Abstract
All-trans retinoic acid (RA) signals via binding to retinoic acid receptors (RARs α, β, and γ). RA directly influences expression of Pdx1, a transcription factor essential for pancreatic development and β-cell maturation. In this study we follow the differentiation of cultured wild-type (WT) vs. RARβ knockout (KO) embryonic stem (ES) cells into pancreatic islet cells. We found that RARβ KO ES cells show greatly reduced expression of some important endocrine markers of differentiated islet cells, such as glucagon, islet amyloid polypeptide (Iapp), and insulin 1 (Ins1) relative to WT. We conclude that RARβ activity is essential for proper differentiation of ES cells to pancreatic endocrine cells.
Keywords: Retinoic acid, RARβ, Stem cell, Endocrine, Pancreas, Islet cells
Introduction1
In 2011 there were an estimated 366 million cases of diabetes worldwide, according to the International Diabetes Federation, and these cases are estimated to increase to 522 million by 2030 [1, 2]. In the U.S. there were 23.7 million diagnosed cases, with an estimated healthcare cost of $113 billion [2, 3]. Diabetes results when insulin production by pancreatic β-cells does not meet the metabolic demand of peripheral tissues such as liver, fat, and muscle [4]. A reduction in β-cell number and function leads to hyperglycemia in both type 1 and type 2 diabetes [4]. In type 1 diabetes, insulin-producing pancreatic β-cells lose self-tolerance and this gives rise to hyperglycemia [5]. Each year in the United States there are over 30,000 new cases of type I diabetes diagnosed [6]. Patients with type I diabetes can control their blood glucose level with insulin supplements [7]. However, the differentiation of stem cells into pancreatic β-cells could be a long term, better solution [8, 9].
Mouse embryonic stem (ES) cells are pluripotent cells derived from the inner cell mass of blastocyst-stage (day 3.5) embryos [9, 10]. Upon LIF removal, ES cells spontaneously differentiate into all three primary embryonic germ layers: endoderm, mesoderm, and ectoderm [9]. Several research groups have shown that the directed differentiation of ES cells along the endocrine pathway can be achieved by using a wide range of growth/differentiation factors, including retinoic acid (RA) treatment [11–16].
The effects of RA on cells and tissues are known to occur through RA binding and activation of retinoic acid receptors (RARα, RARβ, and RARγ) and their isoforms [17, 18]. Each RAR has some specific functions and activates specific subsets of genes [19–21]. RA signaling is crucial for endocrine pancreatic development in Xenopus [22]. In addition, transgenic mice that express a dominant negative RARα403 mutant, used to ablate all retinoic acid-dependent processes in vivo, lack both dorsal and ventral pancreas, and die at the neonatal stage [23]. Impaired pancreatic islet function was also observed in vitamin A deficiency and repletion rodent models [24, 25]. Another study, focused on the role of CRABP1 and RBP4 in pancreatic differentiation, showed an increase in RARβ expression in early differentiation [10]. While previous studies showed that RARβ is expressed during pancreas development, little is known about the role of RARβ in normal islet maintenance and function in adult animals [26, 27].
The RARβ gene is frequently hypermethylated at CpG islands in human pancreatic adenocarcinoma [28] and this phenomenon could also be associated with other pathologies such as diabetes. We hypothesized that RARβ plays a key role in ES cell differentiation to pancreatic endocrine cells, and that this function of RARβ may be altered in pancreatic physiopathology. In this report, we measured the expression profiles of various pancreatic differentiation and retinoid signaling markers in WT and RARβKO ES cells. We show that the lack of all three isoforms of RARβ impairs the differentiation of cultured ES cells to pancreatic β-like cells.
Materials and Methods
Cell culture and isolation of RARβ homozygous ES cell lines
Murine J1 wild-type ES cells were cultured as described previously [29]. 129; C57BL/6 RARβ homozygous null mice were provided by Dr. Pierre Chambon (Strasbourg-Cedex, France) [27]. Mice were housed and treated according to appropriate WCMC IACUC guidelines. Blastocysts were harvested on day E3.5 and individually cultured in ES cell medium as previously described [29] to generate RARβ KO ES cells by homologous recombination. These RARβ KO ES cells were karyotyped and shown by Southern analysis to possess two RARβ KO alleles (not shown).
Pancreatic endocrine differentiation protocol
A slightly modified version of the established protocols published by Borowiak [13] and D’Amour [14] was used to carry out differentiation of hormone expressing endocrine cells from mouse ESCs. Prior to differentiation, ESCs were seeded at 5 × 105 on 30 mm gelatin-coated plates. After overnight culture, cells were exposed to 250 nM BIO-Acetoxime (EMD Bioscience, San Diego, CA) + 50 ng/ml activin A (R&D Systems, Minneapolis, MN) in Advanced RPMI (GIBCO, Grand Island, NY) supplemented with 1X L-Glu and 0.2% FBS (GIBCO) for 1 day, and then to activin A alone in the same media. Cells were then cultured for 4 days to induce endoderm differentiation. For pancreatic progenitor induction, the cells were transferred to 50 ng/ml FGF10 (R&D Systems), 7.5 μM cyclopamine (Calbiochem, San Diego, CA) in DMEM supplemented with 1X L-Glu, 1X Pen/Strep, and 1X B27 (Invitrogen, Grand Island, NY) for 2 days. At day 7, cells were transferred to FGF10, cyclopamine, and 2 μM all-trans RA (Sigma, St. Louis, MO) in DMEM supplemented with 1X L-Glu, 1X Pen/Strep, and 1X B27 (Invitrogen) for 4 days. At day 11, cells were cultured in the presence of DMEM supplemented with 1X L-Glu, 1X Pen/Strep, and 1X B27 for 3 days. At day 14, CMRL (Invitrogen) medium was added and supplemented with 1X L-Glu, 1X Pen/Strep, 1X B27, 50 ng/ml IGF-1 (R&D Systems), 50 ng/ml HGF (R&D Systems), and, in some experiments, 10 mM nicotinamide (Sigma) for 3 more days. All stock compounds were made in either PBS or ethanol.
