Summary
We investigated 67 breakpoint junctions of gene copy number gains (CNVs) in 31 unrelated subjects. We observed a strikingly high frequency of small deletions and insertions (29%) apparently originating from polymerase-slippage events, in addition to frameshifts and point mutations in homonucleotide runs (13%), at or flanking the breakpoint junctions of complex CNVs. These simple nucleotide variants (SNV) were generated concomitantly with the de novo complex genomic rearrangement (CGR) event. Our findings implicate a low fidelity error-prone DNA polymerase in synthesis associated with DNA repair mechanisms that leads to a local increase in point mutation burden associated with human CGR.
Keywords: MMBIR, FoSTeS, MECP2, duplication, complex rearrangements, triplication, CNVs
Introduction
Complex genomic rearrangements (CGRs) are those that consist of more than one simple rearrangement, and have two or more breakpoint junctions formed during the same mutational event 1,2. The frequency of formation of complexities in the human genome, particularly for copy-number gains, is still largely unknown due to the challenges in obtaining the precise sequence and structure at breakpoint junctions. Breakpoint junction sequencing is an experimental approach that usually requires assumptions about both the structure of the variant and the structure of the personal genome in which it occurred, the interpretation of which often depends upon the limitations of a consensus reference haploid human genome.
Genome-wide studies of human germline copy-number variants (CNVs) using capture arrays and next-generation sequencing technologies 3 found complexities in about 5% of the breakpoint junctions sequenced. Another genome-wide study analyzed the breakpoints of 1054 structural variants based on capillary sequencing of clone inserts 4 and observed that a fraction of those variants, 16% (153/973) of the insertion and deletion variants and 9% (7/81) of the inversions, showed additional sequences inserted at the junctions.
Locus-specific studies of CNV causing genomic disorders including duplications and triplications of MECP2 5–7, duplications of PLP1 8,9, duplications of 17p11.2 10–12, duplications of LIS1 13, duplications of STS 14, deletions and duplications of γ-globin genes 15, deletions involving the α-globin gene cluster 16,17, duplications of MARS2 that causes Autosomal Recessive Spastic Ataxia 18 and rearrangements involving the DMD gene 19, have shown the presence of short segments of distantly located DNA sequence at the breakpoint junctions, most apparently originating from genomic regions flanking the breakpoint by an apparent template driven mechanism. Notably, the frequency of such events was estimated based on a limited number of sequenced junctions (reviewed in 1). Interestingly, in vitro mammalian cells subjected to induced double-strand breaks (DSBs) seem prone to capture DNA sequences from various sources, including microsatellites, retrotransposable elements and exogenous DNA by a mechanism that remains to be defined (20,21 and references therein).
We hypothesized that a replication-based mechanism involving template switching, such as Fork Stalling and Template Switching (FoSTeS) 9,22 or Microhomology-Mediated Break Induced Replication (MMBIR) 23,24 following a duplication formed by template-switch between paralogous inverted repeats might underlie the formation of CGRs including triplications and inversions. The key observations underlying the hypothesized replicative mechanism include templated insertions and microhomologies at the breakpoint junctions; proposed ‘signature variant sequences’ representing products of the replicative event. In this present work we studied 31 patients with MECP2 duplication syndrome, 21 novel patients and 10 others previously studied using only aCGH 6. We used both aCGH and breakpoint junction sequencing approaches for analysis of all subjects. We confirmed our previous results that high-resolution aCGH detected ~ 26% of complex rearrangements in MECP2 duplication patients6. Surprisingly, with the higher resolution afforded by DNA sequencing of the breakpoints, we found that an even more substantial percentage (52%) of events were complex. Most complexities consist of insertions of nearby sequence at the junctions, but interchromosomal insertions were also observed in a few rearrangements. Therefore, an apparent single breakpoint can include multiple novel DNA junctions.
The most striking observation for human CGR, however, was the high frequency of concomitant nucleotide variation (i.e. de novo frameshifts and substitution mutations) associated with the CGR event indicating that apparently simple rearrangements might have a higher mutational complexity than previously anticipated and, further, that this mutational load, in terms of novel DNA sequence variation generated, is not confined to the breakpoint junctions.
Results
Complex MECP2 duplication rearrangements detected by genomic arrays
Thirty-one DNA samples from unrelated male patients with MECP2 duplication syndrome were analyzed using high-resolution custom aCGH. Twenty-two samples showed an aCGH pattern consistent with a “simple” non-recurrent rearrangement whereas nine revealed a pattern indicative of complex rearrangements of two general types: four samples had duplicated segments interspersed with stretches of non-altered copy number (i.e. DUP-NML-DUP) whereas five samples had triplicated segments embedded in duplications consistent with a recently described complex structure of DUP-TRP/INV-DUP 7 (Supplementary Fig. 1). Duplications visible by aCGH varied in size from 5.3 kb to 3.8 Mb; triplications varied from 13.8 kb to 211 kb; none of the latter included the entire MECP2 gene.
Further sequencing of breakpoint junctions confirmed the occurrence of a complex rearrangement in eight of these nine CGR cases, except for BAB2806 for which sequencing results indicated that the apparent DUP-NML-DUP structure was likely a result of a simple duplication that occurred on an ancestral chromosome carrying the LCRK1/LCRK2 inverted haplotype, a structural variant that can be found in 18% of individuals of European-descent 25. In summary, visual inspection of high-resolution aCGH revealed complex rearrangements in eight out of thirty-one patients (26%) with MECP2 duplication syndrome.
