Abstract
A new avian-origin influenza virus A (H7N9) recently crossed the species barrier and infected humans; therefore, there is an urgent need to establish mammalian animal models for studying the pathogenic mechanism of this strain and the immunological response. In this study, we attempted to develop mouse models of H7N9 infection because mice are traditionally the most convenient models for studying influenza viruses. We showed that the novel A (H7N9) virus isolated from a patient could infect inbred BALB/c and C57BL/6 mice as well as outbred Institute of Cancer Research (ICR) mice. The amount of bodyweight lost showed differences at 7 days post infection (d.p.i.) (BALB/c mice 30%, C57BL/6 and ICR mice approximately 20%), and the lung indexes were increased both at 3 d.p.i. and at 7 d.p.i.. Immunohistochemistry demonstrated the existence of the H7N9 viruses in the lungs of the infected mice, and these findings were verified by quantitative real-time polymerase chain reaction (RT-PCR) and 50% tissue culture infectious dose (TCID50) detection at 3 d.p.i. and 7 d.p.i.. Histopathological changes occurred in the infected lungs, including pulmonary interstitial inflammatory lesions, pulmonary oedema and haemorrhages. Furthermore, because the most clinically severe cases were in elderly patients, we analysed the H7N9 infections in both young and old ICR mice. The old ICR mice showed more severe infections with more bodyweight lost and a higher lung index than the young ICR mice. Compared with the young ICR mice, the old mice showed a delayed clearance of the H7N9 virus and higher inflammation in the lungs. Thus, old ICR mice could partially mimic the more severe illness in elderly patients.
Keywords: avian influenza virus, H7N9, mice
INTRODUCTION
Avian influenza viruses are a persistent threat to global public health. Recently, a novel avian-origin influenza A (H7N9) virus that originated from three existing avian influenza A viruses (with hemagglutinin from H7N3, neuraminidase from H7N9 and six internal genes from H9N2) has caused over 127 cases in China, 26 of which resulted in deaths.1,2 As previous pandemic viruses have been derived from avian host origins,3,4 further adaptation of the novel H7N9 to mammals in the future could make H7N9 a pandemic virus.5,6,7
To predict whether this virus can become pandemic, it is important to understand the high pathogenicity and potential risk of transmission of the novel H7N9 avian influenza viruses among humans by utilizing mammalian models. Clinical and epidemiological data collected from human cases could partially reveal features of infection with the novel H7N9 avian influenza virus, but such data are insufficient to dissect the biological or molecular mechanisms of virus pathogenicity in the host. Mammalian models are urgently needed to study the pathogenic and immunological mechanisms of H7N9 infection and to establish efficient vaccines and therapeutic drugs.
Several mammalian species can support influenza replication and are employed as infectious models, including mice, ferrets, guinea pigs, cats, pigs and non-human primates.8 Very recently, Zhu et al.7 reported that the human H7N9 virus infected ferrets and pigs, and those models were employed to study the infectivity and transmission of this novel virus, as well as the pathology that ensues. Although infections in ferrets and pigs can mimic the typical signs of human symptoms, a major limitation of those models is the lack of specific reagents to study the immune responses; additionally, these species lack the abundant resources of transgenic, gene-knockout or knockin strains to analyse the function of individual genes, which are characteristics present in mice.
Mice are one of the most common and convenient mammalian models for studying the pathogenesis and immunology of many influenza viruses9,10,11,12 because of their well-characterized genome, advanced techniques for genomic manipulation, the convenient availability of mouse-specific immunological reagents, easy husbandry and low cost. Those features have allowed mice to become one of the best tools for drug evaluation and vaccine studies for the influenza virus. However, not every influenza virus can infect mice easily without adaptation by the virus. With respect to this new avian-origin virus that recently crossed the species barrier and infected humans, it is not yet known whether it can infect mice.
MATERIALS AND METHODS
Viral isolation
The prototype virus, A/Shanghai/4664T/2013(H7N9) (GenBank No. KC853225-KC853232), was isolated from throat swab specimens obtained from an H7N9 influenza patient whose status was confirmed by the Chinese Center for Disease Control and Prevention. This case was a 27-year-old male who was admitted to the Fifth People's Hospital of Shanghai with symptoms of dizziness and chills on 2nd March 2013. The specimen was collected before the patient received oseltamivir. Although he received supplemental oxygen and symptomatic and supportive treatment, his symptoms were not relieved and his chest computed tomography indicated ‘white lung'. He died of pneumonia and respiratory failure on 9th March 2013.