RT-PCR analysis
Various markers for endodermal (day 5), pancreatic progenitor (day 11), endocrine progenitor (day 14) and endocrine (day 17) differentiation were analyzed by semi-quantitative RT-PCR in J1 WT and RARβ KO ESCs. Specific primers used and amplification conditions are listed in Table 1. Total RNA extraction, semi-quantitative, and quantitative PCR reactions were performed as previously described [30]. Amplified PCR products were resolved on 1.5% agarose gels and visualized by staining with ethidium bromide. PCR bands were sequenced for verification of the correct amplicon. Quantitation was performed using ImageJ software (National Institutes of Health) from three experimental, independent biological repeats.
Table 1.
Primer sequences used for RT-PCR
| Primer | Application | Forward sequence (5′-3′) | Reverse sequence (5′-3′) | Product size (bp) |
|---|---|---|---|---|
|
| ||||
| mIns1 | RT-PCR | TAGTGACCAGCTATAATCAGAG | ACGCCAAGGTCTGAAGGTCC | 289 |
| mGcg | RT-PCR | CCGCCGTGCCCAAGATTTT | CCTGCGGCCGAGTTCCT | 232 |
| mSst* | RT-PCR | GAGCCCAACCAGACAGAGAA | GAAGTTCTTGCAGCCAGCTT | 150 |
| mNgn3* | RT-PCR | CTGCGCATAGCGGACCACAGCTTC | CTTCACAAGAAGTCTGAGAACACCAG | 233 |
| mRARβ | RT-PCR | GATCCTGGATTTCTACACCG | CACTGACGCCATAGTGGTA | 248 |
| mRARγ2 | RT-PCR | ATGTACGACTGCATGGAATCGT | GATACAGTTTTTGTCACGGTGACAT | 366 |
| mNanog | RT-PCR | AAAGGATGAAGTGCAAGCGGTGG | CTGGCTTTGCCCTGACTTTAA | 520 |
| mRex1 | RT-PCR | GAAAGCAGGATCGCCTCACTGTGC | CGATAAGACACCACAGTACACAC | 641 |
| mCyp26a1 | RT-PCR | GAAACATTGCAGATGGTGCTTCAG | CGGCTGAAGGCCTGCATAATCAC | 272 |
| mPax6 | RT-PCR | GCAACCCCCAGTCCCCAGTCAGA | AGTCCATTCCCGGGCTCCAGTTCA | 399 |
| mIsl1* | RT-PCR | CCCGGGGGCCACTATTTG | CGGGCACGCATCACGAA | 397 |
| mIapp* | RT-PCR | TGGGCTGTAGTTCCTGAAGC | GCACTTCCGTTTGTCCATCT | 199 |
| mPdx1 | RT-PCR | CTTTCCCGTGGATGAAATCC | GTCAAGTTCAACATCACTGCC | 205 |
| mNkx6.1 | RT-PCR | AGAGAGCACGCTTGGCCTATTC | GTCGTCAGAGTTCGGGTCCAG | 215 |
| HPRT1 | RT-PCR | TGCTCGAGATGTGATGAAGG | TCCCCTGTTGACTGGTCATT | 192 |
All primers for RT-PCR are designed around introns, except those marked with *.
Indirect immunofluorescence
Immunofluorescence assays on cells and tissue sections were performed as previously described [31]. Briefly, differentiated samples were fixed using 4% (w/v) paraformaldehyde and membrane permeabilization (for cells only) was done with 0.3% (w/v) Triton-X 100 (Sigma). Unspecific sites were blocked using 2% BSA for 30 min prior to incubation with rabbit polyclonal anti–Pdx1 (Millipore, 06-1379, 1:1000), rabbit anti-C-Peptide (Cell Signaling, 4593, 1:500, Danvers, MA) and mouse monoclonal anti-Glucagon (Abcam, ab10988, 1:200) primary antibodies. Phalloidin-TRITC (Millipore, FAK100, 1:1000, Billerica, MA) was used to stain the actin stress fibers network (F-actin). Nuclei were stained using DAPI contained in Vectashield® mounting medium for fluorescence (Vector labs, Burlingame, CA). Quantitation of C-peptide positive stained cells and islet surface area was performed using NIS-Elements Advanced Research software (Nikon).