Breakpoint junction sequencing reveals increased genomic complexity
We designed outward-facing sets of primer pairs for long-range PCR in which amplification was predicted to span the transitions from an unchanged copy-number state to gains of genomic sequence for each patient in this cohort (Supplementary Fig. 2). Most rearrangements (87%) have centromere distal breakpoints that show, by aCGH, an apparent grouping because they are located within LCRs that flank MECP2, particularly LCRJ, which is involved in 48% of the centromere distal duplicated breakpoints, and LCRK which is involved in 80% of the breakpoint junctions of cases with triplication 5,6,26. Proximity to these LCRs makes breakpoint junction sequencing challenging, because the paralogous sequences hamper the ability to specify the breakpoint transition uniquely. We overcame this obstacle by designing several primers spanning LCRJ and selecting those that would match more than one unit (the “Opsin panel”, Supplementary Fig. 3 and Supplementary Table 1). With this design, every sample with distal breakpoints mapping within LCRJ was screened by the Opsin panel primer paired to sample-specific primers located proximally to the centromere which enabled us to obtain breakpoint junctions for the rearrangements in all subjects included in this study.
Surprisingly, sequencing of individual breakpoint junctions revealed far greater complexity than was predicted. About 35% of the samples (11 out of 31 cases) showed evidence for insertion of small segments (3 to 80 bp) at the junctions; in 83% of cases (except BAB3204 and BAB3241) the origins of the insertions could be identified from genomic regions flanking the breakpoints, either upstream or downstream from the patients’ large rearrangement (Supplementary Fig. 5, Supplementary Table 2). The distances to the genomic origin of inserted templated sequences varied from 5 bp (BAB2799 and BAB3027) to 26,931 bp (BAB2991). In two cases, BAB3204 and BAB3241, the sequence of the genomic segments originated from a different chromosome (6 and 16, respectively, Supplementary Table 2).
Importantly, microhomologies of from 1 to 16 nucleotides, a signature sequence for possible involvement of a replicative process, were observed in all but four of the 67 breakpoints sequenced (Table 1 and Supplementary Table 2, Supplementary Fig. 4 and Supplementary Fig. 5). These four consisted of joining events observed in patients BAB2626/BAB2628 (brothers, same event noted as expected), BAB2799 and BAB3259 who had insertions of small sequences (4–10 bp) of unknown origin and BAB3204 who presented a blunt breakpoint junction. In all these cases there was more than one insertion event in which we were able to identify the likely genomic origin of inserted sequence from the haploid reference genome (Fig. 1, Supplementary Table 2, Supplementary Fig. 5).
Table 1.
Summary of Xq28 rearrangements from 31 patients with MECP2 duplication
| Patient BAB# | Inheritance | Sequenced segment (bp) | Insertions a at brkpt jct | Insertion unknown origin | TS same Fork (< 290bp) | TS to a different Fork (> 290 bp) | FM | SM |
|---|---|---|---|---|---|---|---|---|
| 2616 | unknown | 200 | N | N | N | N | N | N |
| 2618 | de novo | 640 | N | N | N | N | N | N |
| 2619 | unknown | 930 | N | N | N | N | N | N |
| 2622 | Maternal | 1650 | N | N | N | Y | N | N |
| 2623 | Maternal | 350 | N | N | Y | N | N | N |
| 2624* | unknown | 750 | N | N | N | Y (DUP-NML-DUP-NML-DUP) | N | N |
| 2626 | unknown | 850 | Y | aaag | N | Y | N | Y |
| 2628 | ||||||||
| 2771 | Maternal | 260 | N | N | N | N | N | N |
| 2799 | unknown | 870 | Y | gccaacc | Y | Y | N | N |
| 2800 | Maternal | 290 | Y | N | Y | Y | N | N |
| 2806 | unknown | 710 | N | N | N | N | N | N |
| 2991 | Maternal | 1100 | Y | N | Y | Y | N | N |
| 3027 | Maternal | 920 | Y | N | Y | Y | Y | N |
| 3147 | Maternal | 250 | N | N | N | Y (DUP-TRP/INV-DUP) | N | N |
| 3154 | Maternal | 450 | N | N | N | N | Y | N |
| 3158 | Maternal | 350 | Y | N | N | Y | N | N |
| 3159 | ||||||||
| 3161 | de novo | 1880 | Y | N | N | Y (DUP-NML-DUP) | N | N |
| 3172 | Maternal | 990 | N | N | N | N | N | N |
| 3174 | Maternal | 160 | N | N | N | N | N | N |
| 3204 | Maternal | 1000 | Y | N | N | Y (ChrX-Chr6) | N | N |
| 3216 | unknown | 365 | Y | N | Y | Y (DUP-TRP/INV-DUP) | N | N |
| 3238 | de novo | 960 | N | N | N | N | N | N |
| 3241 | unknown | 420 | Y | N | Y | Y (ChrX-Chr16) | N | N |
| 3247 | unknown | 850 | N | N | N | N | N | N |
| 3255 | Maternal | 1000 | N | N | N | Y (DUP-TRP/INV-DUP) | N | N |
| 3259* | Maternal | 900 | Y | ctcgtttgtt | N | Y | N | N |
| 3261 | Maternal | 910 | N | N | N | N | N | N |
| 3267 | Maternal | 670 | N | N | Y | N | N | N |
| 3268 | ||||||||
| 3273 | Maternal | 910 | N | N | N | N | Y | N |
| 3274 | Maternal | 820 | N | N | Y | Y (DUP-TRP/INV-DUP) | N | N |
| 3275 | ||||||||
| 3325 | Maternal | 860 | N | N | N | N | N | N |
| Total 31 | 23265 | 11 | 3 | 9 | 16 | 3 | 1 |
only one junction analyzed;
Duplicated and triplicated segments visible by aCGH were not considered as “insertions” in this table; Y: Yes; N: No; TS: Template Switching; brkpt jct: breakpoint junction; DUP: duplication; TRP; triplication; INV: inversion; NML: normal; FM: frameshift mutation; SM: substitution mutation
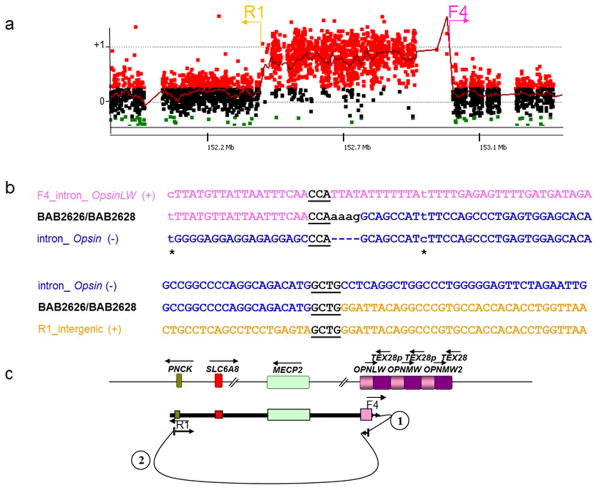
Figure 1. Patient BAB2626 and BAB2628 breakpoint junction mutation load.