Madin–Darby canine kidney (MDCK) cells were maintained in complete minimum essential medium (HyClone, Utahi, USA) that was supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin and foetal bovine serum to a final concentration of 10% (Hyclone) for 24 h before viral inoculation. Subsequently, 400 µL of the swab sample was incubated with the MDCK cells for one hour, washed with phosphate-buffered saline (PBS), and further cultivated in the MDCK cells for 96 h. The supernatant from the cell culture was collected and was subjected to titration before animal infection.
All experiments related to the H7N9 virus, including the virus isolation, cultivation, amplification, titration and animal infection, were conducted in the Animal Biosafety Level 3 Laboratory following the standard operating protocols approved by the Institutional Biosafety Committee at the Shanghai Public Health Clinical Center, Fudan University.
Mice
Six- to eight-week-old female BALB/c, C57BL/6 and ICR mice as well as old female ICR mice (60 weeks old) were purchased from the B&K Universal Group Limited (Shanghai, China) and housed under specific pathogen-free conditions at the animal facilities of the Shanghai Public Health Clinical Center, Fudan University (Shanghai, China). The mice were transferred to the Animal Biosafety Level 3 Laboratory before infection. All mouse-related procedures were performed in accordance with the Institutional Animal Care and Use Committee-approved protocols.
Infection and sample collection
Female mice of the BALB/c, C57BL/6 and ICR strains (B&K Universal Group Limited, Shanghai, China) were used in this study. The young mice were 6–8 weeks old, and the old ICR mice were 60 weeks old. Twelve or fourteen mice of each strain were divided into experimental and control groups. The mice were inoculated with 5×104 TCID50 of the virus or with PBS after sevoflurane (Hearem, Osaka, Japan) inhalation anaesthesia. The mice were monitored for clinical signs and survival, and bodyweight was measured daily during the 7-day observation period. Bodyweight loss was calculated using the formula: (weightinfected day − weight0 day)/weight0 day×100%. Three or four mice in each of the experimental groups of BALB/c, C57BL/6, ICR and old ICR mice were euthanized by cervical dislocation at 3 and 7 days post inoculation (d.p.i.); three mice from each control group were also euthanized at the same time. The lungs, livers, hearts and brains were removed under aseptic conditions and washed three times in sterile PBS. A portion of the lungs was removed for pathology, and the remaining organs were mechanically and ultrasonically homogenized (Jxfstprp-24, Shanghai, China) in sterile PBS (500 µL PBS for 1 g of lung tissue, and 800 µL PBS for 1 g of other organs). The centrifuged supernatant was aliquoted and stored at −80 °C until use. The whole lung was weighed to calculate the lung index (LI), using the following formula: LI=weight of lung ×100/bodyweight. The lung index increase rate was also calculated, using the following formula: (LIinfected−LImock)/LImock×100%.
TCID50 detection
The MDCK cells were seeded in 96-well plates at 5000 cells/well in 100 µL of Dulbecco's modification of Eagle's medium with 10% foetal bovine serum and antibiotics (penicillin and streptomycin) for approximately 24 h. The viruses or samples were diluted 1:10 to a final concentration of 1:1 000 000 in a separate V-bottom 96-well plate with serum-free Dulbecco's modification of Eagle's medium medium. Ten microlitres of each dilution was transferred to the MDCK cells mixed with 90 µL Dulbecco's modification of Eagle's medium containing bovine serum albumin Fraction V. The plates were placed in an incubator at 37 °C with 5% CO2 for approximately 5 days, and the cytopathic effect was observed daily in each well using a microscope (Olympus, Shinjuku, Japan). The supernatants were collected and subjected to viral load quantitation. The TCID50 values were calculated using the Reed–Muench method.
Extraction of total RNA and quantitative RT-PCR for viral load
Total RNA was extracted from whole homogenized tissue using an extraction kit (Qiagen, Stow Kark, German) according to the manufacturer's instructions. The quality of the RNA samples was measured by detecting the optical density (280/260); the absorption ratios ranged from 1.9 to 2.25. The total RNA samples were stored at −80 °C until use. To remove any residual DNA contamination, the RNA samples were incubated with RNase-free DNase I at 37 °C for 20 min, followed by stopping the reaction at 65 °C for 10 min.