Microarray analysis
Total cellular RNA was isolated with Trizol reagent (Invitrogen), and concentration and integrity were assessed using Nanodrop® technology (Thermo Scientific). Preparation of cRNA, chip hybridization and scanning were carried out by the Microarray Core Facility at Weill Cornell Medical College (WCMC). The microarray analyses were performed following the Affymetrix Genechip expression analysis technical manual. The fragmented cRNA was hybridized to the microarray chips (MG-430.2, Cat. #900496, Affymetrix, CA, USA), which include over 45 000 transcripts representing 34 000 substantiated mouse genes. Data analysis was performed using Genespring v7.0 software (Agilent Technologies) as previously described [30].
Statistical analyses
All experiments were performed at least 3 times using independent biological triplicates. Results are presented as means ± SEM. All statistical tests were performed using GraphPad InStat software version 3.10. A p-value of ≤0.05 indicates statistical significance.
Results
Pancreatic differentiation and assessment of pancreatic markers in WT murine ES cells
We selected the endocrine differentiation protocol of D’Amour et al [14] because this protocol resulted in expression of later stage endocrine markers using human ES cells [14]. We modified the D’Amour et al protocol by replacing Wnt3a with BIO-acetoxime. Wnt3a has been documented as being important for mesendoderm specification and BIO-acetoxime acts in the same signaling pathway [32, 33]. Second, we included nicotinamide during the last stage of differentiation; various published protocols included this reagent because of its reported effectiveness in supporting pancreatic differentiation [34, 35] (Fig. 1A, B).
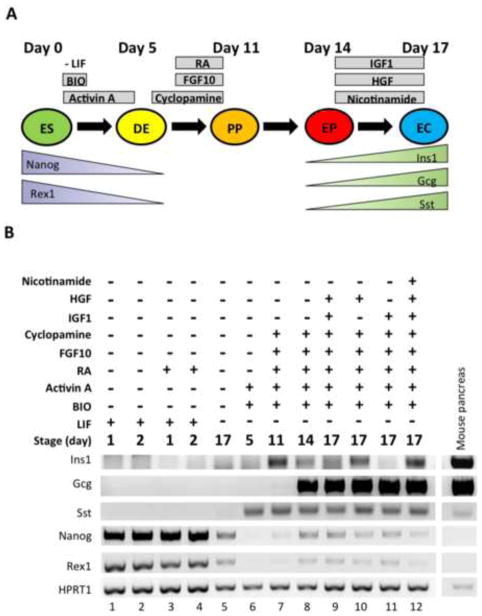
Figure 1. Differentiation of WT ES cells to pancreatic endocrine cells.
(A) Schematic representation of the endocrine differentiation protocol (14). WT murine ES cells are treated with different growth factors to differentiate the cells into definitive endoderm (DE), pancreatic progenitor (PP), endocrine progenitor (EP), and endocrine cells (EC). (B) WT ES cells were subjected to the 17-day differentiation protocol. Each lane represents a different condition at a specific time point. RT-PCR analyses were performed to monitor the expression of pancreatic differentiation markers such as insulin-1 (Ins1), glucagon (Gcg), somatostatin (Sst), neurogenin-3 (Ngn3), Pdx1, and Sox17, as well as the ES cell markers Nanog and Rex1. HPRT1 was used as a loading control. Pancreas extracts from C57BL/6 WT mice were used as a positive control (far right lane). Sample lanes are labeled 1–12 at the bottom. This experiment was performed 7 times, starting with fresh cells, with similar results.
To characterize the differentiation of WT ES cells, we harvested cellular extracts at various time points (Fig. 1B) and assessed the mRNA levels of various differentiation markers by RT-PCR. LIF removal combined with BIO and Activin A caused a major decrease in transcripts of the stem cell markers Nanog [36] and Rex1 (Zfp42) [37] (Fig. 1B, lane 6) compared to the levels in ES cells cultured with LIF (Fig. 1B, lanes 1–4). Nanog and Rex1 transcript levels remained low for the remainder of the differentiation process (Fig. 1B, lanes 7–12). While we observed robust expression of transcripts for glucagon (Gcg), a functional marker of pancreatic α-cells (37), by day 14 (Fig. 1B, lane 8), somatostatin (Sst), a hormone secreted by δ-cells [38], was detectable as early as day 5 (Fig. 1B, lane 6). Insulin-1 (Ins1), a β-cell marker [38], was detected by day 11 of the differentiation process and its expression fluctuated depending on the presence of HGF, IGF1, or both factors together (Fig. 1B, lanes 9 to 11). The most consistent expression of all three pancreatic endocrine differentiation markers tested was observed in the presence of HGF, IGF1, and nicotinamide from days 14 to 17 (Fig. 1B, lane 12). As another control we cultured WT ES cells in absence of LIF for 17 days but with no other factors added; we observed a decrease in Nanog and Rex1 transcripts, but no induction of the differentiation markers tested (Fig. 1B, lane 5). For comparison, RNA from adult mouse pancreas is shown (Fig. 1B, right panel).
Thus, we demonstrated the differentiation of WT ES cells to endocrine cells capable of expressing pancreatic hormone-encoding genes. This differentiation protocol was then employed to investigate the role of RARβ at specific stages of pancreatic endocrine differentiation.