These patients have at least three mutations at and flanking the CGR breakpoint junction that were likely produced in the same event: two point mutations (transitions) before and after the breakpoint junction, one insertion (AAAG) for which the origin could not be defined, and two long-distance template-switches (1.6 kb and 472.9 kb, respectively).
(a) BAB2626/BAB2628 aCGH result and approximate location of the primers (F and R) used to obtain patient specific breakpoint junctions.
(b) Breakpoint junction sequence is aligned to the proximal and distal genomic references and color-matched. Strand of alignment (+ or −) is indicated in parenthesis. Microhomology at the breakpoint is indicated by black bold underlined letters. Dashed lines represent nucleotides that did not align to the reference sequence; asterisks indicate point mutations flanking the breakpoint junction.
(c) Representation of the genomic structure for the reference genome (top) and for the surmised genomic structure of BAB2626 and BAB2628 (bottom), showing predicted order, origins, and relative orientations of duplicated sequences. Arrows show orientation of DNA sequence relative to the positive strand; filled arrows with circled numbers below represent a template switch that resulted in insertion of segments. The last arrow signifies resumption of replication on the original template that produced the CGR identified by aCGH. Approximate location of primers used to obtain the breakpoint junctions are shown on the bottom.
In addition to insertion of flanking genomic segments, we identified other nucleotide variation such as small deletions from 4 to 17 bp (BAB2623, BAB2991, BAB3027, BAB3267, BAB3273, BAB3274/BAB3275), frameshift mutations (BAB3027 delA, BAB3154 delG, BAB3273 delT) and two events of C to T transition in one case (BAB2626/BAB2628). These nucleotide variations were all found in proximity to the breakpoint junctions (from 0 to 45 bp distance) (Figs. 1–3, Supplementary Table 2, Supplementary Fig. 4 and Supplementary Fig. 5).
Figure 3. Patient BAB3027 breakpoint junction mutational load.
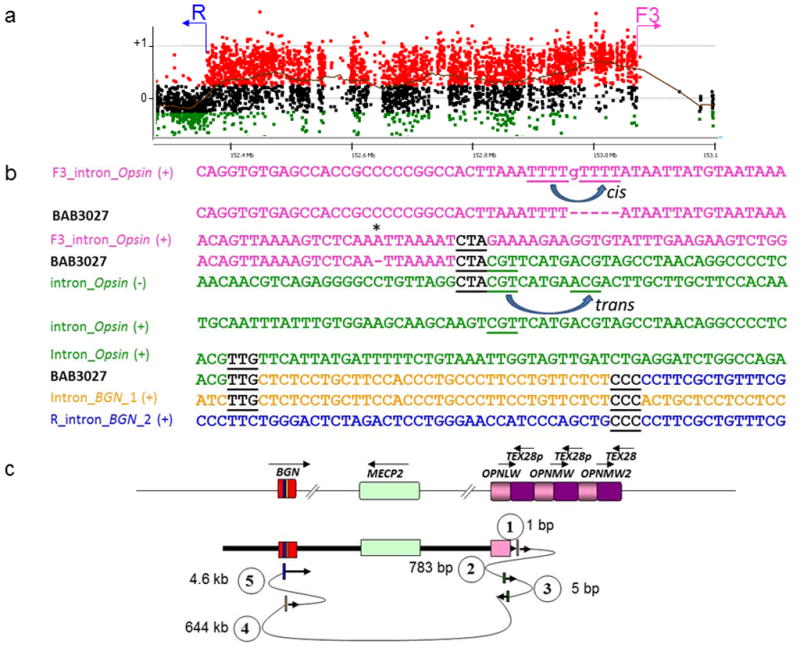
Patient BAB3027 presented at least three mutations at and flanking the CGR breakpoint junctions: a frameshift before the breakpoint junction, and multiple template-switch events. (a) BAB3027 aCGH result and approximate location of the primers (F and R) used to obtain patient specific breakpoint junctions. (b) Breakpoint junction sequence is aligned to the proximal and distal genomic references and color-matched. Strand of alignment (+ or −) is indicated in parenthesis. Microhomology at the breakpoint is indicated by black bold underlined letters. Dashed lines represent nucleotides that did not align to the reference sequence; asterisks indicate frameshifts flanking the breakpoint junction. Misalignment and re-annealing of short repeats present in the primer strand and template strand in cis can produce deletion in the newly synthetized strand (forward slippage) or insertion (backward slippage) 42. In addition, misalignment and re-annealing in trans would produce small inversion at the junctions 41. (c) Representation of the genomic structure for the reference genome (top) and for the surmised genomic structure (bottom), showing predicted order, origins, and relative orientations of duplicated sequences. Arrows show orientation of DNA sequence relative to the positive strand; filled arrows with circled numbers below represent a template switch that resulted in deletion or insertion of segments. Distance between the template switches are shown in bp or kb. The last arrow signifies resumption of replication on the original template which produced the CGR identified by aCGH.