The TaqMan real-time quantitative PCR amplification reactions were carried out in an Eppendorf Mastercycler ep realplex RT-PCR system. The RT-PCR was carried out with the One-Step RT-PCR kit (TaKaRa, Tokyo, Japan). A 25 µL reaction mixture consisting of a 12.5 µL TaqMan Universal PCR Master Mix, 400 nM primers and a 300 nM TaqMan probe was used for each of the target genes. A master mix (22.5 µL) was prepared into the wells of a 0.2 mL optical-grade 96-well PCR plate. Then, 2.5 µL of an RNA template was added to a final volume of 25 µL. Hemagglutinin gene fragments, amplified with primers designed according to the published sequence of the H7 subtype (GenBank No. KC853228), were inserted into the recombinant T plasmids (TaKaRa) and were used as a quantitative standard. We used the housekeeping gene glyceraldehyde phosphate dehydrogenase as a reference gene (sense primer: 5′-CAA TGT GTC CGT CGT GGA TCT-3′ antisense primer: 5′-GTC CTC AGT GTA GCC CAA GAT G-3′ probe primer: 5′-(6-FAM)-CGT GCC GCC TGG AGA AAC CTG CC-(TAMRA)-3′). All of the reactions were carried out in triplicate without a template control as well as with the influenza A virus (A/Shanghai/4664T/2013(H7N9)) as a positive control. The thermal cycle conditions were as follows: 42 °C for 10 min (AmpErase activation) followed by 95 °C for 1 min and 45 cycles of 95 °C for 15 s and 60 °C for 45 s. All of the data were analysed using the REALPLE×2.2 software (Mastercycler reprealplex real-time PCR system, Eppendorf, German). The quantification of the H7N9 virus was shown as gene copies per µg total RNA of tissue, which was further normalized by the expression level of the housekeeping gene glyceraldehyde phosphate dehydrogenase.
Pathology and immunohistochemistry
The mouse lungs were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin and cut into 5-mm-thick sections, which were stained with haematoxylin and eosin (H&E). For the immunohistochemistry (IHC) assay, the slides were incubated with serum collected from a male H7N9 patient on day 15 of hospitalisation, which was used as the primary antibody (1:100, titred by immunofluorescent assay). Serum from a healthy male donor was used as the control. Rabbit anti-human immunoglobulin G (Dako Corp., Carpinteria, CA, USA) was used as the secondary antibody for the IHC analysis in accordance with the manufacturer's instructions. The slides were viewed using an Olympus BX51 microscope, and the images were captured and analysed by the corresponding acquisition software (DP controller; Olympus). Only the cells with red-stained nuclei were considered positive in the IHC assay.
Data analysis
All of the statistical analyses were performed using GraphPad Prism Software (Version 5.0; GraphPad, La Jolla, CA, USA). Normality tests were conducted, after which parametric (unpaired t-test) or nonparametric (Mann–Whitney U test) tests were used to analyse the differences between the control and experimental groups. The statistical significance was set at P≤0.05.
RESULTS
General appearance, body weight and lung index
The three mouse strains were intranasally inoculated with influenza virus A (H7N9). No change in the general appearance of the mice was observed during the first 2 days after inoculation. However, after the third day, all three strains of mice showed reduced activity, ruffled fur and difficulty breathing (tachypnea and laboured respiration) accompanied by reduced food and water intake and obvious weight loss.
From 3 to 7 d.p.i., bodyweight loss and a lung-index increase were observed. A similar trend of bodyweight loss occurred in all three strains of mice (Figure 1A). However, the extent of the bodyweight loss in the BALB/c mice was greater than that in the C57BL/6 and ICR mice. The percentage of bodyweight loss reached 30% in the BALB/c mice and 20% in the C57BL/6 and ICR mice at 7 d.p.i. (Figure 1A). The rate of increase in the lung index of the BALB/c mice (70%±17.76%) was higher than those of the C57BL/6 (40%±1.34%) and ICR mice (5%±1.04%) at 3 d.p.i. However, the lung index of the C57BL/6 mice increased more than that of the ICR and BALB/c mice at 7 d.p.i.; the lung index increase rates were approximately 160%±10.07%, 100%±45.47% and 70%±10.01%, respectively (Figure 1B).
Figure 1.