The Lack of RARβ delays Pdx1 expression during pancreatic endocrine differentiation of ES cells
We next subjected WT and RARβKO ES cells to the endocrine differentiation protocol described above. We confirmed the absence of RARβ transcripts in the RARβKO ES cells by RT-PCR (Fig. 2A). We detected an RA-associated increase in Cyp26a1 mRNA level in both WT and RARβKO ES cells, indicating that the RARβKO ES cells can still respond to RA via RARγ [39] (Fig. 2A) to activate Cyp26a1 transcriptionally.
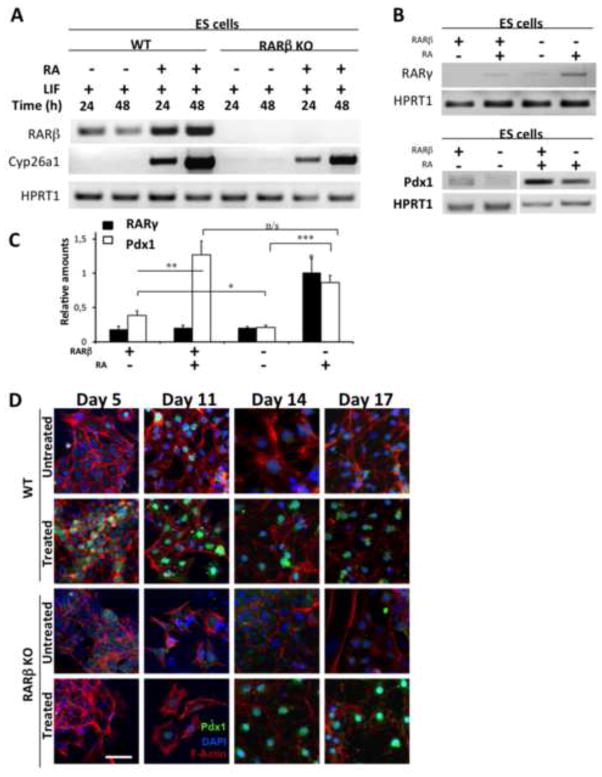
Figure 2. Impact of RARβ deletion on Pdx1 protein expression.
(A) RT-PCR analysis confirming the lack of RARβ transcripts in RARβKO ES cells. Analysis of Cyp26a1, a RA-responsive gene, demonstrates the presence of RA signaling activity via other receptors in RARβKO cells. HPRT1 was used as a loading control. (B) Representative RT-PCR analysis of Pdx1 and RARγ2 expression in WT and RARβ KO ES cells, control untreated (NT) and RA treated (1 μM, 48 h). (C) Relative amounts of Pdx1 and RARγ2, normalized to HPRT1 levels, are shown in the histogram (n=3; *: p≤0.047; **: p=0.001; ***: p≤0.0001). (D) Indirect immunofluorescence staining for Pdx1 protein (green) in WT and RARβKO cells at 5, 11, 14, and 17 days in the absence (untreated) or in the presence (treated) of growth factors used in the differentiation protocol. Cells were counterstained using rhodamine-conjugated phalloidin, which binds to F-actin (red), and nuclei were stained with DAPI (blue) (Bars = 50 μm).
We evaluated the expression profile of Pdx1, a master regulator of pancreatic development [40], in WT and RARβKO ES cells, in the presence or absence of RA treatment (Fig. 2B). RT-PCR experiments revealed lower levels of Pdx1 in RARβKO (55.2%, p=0.047) cells compared to WT in untreated conditions (Fig. 2B, C). Interestingly, RA treatment caused a remarkable increase in Pdx1 expression in both WT (~3.3-fold, p=0.0012) and RARβKO (~4.1-fold, p≤0.0001) cells compared to untreated conditions (Fig. 2B, C). Microarray-based transcriptome analyses performed on WT and RARβKO ES cells showed an increase in RARγ gene expression in RARβ-null cells following RA treatment (Suppl. Table S1). Although we observed no statistically significant changes in RARα expression in RARβKO ES cells compared to WT, RT-PCR analysis confirmed the induction of RARγ2 expression in RA-treated RARβKO ES cells (Fig 2B). The absence of RARβ, combined with low RARγ2 expression, lead to reduced Pdx1 levels (Fig. 2C). Moreover, the RA-dependent RARγ2 induction observed in RARβ-null cells (~5-fold, p=0.0162) correlates with a restoration of Pdx1 expression (Fig. 2C).
WT and RARβKO ES cells were differentiated into pancreatic endocrine cells, as described in Figure 1, and we performed indirect immunofluorescence staining at different time points to determine the Pdx1 protein expression profile (Fig. 2D). We detected Pdx1 protein in differentiating WT cells by day 5, and Pdx1 was detectable at all of the other differentiation stages tested (Fig. 2D). In contrast, Pdx1 protein was absent from nuclei of differentiating RARβKO cells at days 5 and 11, was only detected at day 14, and was only detected at a low level (Fig. 2D). These data indicate that the lack of RARβ results in a delay in the induction of Pdx1, which could potentially affect subsequent steps in endocrine specialization.