Remarkably, almost all small deletions were flanked by 2 to 3 bp of microhomology in the reference genome and all frameshift and point mutations occurred in homonucleotide runs (≥ 2 bases) (Table 2). Importantly, none of the observed breakpoint-associated nucleotide sequence alterations is present in the current dbSNP database (build 137) documenting that they do not represent common polymorphisms.
Table 2. De novo single-nucleotide variants observed flanking genomic rearrangement breakpoint junctions.
| Patient BAB# | Type | Distance from junction | Context | Original copy tested? |
|---|---|---|---|---|
| 2626/2628 | C>T | 19 bp | Poly T run | Yes |
| C>T | 9 bp | Poly T run | Yes | |
| 3027 | Del A | 8–10 bp | Poly A run | Yes |
| 3154 | Del G | 1–3 bp | Poly G run | Yes |
| 3273 | Del T | 40–42 bp | Poly T run | Yes |
In summary, mutations in homonucleotide runs were observed in 13% (4 out of 31) of CGR examined, and deletions mediated by microhomology were observed in 16% (5 out of 31). Insertions of small segments (< 100 bp) at the junctions were observed in 35% (11 out of 31). If these breakpoint insertional events are summed with the gross alterations detected by aCGH (DUP-NML-DUP and DUP-TRP/INV-DUP), then we can discern experimentally that at least 52% (16 out of 31) of MECP2 duplication rearrangements show sequence complexities at their junctions (Table 1).
Duplicated and triplicated segments originate from the same chromosome
To examine for potential interchromosomal exchanges between different X-chromosomes during rearrangement formation, we evaluated marker haplotypes from the genomic interval spanning the CGR using either an Illumina HumanOmni1-Quad or HumanOmni2.5–8v1 genotyping microarray. Interestingly, and confirming our previous observations for DUP-TRP/INV-DUP rearrangements 7, all 27 subjects for whom there was available biological material were notable for an absence of heterozygosity throughout the duplicated or triplicated regions for all SNPs tested using these platforms. The absence of heterozygosity observed for all SNP markers (N=66 to 992 SNPs analyzed for each sample depending on the size of the rearranged genomic interval) in 100% of the cases (27 of 27) examined is most consistent with the substrate(s) for these alterations originating from a single chromosome, i.e. they represent intrachromosomal events. Patients BAB2616, BAB2618, BAB2624 and BAB2799 were not analyzed by SNP array due to lack of biological material.
As an independent assessment of marker genotype segregation, we developed a microsatellite PCR assay (Supplementary Fig. 6a). This approach also supported an interpretation of a de novo intrachromosomal event in BAB2618, from whom we did not have enough biological material to perform SNP array experiments (Supplementary Fig. 6b). Furthermore, this microsatellite genotyping assay, based on a marker with greater informativeness than SNP marker genotypes, revealed a single allele in all duplications examined in this cohort of males, again consistent with an intrachromosomal event (data not shown).
Breakpoint complexities and SNV occur de novo
Our analysis revealed a high frequency of insertions, deletions and point mutations near or at the breakpoint junctions associated with CNV formation, but a remaining question was whether such variations were generated concomitantly with the CGR event. To answer this question we first examined de novo cases that presented small insertions and deletions or SNVs at the breakpoint junction (Table 1). Two appropriate de novo cases were identified: patient BAB3161 and BAB3155, the latter is the carrier mother of subject BAB3154. Using genome-wide SNP arrays we were able to surmise the origin of both duplications to either the maternal X-chromosome or the maternal grandfather’s X-chromosome, respectively (data not shown).
BAB3161 has a complex DUP-NML-DUP rearrangement in addition to an insertion of 12 nucleotides apparently originating from a region 7 kb distal to the telomere proximal junction (Supplementary Fig. 5). None of the breakpoint junctions that we detected in patient BAB3161, including the one with the 12 nucleotide insertion, were observed in his mother, BAB3162 (breaks termed “FD_intergenic” and “2F3_intron_VAMP7” in Supplementary Fig. 5). These results support the hypothesis that the breakpoint associated with the insertion mutation was formed concomitantly with the occurrence of the complex duplication. Also by PCR and sequencing we confirmed that the 12 nucleotide segment was present in patient BAB3161 at its expected genomic position, based on the human reference, in addition to being present at the breakpoint, which supports a replication mechanism underlying its formation as opposed to it being generated by a non-homologous end joining (NHEJ) or other nonreplicative mechanism.
BAB3155 (and BAB3154) have a frameshift deletion (delG) that has occurred in a mononucleotide run, GGG, at or nearby the junction (Supplementary Fig. 4). PCR and sequencing of the loci involved in the breakpoint junction in her father’s DNA sample indicated that the rearrangement and frameshift deletion were generated de novo and concomitantly.
