTR vectors into pDEST vectors using the Gateway LR reaction (pDEST-His6-MBP for HK853 and PhoR; pDEST-His6-TRX for PhoB; pDEST-His6 for RR468). For crystallization assays, the complete cytoplasmic portion of HK853* (232–489) was recloned from pDEST-His6-MBP into pET24b and full-length RR468* (1–122) was recloned from pDEST-His6 into pET22b, using in both cases, the In-Fusion HD cloning technology (Clontech) (Table S3). Expression and purification was carried out as described previously (Casino et al., 2009; Skerker et al., 2005).
Phosphotransfer assays
Autophosphorylation and phosphotransfer assays were performed as described previously (Capra et al., 2010). Histidine kinases diluted to 5 µM in HKEDG buffer (10 mM HEPES-KOH pH 8.0, 50 mM KCl, 10% glycerol, 0.1 mM EDTA, 2 mM DTT) supplemented with 5 mM MgCl2 were autophosphorylated with 500 µM ATP and 0.5 µCi [γ32P]-ATP (from a stock at ~6000 Ci/mmol, Perkin Elmer). PhoR was autophosphorylated for 1 hour at 30°C; HK853 and all HK853 mutants were autophosphorylated for 20 minutes at room temperature (see Figure S2B for autophosphorylation time course). The autophosphorylated kinase mixture was added directly to response regulator (at 5 µM in HKEDG buffer supplemented with 5 mM MgCl2). Reactions were quenched with 4× loading buffer (500 mM Tris-HCl pH 6.8, 8% SDS, 40% glycerol, 400 mM β-mercaptoethanol) and analyzed by SDS-PAGE and phosphorimaging. For reactions carried out at 4°C, the autophosphorylated histidine kinase was incubated at 4°C for 5 minutes prior to addition of chilled response regulator. Radiolabeled bands were quantified with ImageJ software.
Phosphatase assays
To perform the phosphatase assays in Figure S2C, [32P] acetyl-phosphate was freshly synthesized as described previously (Jagadeesan et al., 2009). RR468 (at 10 µM in HKEDG buffer supplemented with 5 mM MgCl2) was mixed 1:1 with [32P] acetyl-phosphate and incubated for 1 hour at room temperature. The mixture was washed three times with cold HKEDG buffer and the concentration of MgCl2 subsequently adjusted to 5 mM. The phosphorylated regulator was chilled at 4°C for 5 minutes and then incubated with buffer or with an equimolar amount of kinase (both pre-chilled) for the times indicated. Because RR468* autophosphorylates more poorly than RR468 using this method, both regulator and kinase concentrations were doubled to 20 µM starting concentration to measure RR468*~P dephosphorylation.
Crystallization, data collection, and model building
Crystallization of HKf* and RRf* proteins, HK*-RR* and HK-RR* complexes was achieved by the vapor diffusion method, using the sitting drop technique, mixing 0.6 µL of protein and 0.6 µL of reservoir solution. Crystals of HKf* were obtained in 8% PEG4000, 0.8 M LiCl and Tris pH 8.5 by mixing 10 mg/mL of protein, 4 mM ADP and 4 mM MgCl2. Crystals of RRf* in complex with BeF3− were obtained in 50% PEG400, NaAc pH 4.6 and 0.2 M Li2(SO4) mixing 15 mg/mL of protein, 30 mM NaF, 5 mM BeSO4 and 7 mM MgCl2. Crystals of the complexes HK*-RR* and HK-RR* were obtained in 2.2 M (NH4)2SO4 and Bis-Tris pH 5.5 by cocrystallization mixing 10 mg/mL of HK853* or HK853, 7.5 mg/mL of RR468*, 4 mM ADP, 30 mM NaF, 5 mM BeSO4 and 7 mM MgCl2. Crystals of HKf* were cryoprotected by increasing PEG4000 to 16% and by addition of 20% sucrose while crystals of the complexes were cryoprotected by addition of 35% sucrose. X-ray diffraction data was collected at Diamond Light Source I04-1 (Oxfordshire, UK) for HKf*, at European Synchrotron Facility ID23-2 (ESRF, Grenoble, France) for RRf*, and at ESRF ID23-1 and ID23-2 for the complexes HK*-RR* and HK-RR*, respectively. Data reduction was performed using XDS, Pointless and Scala to a Bragg space of 2.7 Å for HKf*, 1.8 Å for RR*, 3.0 Å for HK*-RR* complex and 3.1 Å for HK-RR* complex. Phases were obtained by molecular replacement using Phaser and the final models were obtained by subsequent cycles of refinement with Refmac5 and model building with the program Coot (Emsley and Cowtan, 2004). Despite the limited resolution for the complexes, the quality of the maps (Figure S6) allowed model building and unambiguous side chain assignments except for the first and last residues in HK* (232 to 243; 481 to 490) and HK (chain A 232 to 233; 480 to 490 and chain B 232 to 235; 480 to 490) where electronic density was absent, which reflects the elevated flexibility of these regions. Crystallographic data and refinement statistics are in Table 1. The programs Pointless, Scala, Phaser and Refmac are contained in CCP4 Suite. Figures 3–5, and Figures S4–S6 were produced using PyMOL (http://www.pymol.org). Superimpositions were carried out with Superpose from CCP4 Suite. Movement analysis was performed using the program Dyndom (Lee et al., 2003). In the HK-RR and HK-RR* complexes, where the asymmetric unit is formed by HK2-2RR, HK chain A and RR chain C were chosen for the structural comparisons. The 3D structures are deposited with the following PDB accession codes: 4JAU for HKf*, 4JA2 for RRf*, 4JAS for HK*-RR* and 4JAV for HK-RR*. PDB accession codes for additional structures analyzed in the manuscript were 2C2A for HKf and 3DGE for HK-RR.