The Absence of RARβ expression impairs the pancreatic endocrine differentiation process
Decreased expression of pluripotency factors, including Nanog, in ES cells is required for proper ES cell differentiation [41]. By comparing Nanog transcript levels in WT and RARβKO ES cells, we observed the sustained expression of Nanog mRNA in RARβKO cells, while in WT Nanog mRNA levels declined over time (Fig. 3A). A similar phenomenon was also observed for other pluripotency factors, such as Pou5f1 and Sox2, in microarray experiments comparing WT and RARβKO ES cell transcriptomes (Suppl. Table S1 and Fig. S2). Furthermore, Neurogenin-3 (Ngn3), a master transcriptional regulator during the onset of pancreatic islet differentiation [40, 42, 43], displayed a transient induction pattern in WT but was not induced in RARβKO cells (Fig. 3A). The RARβ2 isoform mRNA was expressed during this ES differentiation protocol in WT cells, but not in RARβ KO cells (Fig. 3A–C). Hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNA levels did not change during the ES differentiation, and thus this transcript was used as a loading control in RNA expression assays (Fig. 3A–C).
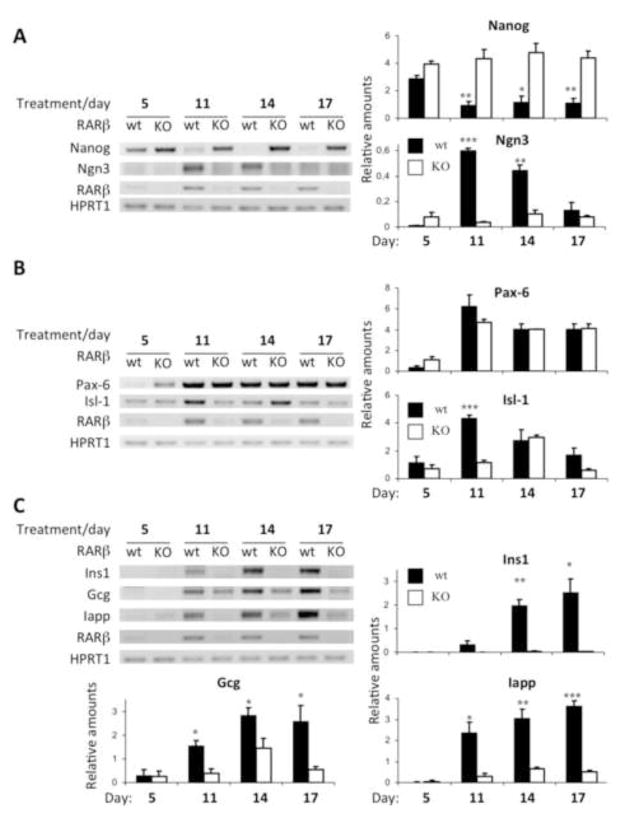
Figure 3. Expression of pancreatic differentiation markers in WT and RARβKO ES cells.
Transcript expression analyses of (A) early, (B) mid, and (C) late stage endocrine pancreatic differentiation markers in WT and RARβKO ES cells. RT-PCR analyses of (A) Nanog, Ngn3, (B) Pax6, Isl1, and (C) Ins1, Gcg, and Iapp mRNA were performed in both cell lines at 5, 11, 14, and 17 days of the differentiation protocol (Fig. 1A). Relative levels, normalized to HPRT1 levels for each marker tested, are shown in histograms (n=3; *: p≤0.05; **: p≤0.0079; ***: p≤0.0003).
Like Ngn3, Paired-box 6 (Pax6) and Islet1 (Isl1) are two transcription factors which are expressed from the intermediate (“mid”) to the terminally differentiated (“late”) stages [43–46]. While we noted no differences in Pax6 expression between WT and RARβKO, we detected the delayed expression of Isl1 in RARβKO as compared to WT cells (day 14 versus day 11) (Fig. 3B).
We also analyzed the expression of different, functional, endocrine differentiation markers, such as glucagon (Gcg; α-cells), insulin-1 (Ins1; β-cells) and islet amyloid polypeptide (IAPP; β-cells) [14, 47, 48] in WT and RARβ KO cells (Fig. 3C). RARβ KO cells exhibited reduced expression of these three transcripts as compared to WT (Fig. 3C). By day 17 Gcg, Ins1, and Iapp transcripts, respectively, were higher by ~5-fold (p=0.04), ~120-fold (p=0.013), and ~7-fold (p=0.0002) in WT as compared to RARβKO (Fig. 3C).
Taken together, these data indicate that retinoid signaling through RARβ plays a central role in ES differentiation to pancreatic endocrine cells by regulating the expression of certain master genes at early and intermediate stages of the differentiation process. The absence of RARβ results in impaired expression of functional markers of islet cells.
Discussion
By using an ES cell-based directed differentiation system we demonstrate a crucial role for RARβ in proper pancreatic endocrine cell differentiation. The absence of RARβ leads to a decrease in terminal pancreatic differentiation and to a great reduction in expression of functional markers, such as insulin1 and glucagon. We also conclude that Pdx1 expression during the pancreatic differentiation process is delayed in the absence of RARβ (Fig. 2). Pdx1 is a key transcription factor in the early determination of pancreatic progenitors and bud expansion [43, 44, 49, 50]. RA directly induces Pdx1 expression in WT ES cells [49]. Moreover, a putative retinoic acid response element (RARE) is located at ~3 kb upstream of the transcription start site of Pdx1 in F9 teratocarcinoma cells by ChIP-chip analyses (unpublished data). We speculate that RARα and RARβ together participate in the Pdx1 biphasic expression pattern during endocrine pancreatic differentiation, as reviewed by Soria [43]. However, the lack of RARβ would need to be compensated by RARγ, which is induced by RA treatment from day 8 through day 11 (Figure 1A and 2B, C). This would explain the delayed Pdx1 expression we observe in the RARβKO cells (Fig. 2D).