Intrachromosomal origin of duplications allows study of the ancestral state
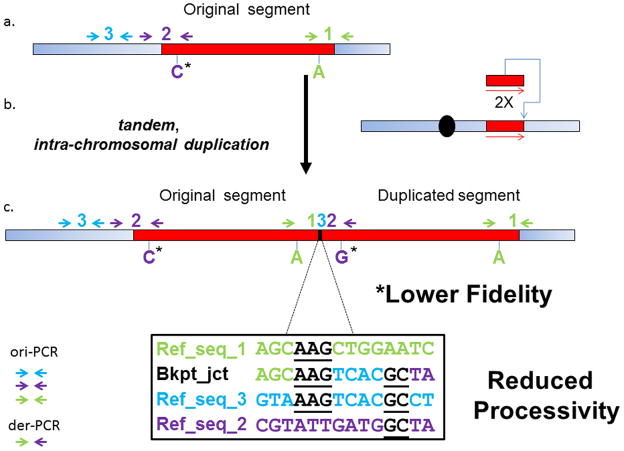
Because the CNVs and single nucleotide variations (SNVs) observed in the subjects reported here were inherited from carrier mothers in 86% of the cases (Table 1), direct examination of the de novo mutational event in the ancestral chromosome from the parent or grandparent with a non-rearranged chromosome is precluded. Nonetheless, all of the rearrangements occurred by an intrachromosomal event, as experimentally evidenced by both SNP array and microsatellites spanning the rearrangements. To our experimental advantage, this latter observation indicates that both the original templated segments, as well as those novel duplicated and triplicated generated segments, are contained within the same derived X-chromosome in carriers. Using this idea we designed PCR-specific assays followed by Sanger sequencing of both the original templated segment (ori-PCR) and the newly generated duplication/triplication breakpoint junction segments (derivative or der-PCR) in order to be able to assay the status of specific genomic regions before and after the formation of the CNV (Fig. 4). Using this approach, ori-PCR and der-PCR provided us with a powerful tool to distinguish whether or not the different types of mutations observed near to the breakpoint junctions of patients with MECP2 duplication were present in the ancestral chromosome of the subject’s personal genome.
Figure 4. Representational figure of the types of mutations that can be observed at and flanking the breakpoint junctions of MECP2 duplications.
a) Wild type Xq28 segment; b) SNP markers and breakpoint junction analysis indicated that duplications involving MECP2 are frequently intrachromosomal head-to-tail duplications; c) Representational genomic structure of the derivative chromosome and the strategies used to uncover the increased mutational load at the breakpoint junctions such as small templated-insertions, frameshifts and point mutations (ori-PCR and der-PCR, please see main text for further details). Templated insertions suggest reduced processivity whereas presence of SNVs suggests lower fidelity of the replicational process. Blue rectangle represents proximal and distal regions flanking the duplication; red rectangle represents the region that will undergo duplication in (b) #1 and #2 represent proximal and distal breakpoints of the duplication; #3 represents a copy of a short local segment inserted at the breakpoint junction of the duplication. Arrows represent forward and reverse primers used to amplify each one of the involved segments in either original or duplicated copy.
We performed ori-PCR and der-PCR in cases BAB2623, BAB2626/BAB2628, BAB2991, BAB3158/BAB3159, BAB3216, BAB3259, BAB3267, BAB3274/BAB3275. For samples BAB3204 and BAB3241 we tested only those alterations that involved chromosome X (Supplementary Table 2). In every case the apparent novel breakpoint junction-associated nucleotide variations, deletions and insertions (i.e. all the simple nucleotide variation or SNV) were present only in the duplicated copy, demonstrating that these nucleotide variations were generated de novo in association with the de novo rearrangement event.
Elevated SNV mutation rate associated with rearrangement breakpoint junctions
The estimated human intergeneration rate of spontaneous mutations has been calculated using different approaches including indirect measurements from databases of de novo mutations for monogenic disorders 27, and direct experimental observations using whole-genome sequences of families and parent-offspring trios. This rate varies from ~1.1 to 1.28 × 10−8 per base pair per haploid genome 28–31 which is 2–4 times lower than direct measurements of single cell analysis of de novo mutation rates in sperm (2–4 × 10−8) 32. These experimentally derived values are of the same order of magnitude as that obtained with the indirect estimate ratio of 2.5 × 10−8 comparing pseudogenes between humans and great apes 33,34.
Here in our studies of CGR we observed five single nucleotide variants (Table 2) in a total of 23 kb of analyzed sequence (Table 1), which represents a de novo point mutation rate of ~ 2.1 × 10−4 mutations/bp. From this we infer that the mutation rate of SNVs associated with CGRs is ~104 fold greater than spontaneous SNVs generated during human gametogenesis. This observation suggests that the replication process involved in the formation of CGRs is highly error prone, possibly utilizing DNA polymerase(s) of low fidelity or a replisome with reduced fidelity in comparison with those involved in intergenerational DNA sequence inheritance.
We also calculated the rate of de novo formation of small insertions and deletions (INDELs), as defined by Mills and colleagues 35, that were observed in our cohort. Mills et al. have considered as INDELs those variants in the 1 bp to 10,000 bp range. In our study we observed 41 of such events (35 insertions and deletions events < 10,000 bp in size + 3 insertions of unknown origin + 3 frameshift mutations, Table 2 and Supplementary Table 2) which represents ~ 1.7 × 10−3 events/bp in 23 kb of total length of analyzed sequence. This ratio is 10 fold greater than the SNV mutation rate calculated above from our experimental observations at CGR breakpoint junctions and 10 to 1,000 fold higher than the de novo locus-specific mutation rate for genomic rearrangements, 10−6 to 10−4 (ref 33) and also higher than the microsatellite mutation rates of ~2.73 to 10.01 × 10−4 mutations per locus per generation as recently inferred from 2,477 dinucleotides and tetranucleotides microsatellites genotyped in Icelanders 36. These observations support the idea that misalignments during replication contribute to the mutational load in patients with CGR. Moreover, such INDEL formation is consistent with a poor processivity DNA polymerase used in the replisome generating CGR as anticipated by the MMBIR model.
Discussion
We observed two types of events at or flanking the breakpoint junctions of our patient cohort in addition to the large duplications visible by aCGH, i) misalignment events (likely reflecting both short and long distance template switches) and ii) presence of new SNVs. Misalignments were observed between segments with very short similarity (microhomologies) that produced short deletions and insertions of flanking sequences at their site of occurrence.
Misalignment or replication slippage between templates located nearby (from 5 bp to 136 bp, Supplementary Table 2) were observed in 29% (9 out of 31) and on both sides of the junctions, in either cis intrastrand or in trans interstrand configurations producing deletions, insertions and inversions at the junctions (Figs. 1–3 and Supplementary Fig. 5). The distances from the slippage events to the breakpoint junction of the gross rearrangements varied from 0 to 41 bp, which is consistent with replication slippage within the same Okazaki initiation zone defined as ~290 bp of the lagging strand that is single stranded in the replication fork 37.