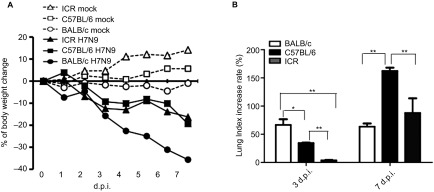
Characterisation of bodyweight lost and lung index post influenza virus A (H7N9) infection in mice. Inbred BALB/c, C57BL/6 and outbred ICR mice were intranasally infected (i.n.) with 4×105 TCID50 influenza A virus (A/Shanghai/4664T/2013(H7N9)). The bodyweight loss was monitored. Lungs were collected at day 3 d.p.i or 7 d.p.i.. The results of lung index are expressed as the mean±s.d., *P<0.05, **P<0.01, n=3. (A) Bodyweight change. (B) Lung index.
Viral infection in the lungs of the three mouse strains
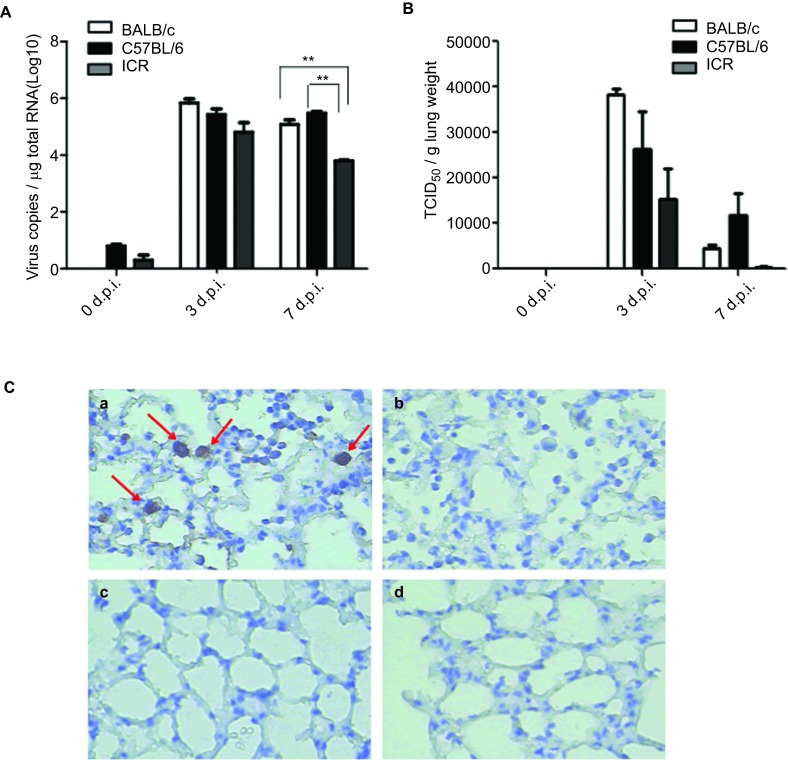
Lung samples from the infected mouse groups were tested by viral-specific RT-PCR, viral titration on the MDCK cells and IHC to confirm whether the H7N9 virus could successfully establish an infection in mice. Evidence of a viral infection could be found in the lungs of all three strains in the form of a viral-specific gene that was detected in the supernatant of the homogenized lung tissues by RT-PCR (Figure 2A); moreover, the virus could be recovered when inoculating the supernatant onto MDCK cells (Figure 2B). The IHC of the lung tissue also showed viral antigen in the pulmonary tissues of the infected mice (Figure 2C), and virus inclusion bodies could be found in the cytoplasm of macrophages and alveolar epithelial cells (data not shown). The peak of the viral load in the lungs was at 3 d.p.i. in all three strains of mice, but the BALB/c mice had a higher viral load than the other two strains (log virus copies were 5.839±0.2367/ µg total RNA in the BALB/c mice, 5.427±0.3576/ µg total RNA in the C57BL/6 mice and 4.809±0.5772/ µg total RNA in the ICR mice). Similar TCID50 values were observed among the three strains in the in vitro assay ((3.83±0.22)×104 TCID50/g lung weight in the BALB/c mice, (2.62±1.45)×104 TCID50/g lung weight in the C57BL/6 mice and (1.22±1.19)×104 TCID50/g lung weight in the ICR mice). The virus in the lungs of the BALB/c and ICR mice were cleared at 7 d.p.i. (log virus copies were 5.074±0.2850/ µg total RNA in the BALB/c mice, 5.477±0.08649/ µg total RNA in the write C57BL/6 in one line mice and 3.798±0.04081/ µg total RNA in the ICR mice), whereas the C57BL/6 mice maintained a level similar to the 3 d.p.i. level.