Pdx1 mis-expression was previously associated with severe β-cell dysfunction and increased cell death [51]. Accordingly, we show that the lack of RARβ causes a reduction in pancreatic β-cell terminal differentiation in this cell culture system, as assessed by markers such as Ins1 and Iapp (Fig. 3C). Recent findings by Dalgin et al. [52] in zebrafish also link RA signaling and endocrine cell fate. Our data suggest that the lack of RARβ also results in a decrease in α-cell differentiation, characterized by reduced expression of glucagon in our cell culture system (Fig. 3C).
Like Pdx1, the bHLH transcription factor Neurogenin3 (Ngn3) is a key protein in the commitment of endoderm cells to become pancreatic precursors [42, 44, 48]. Among the transcription factors involved in pancreas development, Ngn3 is the earliest to be expressed in the endocrine differentiation pathway [44, 53]. We found that the RARβKO ES cells displayed decreased levels of Ngn3 transcripts during pancreatic differentiation (Fig. 3A). Pax6 and Isl1 are two transcription factors that play roles in endocrine lineage specification after bud formation [46, 54]. Since Pax6 and Nkx6.1 transcript levels are not altered in the RARβKO cells (Fig. 3B and Fig. S1B) and Isl1 expression is only delayed (Fig. 3), we speculate that the absence of RA signaling through RARβ is not sufficient to abrogate completely endocrine differentiation, but may lead to significant defects in islet cell function. Moreover, the low, residual expression of Ngn3 mRNA detected in the RARβKO ES cells could account for the low levels of Gcg and Iapp mRNA observed in these cells (Fig. 3C). According to our observations, the directed-differentiation protocol used in our experiments gives raise to a mixed population of α, β, and δ-cells, considering the simultaneous detection of their respective markers Gcg, Ins1, and Sst (Figs. 1 and 3). Interestingly, the absence of RARβ in our model seems to specifically impair α and β-cell terminal differentiation, while Sst expression (δ-cell marker) is unchanged in RARβKO cells (Fig. S1A).
Another aspect of cell differentiation that is altered by the absence of RARβ is the pluripotency marker Nanog; Nanog expression persists in the mutant RARβ cells throughout the endocrine pancreatic differentiation protocol (Fig. 3A). Microarray analyses also highlighted the diminished capacity of RARβKO cells to repress other pluripotency genes, such as Pou5f1 and Sox2, upon RA treatment (Fig. S2). This was accompanied by the reduced induction of early pancreatic endoderm markers, such as Gata4, Gata6, and FoxA1 (Fig. S2) [44, 55]. Thus, our data suggest that RARβ, in addition to its role in Pdx1 transcriptional regulation, affects other aspects of ES cell differentiation in this cell culture system.
RARβ expression is known to depend on epigenetic regulation [56, 57]. Aberrant hypermethylation of the RARβ2 promoter was reported in different pancreatic disorders, such as cancer, diabetes, and chronic pancreatitis [58–60]. Therefore, we suggest that the epigenetic silencing of RARβ, combined with partial vitamin A deficiency, could play a causal role in various diseases involving the pancreas, including diabetes and pancreatic adenocarcinoma.
Conclusions
The production of insulin secreting endocrine cells from ES cells using RA-based protocols is a promising tool for diabetic therapy. Our findings provide new insights into the role of RARβ in pancreatic endocrine differentiation.
Supplementary Material
Highlights.
Retinoic Acid Receptor β controls aspects of differentiation
Nanog mRNA remains high in RARβ−/− ES cells
Pdx1 Expression is delayed and greatly reduced in RARβ−/− vs WT cells
Acknowledgments
We thank Dr. Lavoisier Ramos for providing INS-1E cells, and Dr. Pierre Chambon for the RARβKO mice. We also thank Drs. Kristian B. Laursen, Steve Trasino, and Naira Rezende for their scientific input, and Tamara Weissman for editorial assistance. This research was supported by funds from Weill Cornell Medical College, and in part by NIH T32 DK07313, NIH T32 CA062948, NCI R01CA043796 (to LJG) and by the Fonds de Recherche en Santé du Québec (PF1-Benoit-25389) (to YDB).