We also observed misalignments between templates located too far away from the breakpoint junctions to have occurred within the same replication fork; classified as long-distance template-switching events (16 out of 31 patients or 52%) (Table 1, Supplementary Table 2). Two distinct entities were observed: those that generated insertions of segments at the breakpoint junctions (35% of the cases or 11 out of 31 patients) that were only revealed by sequencing because of their small size (from 3 bp to 80 bp), and those that generated the CGR visible by high-resolution aCGH (26% of the cases or 8 out of 31 patients). Interestingly, the origin of the small templated insertion could generally be traced to a limited genomic area of up to ~ 27 kb flanking the proximal gross rearrangement breakpoint site (Supplementary Table 2). This observation led us to hypothesize that the gross rearrangements are the final product of an unstable process that involves multiple attempts to reform the replication fork until a stable replisome is established. Multiple misalignments occurred in a few patients (Figs. 1–3, Supplementary Fig. 5), supporting this contention and the existence of low processivity DNA polymerization at the initiation of a CGR event.
In contrast, template switches between substrates located far away (> 27 kb) in the reference genome generally produced gross genomic rearrangements that could be visualized by aCGH. For example, the CGR observed in subject BAB3161 is formed by multiple template switches between genomic regions located distally up to 2.1 Mb away in the reference genome that led to a DUP-NML-DUP pattern of CGR. Such an event produced a final genomic structure in which the distal duplicated segment (1.06 Mb) was inserted in an inverted orientation, potentially facilitated by spatial proximity of templates, among the duplicated copies of the proximal duplication (1.45 Mb) (Supplementary Table 2, Supplementary Fig. 5). We have also reported such an event at the PLP1 locus 8. Interestingly, two patients (BAB3204 and BAB3241) showed a striking pattern of interchromosomal insertions at their breakpoint junctions, suggesting that multiple iterative template switches (8 and 4 events, respectively) can produce very complex structures (Supplementary Fig. 5).
The gross rearrangements in our cohort were characterized as intrachromosomal events, involving the same chromosome X (sister chromatid). This result confirmed our previous studies in cases with MECP2 duplication carrying the DUP-TRP/INV-DUP structure 7 and enabled us to show apodictically that all SNVs and small insertions and deletions detected at or near the breakpoint junctions not only segregate with the CNVs but also were generated de novo, supporting the hypothesis that they were produced concomitantly with the gross rearrangement.
We previously hypothesized that repair of a one ended, double-stranded DNA molecule that can result from a collapsed replication fork, utilizing replication mechanisms, might lead to constitutional rearrangements involving multiple template switches on which widely scattered breakpoints are joined together in a single complex arrangement that leaves their original loci unchanged 2,9,38. The fact that 52% of the rearrangements in our patient cohort have complexities that were not present in the original copy lends further support to our chromoanasynthesis/chromothripsis – hypothesis 38,39.
The presence of both direct and inverted polymerase slippage insertions suggests that slippage occurred within a replication fork so that both leading- and lagging-strand synthesis was occurring, as postulated by the serial replication slippage (SRS) model 40–42, rather than gap-filling synthesis subsequent to resection in the course of two-ended double-strand break-repair which is characteristic of NHEJ. This implicates a break-induced replication (BIR) mechanism - a replication-based mechanism that repairs one-ended double-stranded breaks and involves extensive DNA synthesis in the repair of collapsed forks 43. In yeast, BIR can lead to interchromosomal template switching due to several rounds of strand invasion, DNA synthesis and dissociation within the first 10 kb of the process, after which switching ceases likely due to establishment of a processive mode of DNA replication 44. Recently, Arlt et al. 45 reported that mouse embryonic stem cells defective for NHEJ repair (Xrcc4−/−) and treated with aphidicolin form de novo CNVs with complexities that include the presence of small inserted segments at the junctions, inversions, and microhomologies (mean length: 2.0 bp) at most breakpoint junctions. These observations support the contention that NHEJ is unlikely to be the major repair mechanism underlying formation of such rearrangements.
Moreover, recently, BIR was shown to be a highly inaccurate process in yeast due to the high rate of frameshift mutations that can be observed along the entire replicated segment (2,800-fold compared to spontaneous events originated from S-phase replication) likely due to a combination of diverse causes including an increased dNTP pool during G2/M DNA damage checkpoint response when BIR repair seems to proceed, as well as to an error-prone polymerase along with a less efficient mismatch repair 46. Consistent with the BIR mutation rate reported by Deem et al. 46, we observed a 104-fold increase in mutation rate nearby the breakpoint junctions of the CNVs reported herein. At least two polymerases seem to be involved with the hypermutation rate associated with BIR: Pol Delta, likely due to a less efficient proofreading activity compared to S-phase replication, and to a minor extent, the translesion polymerase Pol Zeta, through a position-dependent error-prone copying of damaged DNA46. Remarkably, Pol Delta is also implicated in increased mutagenesis identified during mitotic gene conversion by synthesis-dependent strand annealing (SDSA) in budding yeast47. In contrast, all three replicative polymerases, alpha, delta and epsilon are implicated in the rate and/or expansion of (GAA)n repeats in a budding model to study the repeat instability causative of Friedreich ataxia in addition to an intriguing phenomenon of repeat-induced mutagenesis (RIM) that is observed 500 bp to 1 kb upstream and downstream of those repeats 48. The role of replicative polymerases or accessory factors involved in the error prone nature of different steps of BIR requires further studies. Iraqui et al. 49, using a system construct based on a polar replication fork barrier in S. pombe, reported that recovery of arrested forks during S-phase is associated with genomic instability that is dependent on homologous recombination: complex rearrangements induced by such events result from occasional ectopic recombination at the site of the arrested fork. In addition, they observed replication slippage mediated by microhomology, as well as base-substitutions and frameshifts if the fork resumes on the appropriate initial template resulting in an error-prone DNA synthesis that resembles the kind of mutations and gross chromosomal rearrangements (GCRs) or CGR described herein.