Figure 2.
Virus loads in the lungs. After intranasally infected (i.n.) with 4×105 TCID50 influenza A virus (A/Shanghai/4664T/2013(H7N9)), lungs were collected and homogenized for virus detection at 3 d.p.i or 7 d.p.i.. Total RNAs in homogenates were extracted and reverse transcribe to cDNA. (A) The virus loads in each mouse lung was determined quantitative PCR to detect the copy number of H7N9 gene. (B) TCID50 assay in MDCK cells were performed to check the titre of H7N9 in infected lung by inoculation of serial diluted supernatant of lung homogenates. The results of RT-PCR are expressed as the average±s.d. (n=3) of the log 10 virus copies per µg total RNA. The results of TICD50 are expressed as the average±s.d. (n=3). *P<0.05, **P<0.01. (C) The lung from infected mice (C-a, C-b) or uninfected mice (C-c, C-d) were fixed and prepared for immunochemistry staining of the H7N9 virus using H7N9 patient's serum (C-a, C-c) and control serum from healthy donor (C-b, C-d) as primary antibody. Images were obtained at magnification is ×400. (C-a) Slide of infected lung detected by H7N9 patient's serum. Alveolar epithelial cells and macrophages with positive signals of H7N9 antigen in slurry and nuclei (arrows). (C-b) Slide of infected lung incubated with control serum from healthy donor. The control lung of non-infected mice incubated with H7N9 patient's serum (C-c) and control serum from healthy donor (C-d).
Histopathological changes in the infected lungs
The majority of the pathological changes in the lungs among all three strains of infected mice consisted of focal inflammation at 3 d.p.i. Compared with the normal lungs in mock-infected mice (Figures 3A, 3B, 3G, 3H, 3M and 3N), most of the lungs from the infected mice showed the infiltration of inflammatory cells, including monocytes/macrophages and lymphocytes, in the pulmonary parenchyma and in the alveolar septa (Figures 3C, 3D, 3I, 3J, 3O and 3P). However, the pathological changes were more severe at 7 d.p.i. and were different among the three strains. These changes presented as a focal pulmonary consolidation in the BALB/c mice (Figures 3E and 3F), pulmonary oedema and haemorrhage in the C57BL/6 mice (Figures 3K and 3L), and the typical pulmonary interstitial inflammatory lesions in the ICR mice (Figures 3Q and 3R) at 7 d.p.i.
Figure 3.
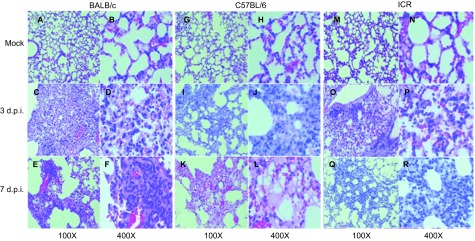
Pathological analysis of the lungs. Lungs were collected from mice infected with influenza virus A/Shanghai/4664T/2013(H7N9) at 3 d.p.i or 7 d.p.i., or from uninfected mice and prepared or histology studies as described in the section on ‘MATERIALS AND METHODS'. Images of H&E-stained slides were observed under microscopy at magnitudes of ×100 and ×400. (A) BALB/c mice PBS negative control (×100). (B) BALB/c mice PBS negative control (×400). (C) BALB/c mice at 3 d.p.i (×100). (D) BALB/c mice at 3 d.p.i (×400). (E) BALB/c mice at 7 d.p.i (×100). (F) BALB/c mice at 7 d.p.i (×400). (G) C57BL/6 mice PBS negative control (×100). (H) C57BL/6 mice PBS negative control (×400). (I) C57BL/6 mice at 3 d.p.i (×100). (J) C57BL/6 mice at 3 d.p.i (×400). (K) C57BL/6 mice at 7 d.p.i (×100). (L) C57BL/6 mice at 7 d.p.i (×400). (M) ICR mice PBS negative control (×100). (N) ICR mice PBS negative control (×400). (O) ICR mice at 3 d.p.i (×100). (P) ICR mice at 3 d.p.i (×400). (Q) ICR mice at 7 d.p.i (×100). (R) ICR mice at 7 d.p.i (×400).