Supplementary Material
Highlights.
-
-
T. maritima HK853-RR468 was rewired to harbor alternative specificity residues
-
-
the rewired proteins dock similarly to the parent proteins, but with a slight rotation
-
-
the rewired interface is repacked to promote binding and catalytic functions
-
-
systematic analyses show limited mutational trajectories for converting specificity
Acknowledgements
We thank A. Keating and members of the Laub laboratory for helpful comments on the manuscript. We acknowledge the European Synchrotron Radiation Facility (ESRF) and Diamond Light Source (DLS) for provision of synchrotron radiation facilities and we would like to thank ID23-1, ID23-2 (ESRF) and I04-1 (DLS) beamlines staff for assistance during data collection. M.T.L is an Early Career Scientist at the Howard Hughes Medical Institute. This work was supported by an NSF CAREER award (MCB-0844442) to M.T.L.; grants BIO2010-15424 from the Ministerio de Ciencia e Innovación and ACOMP2012/044 from Generalitat Valenciana to A.M.; and NSF Graduate Research Fellowship to A.P. P.C. is recipient of a postdoctoral junior grant “Juan de la Cierva” from the Ministry of Economy and Competitiveness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aharoni A, Gaidukov L, Khersonsky O, McQ Gould S, Roodveldt C, Tawfik DS. The 'evolvability' of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Albanesi D, Martín M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci U S A. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenberg O, Keating AE, Laub MT. Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CH, Porter SL, Strawson A, Stuart DI, Armitage JP. Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex. PLoS Biol. 2010;8:e1000306. doi: 10.1371/journal.pbio.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. Epistasis as the primary factor in molecular evolution. Nature. 2012;490:535–538. doi: 10.1038/nature11510. [DOI] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Perchuk BS, Lubin EA, Ashenberg O, Skerker JM, Laub MT. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet. 2010;6:e1001220. doi: 10.1371/journal.pgen.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Perchuk BS, Skerker JM, Laub MT. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell. 2012;150:222–232. doi: 10.1016/j.cell.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. The mechanism of signal transduction by twocomponent systems. Curr Opin Struct Biol. 2010;20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Chen TS, Keating AE. Designing specific protein-protein interactions using computation, experimental library screening, or integrated methods. Protein Sci. 2012;21:949–963. doi: 10.1002/pro.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codoner F, Fares M. Why should we care about molecular coevolution? Evol Bioinform Online. 2008;4:29–38. [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fisher SL, Kim SK, Wanner BL, Walsh CT. Kinetic comparison of the specificity of the vancomycin resistance kinase VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing W, Silhavy TJ. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphris EL, Kortemme T. Prediction of protein-protein interface sequence diversity using flexible backbone computational protein design. Structure. 2008;16:1777–1788. doi: 10.1016/j.str.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Huynh TN, Stewart V. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol Microbiol. 2011;82:275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo MM, Ninfa AJ, Stock JB, Silhavy TJ. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- Jagadeesan S, Mann P, Schink CW, Higgs PI. A novel "four-component" two-component signal transduction mechanism regulates developmental progression in Myxococcus xanthus. J Biol Chem. 2009;284:21435–21445. doi: 10.1074/jbc.M109.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Lee RA, Razaz M, Hayward S. The DynDom database of protein domain motions. Bioinformatics. 2003;19:1290–1291. doi: 10.1093/bioinformatics/btg137. [DOI] [PubMed] [Google Scholar]
- Levin KB, Dym O, Albeck S, Magdassi S, Keeble AH, Kleanthous C, Tawfik DS. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nature Struct & Mol Biol. 2009;16:1049–1055. doi: 10.1038/nsmb.1670. [DOI] [PubMed] [Google Scholar]
- Matsumura I, Ellington AD. In vitro evolution of beta-glucuronidase into a beta-galactosidase proceeds through non-specific intermediates. J Mol Biol. 2001;305:331–339. doi: 10.1006/jmbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazy Y, Wollish AC, Thomas SA, Miller PJ, Collins EJ, Bourret RB, Silversmith RE. Matching biochemical reaction kinetics to the timescales of life: structural determinants that influence the autodephosphorylation rate of response regulator proteins. J Mol Biol. 2009;392:1205–1220. doi: 10.1016/j.jmb.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Kortemme T. Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J Mol Biol. 2008;380:742–756. doi: 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Zapf J, Madhusudan M, Grimshaw CE, Hoch JA, Varughese KI, Whiteley JM. A source of response regulator autophosphatase activity: the critical role of a residue adjacent to the Spo0F autophosphorylation active site. Biochemistry. 1998;37:7725–7732. doi: 10.1021/bi9729615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.