Footnotes
Abbreviations: ES, embryonic stem; Gcg, glucagon; Iapp, islet amyloid polypeptide; Ins1, insulin 1; KO, knockout; Ngn3, Neurogenin3; RA, all-trans retinoic acid; RAR, retinoic acid receptor; RARE, retinoic acid response element; Sst, somatostatin; WT, wild-type.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guariguata L, Whiting D, Weil C, Unwin N. The International Diabetes Federation diabetes atlas methodology for estimating global and national prevalence of diabetes in adults. Diabetes Res Clin Pract. 2011;94:322–332. doi: 10.1016/j.diabres.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 2.Whiting DR, Guariguata L, Weil C, Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract. 2011;94:311–321. doi: 10.1016/j.diabres.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 3.Huang ES, Basu A, O’Grady M, Capretta JC. Projecting the future diabetes population size and related costs for the U.S. Diabetes Care. 2009;32:2225–2229. doi: 10.2337/dc09-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waldron-Lynch F, Herold KC. Immunomodulatory therapy to preserve pancreatic beta-cell function in type 1 diabetes. Nat Rev Drug Discov. 2011;10:439–452. doi: 10.1038/nrd3402. [DOI] [PubMed] [Google Scholar]
- 6.Waldron-Lynch F, von Herrath M, Herold KC. Towards a curative therapy in type 1 diabetes: remission of autoimmunity, maintenance and augmentation of beta cell mass. Novartis Found Symp. 2008;292:146–155. doi: 10.1002/9780470697405.ch14. discussion 155–148, 202–143. [DOI] [PubMed] [Google Scholar]
- 7.Charbonnel B, Penfornis A, Varroud-Vial M, Kusnik-Joinville O, Detournay B. Insulin therapy for diabetes mellitus: Treatment regimens and associated costs. Diabetes Metab. 2011 doi: 10.1016/j.diabet.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weir GC, Cavelti-Weder C, Bonner-Weir S. Stem cell approaches for diabetes: towards beta cell replacement. Genome Med. 2011;3:61. doi: 10.1186/gm277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sui J, Mehta M, Shi B, Morahan G, Jiang FX. Directed Differentiation of Embryonic Stem Cells Allows Exploration of Novel Transcription Factor Genes for Pancreas Development. Stem Cell Rev. 2012;1:1–10. doi: 10.1007/s12015-011-9346-3. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Yehudah A, White C, Navara CS, Castro CA, Ize-Ludlow D, Shaffer B, Sukhwani M, Mathews CE, Chaillet JR, Witchel SF. Evaluating protocols for embryonic stem cell differentiation into insulin-secreting beta-cells using insulin II-GFP as a specific and noninvasive reporter. Cloning Stem Cells. 2009;11:245–257. doi: 10.1089/clo.2008.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blyszczuk P, Czyz J, Kania G, Wagner M, Roll U, St-Onge L, Wobus AM. Expression of Pax4 in embryonic stem cells promotes differentiation of nestin-positive progenitor and insulin-producing cells. Proc Natl Acad Sci U S A. 2003;100:998–1003. doi: 10.1073/pnas.0237371100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormoneexpressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 15.Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D’Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucoseresponsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 16.Micallef SJ, Janes ME, Knezevic K, Davis RP, Elefanty AG, Stanley EG. Retinoic acid induces Pdx1-positive endoderm in differentiating mouse embryonic stem cells. Diabetes. 2005;54:301–305. doi: 10.2337/diabetes.54.2.301. [DOI] [PubMed] [Google Scholar]
- 17.Mongan NP, Gudas LJ. Diverse actions of retinoid receptors in cancer prevention and treatment. Differentiation. 2007;75:853–870. doi: 10.1111/j.1432-0436.2007.00206.x. [DOI] [PubMed] [Google Scholar]
- 18.Niederreither K, Dollé P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 19.Boylan JF, Lohnes D, Taneja R, Chambon P, Gudas LJ. Loss of retinoic acid receptor gamma function in F9 cells by gene disruption results in aberrant Hoxa-1 expression and differentiation upon retinoic acid treatment. Proc Natl Acad Sci U S A. 1993;90:9601–9605. doi: 10.1073/pnas.90.20.9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Means AL, Gudas LJ. The roles of retinoids in vertebrate development. Annu Rev Biochem. 1995;64:201–233. doi: 10.1146/annurev.bi.64.070195.001221. [DOI] [PubMed] [Google Scholar]
- 21.Kashyap V, Gudas LJ, Brenet F, Funk P, Viale A, Scandura JM. Epigenomic reorganization of the clustered Hox genes in embryonic stem cells induced by retinoic acid. J Biol Chem. 2011;286:3250–3260. doi: 10.1074/jbc.M110.157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Pan FC, Brandes N, Afelik S, Solter M, Pieler T. Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Dev Biol. 2004;271:144–160. doi: 10.1016/j.ydbio.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 23.Ostrom M, Loffler KA, Edfalk S, Selander L, Dahl U, Ricordi C, Jeon J, Correa-Medina M, Diez J, Edlund H. Retinoic acid promotes the generation of pancreatic endocrine progenitor cells and their further differentiation into beta-cells. PLoS One. 2008;3:e2841. doi: 10.1371/journal.pone.0002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews KA, Rhoten WB, Driscoll HK, Chertow BS. Vitamin A deficiency impairs fetal islet development and causes subsequent glucose intolerance in adult rats. J Nutr. 2004;134:1958–1963. doi: 10.1093/jn/134.8.1958. [DOI] [PubMed] [Google Scholar]