Old ICR mice showed more severe inflammation and delayed viral clearance than their young counterparts
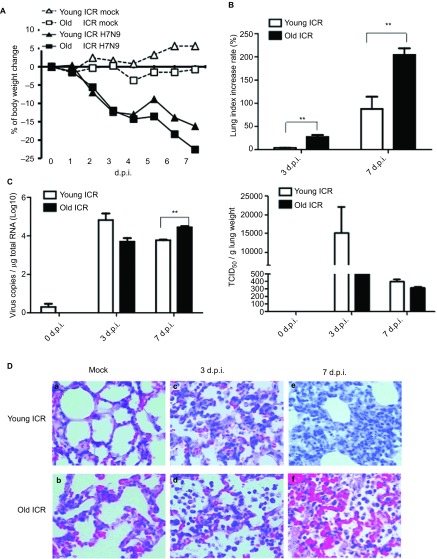
When comparing the young and old ICR mice, the bodyweight loss of the old mice was only slightly greater than that of the young mice (22% vs. 15%) at 7 d.p.i. (Figure 4A). However, the lung index increase rates of the old ICR mice were approximately 27.4%±7.31% and 204%±29.03% at 3 and 7 d.p.i., respectively, which were much higher than those of their younger counterparts (3.8%±1.03% and 87.7%±45.47%, respectively) at the corresponding time points (Figure 4B).
Figure 4.
Comparison of influenza virus A (H7N9) infections between young mice and old ICR mice. Outbred mice ICR young (6–8 weeks) or old (60 weeks) were infected by 4×105 TCID50 influenza virus A (H7N9). Lungs were collected at day 3 d.p.i or 7 d.p.i, and LI was calculated using the formula: LI=weight of lung×100/bodyweight. Lung index percentage=(LIinfected−LImock)/LImock×100%. The virus loads of lung homogenates were determined by RT-RCR and TCID50 detection (R). H7N9 titres in lungs were collected at 3 d.p.i or 7 d.p.i and fixed in 4% paraformaldehyde and stained with H&E. Magnification was ×400. Cytokines/chemokines in lungs were examined. (A) Bodyweight lost curve. (B) Lung index. (C) Virus loads of lungs detected by RT-PCR (left) and TCID50 (right). The results are expressed as the average±s.d. (n=3). *P<0.05, **P<0.01. (D) Slides of lungs with H&E staining (×400). (D-a) Young ICR mice with PBS; (D-b) old ICR mice with PBS; (D-c) young ICR mice at 3 d.p.i; (D-d) old ICR mice at 3 d.p.i.; (D-e) young ICR mice at 7 d.p.i.; (D-f) old ICR mice at 7 d.p.i..
Interestingly, the duration of the viral shedding was different between the old ICR mice and the young ICR mice. The kinetics of the viral loads in the young ICR mice peaked at 3 d.p.i. and decreased at 7 d.p.i. However, in the old ICR mice, the viral load grew slowly at the early stages of infection, but remained higher at 7 d.p.i., which indicated delayed viral clearance in the older ICR mice (Figure 4C). The lung viral loads of the young ICR mice were higher than those of the old ICR mice at 3 d.p.i. (log virus copies were 4.089±0.2850/ µg total RNA in the young ICR mice and 3.700±0.3131/ µg total RNA in the old ICR mice) while less than that of the old ICR mice at 7 d.p.i. (log virus copies were 3.798±0.0408/ µg total RNA in the young ICR mice and 4.443±0.1144/ µg total RNA in the old ICR mice, P=0.0003).
Compared with uninfected mock (Figures 4D-a and 4D-b), both young ICR mice and old ICR mice have inflammatory cells infiltration (Figures 4D-c and 4D-d) at 3 d.p.i.. However, different pathological changes observed between these two groups of mice at 7 d.p.i. Young ICR mice presented the typical pulmonary interstitial inflammatory lesions, while old ICR mice showed many inflammatory cell infiltrations and severe hemorrhages (Figures 4D-e and 4D-f). Therefore, though old ICR mice showed severe inflammation responses than the young mice, it did not seem to contribute to the virus clearance.