In 35% (11 out of 31) of the duplications, no additional complexities nor point mutations flanking the breakpoint junctions were observed; these may constitute simple, in tandem duplications. All show microhomologies at the junctions examined varying from 1 to 17 nt, 2 out of 11 represent Alu/Alu mediated rearrangements, suggesting either MMBIR or microhomology-mediated end joining (MMEJ) as the mechanism for formation 1, 24, 50.
In summary, our data indicate that CGR can be associated with a high mutational load due both to increased de novo SNV and INDEL mutation rates (~ 2.1 × 10−4 mutations/bp and ~ 1.7 × 10−3 events/bp, respectively) at or near the breakpoint junction of the CGR, and to the novel joints generated by rearrangements of the genome. The high frequency of complexities at the breakpoint junctions likely contributes to the challenges inherent to breakpoint mapping for CGR and suggests that copy number changes remain an underexplored source of mutations in the human genome.
Methods
Subjects
Families with genomic rearrangements of Xq28 including the MECP2 gene were identified by physician referral or self-referral. Informed consent for participation and sample collection was obtained using protocols H-26667 and H-20268 approved by the Institutional Review Board for Baylor College of Medicine and affiliated hospitals.
Duplication size and genome content
To determine the size, genomic extent and gene content of each rearrangement, we designed a tiling-path oligonucleotide microarray spanning 4.6 Mb surrounding the MECP2 region on Xq28. The custom 4x44k Agilent Technologies microarray was designed using the Agilent earray website. We selected 22,000 probes covering ChrX: 150,000,000–154,600,000 (NCBI build 36), including the MECP2 gene, which represents an average distribution of 1 probe per 209 bp. Probe labeling and hybridization were performed as described 50. Samples from patients and their biological mothers were collected and analyzed using aCGH.
Long-range PCR amplification
Reverse and forward primer pairs (relative to the reference genome) were designed at the apparent boundaries of each duplicated or triplicated segment as defined by aCGH analysis. Long-range PCR was performed using TaKaRa LA Taq (Clontech, Mountain View, CA). PCR sample-specific products were sequenced by Sanger sequencing methodology. PCR and sequencing results were independently confirmed by repeated experiments. DNA samples from mothers were also tested for the presence of the breakpoint junctions and mutations in all cases.
Genotyping
DNA samples were quantified using Quant-iT PicoGreen dsDNA Reagent (Invitrogen) in a Tecan GENios microplate reader (Tecan Group, Mannendorf, Switzerland). Genotyping was performed on Illumina HumanOmni1-Quad or HumanOmni2.5-8v1 genotyping microarray (Illumina, Inc., San Diego, CA, U.S.A.) following the manufacturer’s instructions. All microarrays had call rates > 0.99. Basic quality control and analysis of the genotyping data were performed on GenomeStudio software, version 2011 (Illumina, Inc., San Diego, CA, U.S.A.). CNV calls were performed using cnvPartition v2.4.4 with default parameters.
As a complementary method to SNP genotyping we developed a microsatellite marker for the same purpose. We selected five simple repeats within the SRO region for which period was > 2 and copy number > 5. After testing them for populational polymorphism using a pool of N = 29 random control female DNA samples, only one presented multiple peaks (Xq28_4). This microsatellite consists of a tetranucleotide repeat with two different sequence unit variation (GATG and GATA). It can be amplified with a standard PCR protocol with primers described in Supplementary Table 1. In our female pool there were six peaks presents in the following order and relative frequency: 551 bp (2%), 555 bp (30%), 559 bp (2%), 563 bp (20%), 567 bp (45%), 571 (1%).
Bioinformatic analyses
Array CGH and coordinates for rearrangements were analyzed using UCSC hg18. Point mutations and small insertions and deletions detected by sequencing were analyzed using the following databases: UCSC hg 19 and dbSNP build 137.
Supplementary Material
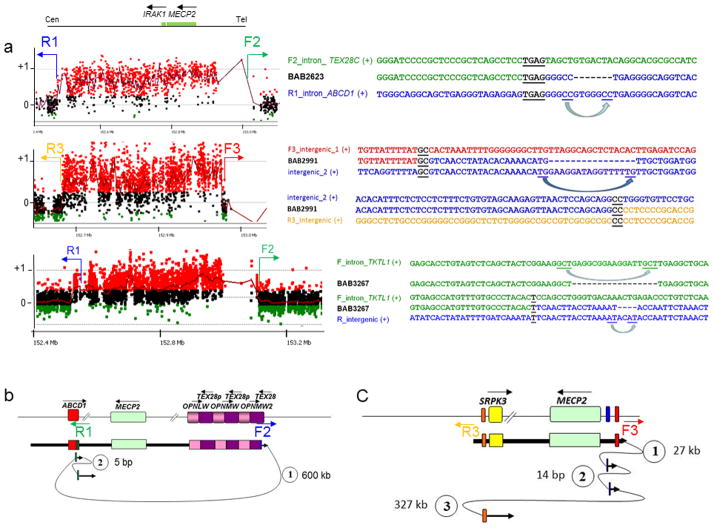
Figure 2. Short and long template-switches can be observed on either or both sides of CGR breakpoint junctions.