DISCUSSION
Gene analysis data indicate that the novel H7N9 viruses may be better adapted to infecting mammals than are other avian influenza viruses.1,13 In contrast to the highly pathogenic avian influenza virus H5N1, these H7N9 viruses are thought to be weakly pathogenic.5 However, recent reports indicate that infection with H7N9 can cause severe respiratory failure in humans.1,14,15 The average age of the patients infected with H7N9 who display severe symptoms is approximately 63 years;15 by contrast, most H5N1-infected patients are young.16,17 The mechanism of H7N9 pathogenicity is not yet understood, and no vaccine is currently available. These gaps motivate us to develop appropriate mammalian models for studying the pathogenesis of the H7N9 virus and evaluating candidate vaccines and antiviral drugs.
Very recently, H7N9 infection models of ferrets and pig have been established.7 The ferrets supported H7N9 virus replication in their upper and lower respiratory tracts and shed the virus for 6–7 days with relatively mild clinical signs. The pigs infected by the H7N9 virus shed the virus for 6 days. However, applications of the ferret model and the pig model were limited because of a lack of immunological research reagents and genetically modified animal resources, as well as high costs.
As a well-characterized mammalian laboratory animal, the mouse is one of the most widely used mammalian models for the studies of influenza virus pathogenesis and immunology, drug evaluations and vaccine developments.8,9,10,11,12 In this study, we aimed to construct mouse models for the H7N9 infection. We chose inbred BALB/c and C57BL/6 mice and outbred ICR mice to verify the infectivity of the novel H7N9 virus in mice.
Our data showed that mice infected with a dosage of 5×104 TCID50/mouse showed different disease manifestations and different in vivo viral replication kinetics depending on the mouse strain background. The BALB/c mice were more sensitive to the H7N9 infection than the other two strains, as evidenced by their bodyweight loss and their high lung index and viral replication in the lungs; additionally, the pathological changes in the lungs were the most severe in BALB/c mice among the three strains, especially at 3 d.p.i. (Figures 1A, 1B, 2A and 2B). Very interestingly, we found that the disease progression was somehow ‘delayed' in the C57BL/6 mice compared to the BALB/c and ICR mice. Most of the viruses in the lungs of the BALB/c and ICR mice were cleared at 7 d.p.i., while the viral loads in lungs of the C57BL/6 mice were maintained at a level similar to that at 3 d.p.i. (Figures 2A and 2B). The pathogenesis of the influenza virus generally correlated with the onset and magnitude of the host innate immune responses, especially excessive cytokine production in the lungs.18,19 Further studies should focus on the cytokine and chemokine production profiles in the infected lungs.
Because most of the clinically severe cases were found in elders,14,15 we compared the outcomes between aged mice (60 weeks old) and their younger counterparts (6–8 weeks old) post H7N9 infection. Although they had lower virus titres in the lungs (Figure 4C), the old mice showed a greater bodyweight loss (Figure 4A) and much more severe tissue damage (Figure 4D) than their younger counterparts. Ageing has been associated with declines in protective immunity against infections, such as those with seasonal influenza viruses; likewise, ageing is also prone to an inflammatory disease state termed inflamm-aging.20,21,22 Hence, it was logical that the old mice took longer to clear the virus than the young mice, although the former had higher inflammatory responses.
Taken our findings together, the novel H7N9 virus was able to replicate and infect inbred strains of mice (BALB/c and C57BL/6) and outbred mice (ICR). The manifestations of disease and the acute host responses were similar to clinical observations. We anticipate that these mouse models will facilitate future study of H7N9 pathogenesis and therapeutic and prophylactic strategy development.
Acknowledgments
This study was supported by grants (2012ZX10004-211 and 2013ZX10004-221-004)) from National Megaprojects of China for Infectious Diseases, grant (KJYJ-2013-01-04) for Special project on human H7N9 from Ministry of Science and Technology of China, grant (11DZ2292900) from Science and Technology Commission of Shanghai Municipality, grant (12140903100) for Special experimental animals project from Shanghai Science and Technology Commission, grant (201316) from Shanghai Public Health Clinical Center, and supported by Shanghai Municipal Health and Family Planning Commission. We appreciate Professor Zhenghong Yuan in Fudan University for his constructive discussions. We thank Dr Ka-Wing Wong in Shanghai Public Health Clinical Center for his critical reading, Mr Zhitong Zhou in Shanghai Public Health Clincal Center for his helps in Animal Biosafety Level 3 lab procedures, the staff in Pathogen Diagnosis and Biosafety Department and the staff in Infectious Animal Department of Shanghai Public Health Clinical Center.
References
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Chinese Center for Disease Control Prevention.The information on epidemic situation of human avian influenza H7N9 in China from 25th April to 1st May, 2013. Beijing; China CDC; 2013. Available at http://www.chinacdc.cn/jkzt/crb/rgrgzbxqlg_5295/rgrqlgyp/201305/t20130503_80728.htm (accessed 1 July 2013). [Google Scholar]
- Reperant LA, Kuiken T, Osterhaus AD. Influenza viruses: from birds to humans. Hum Vaccin Immunother. 2012;8:7–16. doi: 10.4161/hv.8.1.18672. [DOI] [PubMed] [Google Scholar]
- Kuiken T, Fouchier R, Rimmelzwaan G, et al. Pigs, poultry, and pandemic influenza: how zoonotic pathogens threaten human health. Adv Exp Med Biol. 2011;719:59–66. doi: 10.1007/978-1-4614-0204-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeki TM, Cox NJ. Global concerns regarding novel influenza A (H7N9) virus infections. N Engl J Med. 2013;368:1862–1864. doi: 10.1056/NEJMp1304661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P. H7N9 is a virus worth worrying about. Nature. 2013;496:399. doi: 10.1038/496399a. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wang D, Kelvin DJ, et al. Infectivity, transmission, and pathology of human H7N9 influenza in ferrets and pigs. Science. 2013;341:183–186. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- Belser JA, Tumpey TM.H5N1 pathogenesis studies in mammalian models Virus Res 2013. February 28. doi 10.1016/j.virusres.2013.02.003. [DOI] [PMC free article] [PubMed]
- Hien ND, Ha NH, Van NT, et al. Human infection with highly pathogenic avian influenza virus (H5N1) in northern Vietnam, 2004–2005. Emerg Infect Dis. 2009;15:19–23. doi: 10.3201/eid1501.080073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CA, McClellan JL, Maassab HF, et al. Cytokines and the acute phase response to influenza virus in mice. Am J Physiol. 1995;268 1 Pt 2:R78–R84. doi: 10.1152/ajpregu.1995.268.1.R78. [DOI] [PubMed] [Google Scholar]
- Boon AC, deBeauchamp J, Hollmann A, et al. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J Virol. 2009;83:10417–10426. doi: 10.1128/JVI.00514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R, Kiso M, Iwatsuki-Horimoto K, et al. A novel bivalent vaccine based on a PB2-knockout influenza virus protects mice from pandemic H1N1 and highly pathogenic H5N1 virus challenges. J Virol. 2013;87:7874–7881. doi: 10.1128/JVI.00076-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V, Eickmann M, Landt O, et al. Specific detection by real-time reverse-transcription PCR assays of a novel avian influenza A(H7N9) strain associated with human spillover infections in China. Euro Surveill. 2013;18:20461. [PubMed] [Google Scholar]
- Chen Y, Liang W, Yang S, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- Prawira Y, Murniati D, Rusli A, et al. Clinical, laboratory, and radiologic characteristics of confirmed avian influenza (H5N1) Southeast Asian J Trop Med Public Health. 2012;43:877–889. [PubMed] [Google Scholar]
- Hien ND, Ha NH, Van NT, et al. Human infection with highly pathogenic avian influenza virus (H5N1) in northern Vietnam, 2004–2005. Emerg Infect Dis. 2009;15:19–23. doi: 10.3201/eid1501.080073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R, Kiso M, Iwatsuki-Horimoto K, et al. A novel bivalent vaccine based on a PB2-knockout influenza virus protects mice from pandemic H1N1 and highly pathogenic H5N1 virus challenges. J Virol. 2013;87:7874–7881. doi: 10.1128/JVI.00076-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG, Fritz R, Lobo MC, et al. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest. 1998;101:643–649. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, et al. Human immunosenescence: the prevailing of innate immunity, the failing of clonotypic immunity, and the filling of immunological space. Vaccine. 2000;18:1717–1720. doi: 10.1016/s0264-410x(99)00513-7. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S.et al. Inflamm-aging. An evolutionary perspective on immunosenescence Ann NY Acad Sci 2000908244–254. [DOI] [PubMed] [Google Scholar]
- Castle SC. Clinical relevance of age-related immune dysfunction. Clin Infect Dis. 2000;31:578–585. doi: 10.1086/313947. [DOI] [PubMed] [Google Scholar]