(a) For each patient (BAB2623, BAB2991 and BAB3267), the aCGH result along with the breakpoint junction sequences obtained by long-range PCR and Sanger sequencing are shown. Approximate location of the primers (F and R) used to obtain patient-specific breakpoint junctions are represented in the aCGH plot. Breakpoint junction sequence is aligned to the proximal and distal genomic references and color-matched. Strand of alignment (+ or −) is indicated in parenthesis. Microhomology at the breakpoint is indicated by black bold underlined letters. Dashed lines represent deleted nucleotides; blue arrows point to the nucleotides likely involved in the misalignment that generated the deletion. (b) and (c) represent the genomic structure for the reference genome (top) and for the surmised genomic structure (bottom) for patients BAB2623 and BAB2991, respectively, showing predicted order, origins, and relative orientations of duplicated sequences. Arrows show orientation of DNA sequence relative to the positive strand; filled arrows with circled numbers below represent a template switch that resulted in insertion or deletion of segments. Distances between the template-switches are shown in bp or kb. The last arrow signifies resumption of replication on the original template that produced the CGR identified by aCGH. Approximate location of primers used to obtain the breakpoint junctions are shown below the reference genome structure.
Acknowledgments
We thank the families for their participation in the study. We gratefully acknowledge Dr. Grzegorz Ira for helpful discussions. This work was supported in part by US National Institute of Neurological Disorders and Stroke (NINDS) grants R01 NS058529 to J.R.L. and 5K08NS062711 to M.B.R. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or NIH.
Footnotes
URLs
UCSC hg 19 (http://genome.ucsc.edu/cgi-bin/hgGateway)
dbSNP build 137 (http://www.ncbi.nlm.nih.gov/projects/SNP/)
earray website (http://earray.chem.agilent.com/earray/)
Supplementary Information includes six figures and two tables.
Author Contributions
C.M.B.C. conducted high-density CGH arrays, breakpoint sequencing, and data analysis. D.P. assisted with high-density CGH arrays and breakpoint sequencing. C.M.B.C. and B.A. designed, performed and implemented microsatellite repeat experiments. M.B.R. coordinated clinical studies and subject recruitment. L.M.F and J.W.B. conducted SNP genotyping and data analysis. P.F. conducted confirmatory CNV studies. P.J.H. and J.R.L. were involved in research design and data analyses. C.M.B.C., P.J.H and J.R.L. prepared the manuscript.
Competing financial interests
J.R.L. is a paid consultant for Athena Diagnostics, holds stock ownership in 23andMe, Inc. and Ion Torrent Systems, Inc., and is a co-inventor on multiple United States and European patents related to molecular diagnostics. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the Medical Genetics Laboratories (http://www.bcm.edu/geneticlabs/).
References
- 1.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–20. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad DF, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–91. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–47. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauters M, et al. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 2008;18:847–58. doi: 10.1101/gr.075903.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho CM, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum Mol Genet. 2009;18:2188–203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho CM, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074–81. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho CM, et al. Evidence for disease penetrance relating to CNV size: Pelizaeus-Merzbacher disease and manifesting carriers with a familial 11 Mb duplication at Xq22. Clin Genet. 2012;81:532–41. doi: 10.1111/j.1399-0004.2011.01716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–53. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, et al. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am J Hum Genet. 2010;86:462–70. doi: 10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am J Hum Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–77. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu P, et al. Copy number gain at Xp22.31 includes complex duplication rearrangements and recurrent triplications. Hum Mol Genet. 2011;20:1975–88. doi: 10.1093/hmg/ddr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann R, Lawson VE, Jeffreys AJ. Dynamics and processes of copy number instability in human gamma-globin genes. Proc Natl Acad Sci U S A. 2010;107:8304–9. doi: 10.1073/pnas.1003634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholls RD, Fischel-Ghodsian N, Higgs DR. Recombination at the human alpha-globin gene cluster: sequence features and topological constraints. Cell. 1987;49:369–78. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- 17.Rugless MJ, et al. A large deletion in the human alpha-globin cluster caused by a replication error is associated with an unexpectedly mild phenotype. Hum Mol Genet. 2008;17:3084–93. doi: 10.1093/hmg/ddn205. [DOI] [PubMed] [Google Scholar]
- 18.Bayat V, et al. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshima J, et al. Regional genomic instability predisposes to complex dystrophin gene rearrangements. Hum Genet. 2009;126:411–23. doi: 10.1007/s00439-009-0679-9. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Waldman AS. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001;29:3975–81. doi: 10.1093/nar/29.19.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–74. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Small K, Iber J, Warren ST. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet. 1997;16:96–9. doi: 10.1038/ng0597-96. [DOI] [PubMed] [Google Scholar]
- 26.Carvalho CM, Zhang F, Lupski JR. Structural variation of the human genome: mechanisms, assays, and role in male infertility. Syst Biol Reprod Med. 2011;57:3–16. doi: 10.3109/19396368.2010.527427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–8. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell CD, et al. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012;44:1277–81. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–4. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–12. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–7. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 34.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills RE, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–90. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun JX, et al. A direct characterization of human mutation based on microsatellites. Nat Genet. 2012;44:1161–5. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleary JD, Pearson CE. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 2005;21:272–80. doi: 10.1016/j.tig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Liu P, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Complex gene rearrangements caused by serial replication slippage. Hum Mutat. 2005;26:125–34. doi: 10.1002/humu.20202. [DOI] [PubMed] [Google Scholar]
- 41.Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Intrachromosomal serial replication slippage in trans gives rise to diverse genomic rearrangements involving inversions. Hum Mutat. 2005;26:362–73. doi: 10.1002/humu.20230. [DOI] [PubMed] [Google Scholar]
- 42.Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat. 2005;25:207–21. doi: 10.1002/humu.20133. [DOI] [PubMed] [Google Scholar]
- 43.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–35. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 44.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–5. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 45.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–5. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah KA, et al. Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2012;2:1088–95. doi: 10.1016/j.celrep.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iraqui I, et al. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 2012;8:e1002976. doi: 10.1371/journal.pgen.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinawi M, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–41. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.