- 25.Chertow BS, Blaner WS, Baranetsky NG, Sivitz WI, Cordle MB, Thompson D, Meda P. Effects of vitamin A deficiency and repletion on rat insulin secretion in vivo and in vitro from isolated islets. J Clin Invest. 1987;79:163–169. doi: 10.1172/JCI112778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolle P, Ruberte E, Leroy P, Morriss-Kay G, Chambon P. Retinoic acid receptors and cellular retinoid binding proteins. I. A systematic study of their differential pattern of transcription during mouse organogenesis. Development. 1990;110:1133–1151. doi: 10.1242/dev.110.4.1133. [DOI] [PubMed] [Google Scholar]
- 27.Ghyselinck NB, Dupé V, Dierich A, Messaddeq N, Garnier JM, Rochette-Egly C, Chambon P, Mark M. Role of the retinoic acid receptor beta (RARbeta) during mouse development. Int J Dev Biol. 1997;41:425–447. [PubMed] [Google Scholar]
- 28.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 29.Martinez-Ceballos E, Chambon P, Gudas LJ. Differences in gene expression between wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitory factor (LIF) removal. J Biol Chem. 2005;280:16484–16498. doi: 10.1074/jbc.M414397200. [DOI] [PubMed] [Google Scholar]
- 30.Laursen KB, Wong PM, Gudas LJ. Epigenetic regulation by RARalpha maintains ligand-independent transcriptional activity. Nucleic Acids Res. 2012;40:102–115. doi: 10.1093/nar/gkr637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benoit YD, Lussier C, Ducharme PA, Sivret S, Schnapp LM, Basora N, Beaulieu JF. Integrin alpha8beta1 regulates adhesion, migration and proliferation of human intestinal crypt cells via a predominant RhoA/ROCK-dependent mechanism. Biol Cell. 2009;101:695–708. doi: 10.1042/BC20090060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spokoini R, Kfir-Erenfeld S, Yefenof E, Sionov RV. Glycogen synthase kinase-3 plays a central role in mediating glucocorticoid-induced apoptosis. Mol Endocrinol. 2010;24:1136–1150. doi: 10.1210/me.2009-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otonkoski T, Beattie GM, Mally MI, Ricordi C, Hayek A. Nicotinamide is a potent inducer of endocrine differentiation in cultured human fetal pancreatic cells. J Clin Invest. 1993;92:1459–1466. doi: 10.1172/JCI116723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lumelsky N, Blondel O, Laeng P, Velasco I, Ravin R, McKay R. Differentiation of embryonic stem cells to insulin-secreting structures similar to pancreatic islets. Science. 2001;292:1389–1394. doi: 10.1126/science.1058866. [DOI] [PubMed] [Google Scholar]
- 36.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 37.Hosler BA, LaRosa GJ, Grippo JF, Gudas LJ. Expression of REX-1, a gene containing zinc finger motifs, is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol Cell Biol. 1989;9:5623–5629. doi: 10.1128/mcb.9.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchand M, Schroeder IS, Markossian S, Skoudy A, Negre D, Cosset FL, Real P, Kaiser C, Wobus AM, Savatier P. Mouse ES cells over-expressing the transcription factor NeuroD1 show increased differentiation towards endocrine lineages and insulin-expressing cells. Int J Dev Biol. 2009;53:569–578. doi: 10.1387/ijdb.092856mm. [DOI] [PubMed] [Google Scholar]
- 39.Langton S, Gudas LJ. CYP26A1 knockout embryonic stem cells exhibit reduced differentiation and growth arrest in response to retinoic acid. Dev Biol. 2008;315:331–354. doi: 10.1016/j.ydbio.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 40.Bernardo AS, Hay CW, Docherty K. Pancreatic transcription factors and their role in the birth, life and survival of the pancreatic beta cell. Mol Cell Endocrinol. 2008;294:1–9. doi: 10.1016/j.mce.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, Mongan NP. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rukstalis JM, Habener JF. Neurogenin3: a master regulator of pancreatic islet differentiation and regeneration. Islets. 2009;1:177–184. doi: 10.4161/isl.1.3.9877. [DOI] [PubMed] [Google Scholar]
- 43.Soria B. In-vitro differentiation of pancreatic beta-cells. Differentiation. 2001;68:205–219. doi: 10.1046/j.1432-0436.2001.680408.x. [DOI] [PubMed] [Google Scholar]
- 44.Van Hoof D, D’Amour KA, German MS. Derivation of insulin-producing cells from human embryonic stem cells. Stem Cell Res. 2009;3:73–87. doi: 10.1016/j.scr.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Gosmain Y, Katz LS, Masson MH, Cheyssac C, Poisson C, Philippe J. Pax6 is crucial for beta-cell function, insulin biosynthesis, and glucose-induced insulin secretion. Mol Endocrinol. 2012;26:696–709. doi: 10.1210/me.2011-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 47.Naujok O, Francini F, Picton S, Bailey CJ, Lenzen S, Jorns A. Changes in gene expression and morphology of mouse embryonic stem cells on differentiation into insulinproducing cells in vitro and in vivo. Diabetes Metab Res Rev. 2009;25:464–476. doi: 10.1002/dmrr.965. [DOI] [PubMed] [Google Scholar]
- 48.Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci U S A. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai J, Yu C, Liu Y, Chen S, Guo Y, Yong J, Lu W, Ding M, Deng H. Generation of homogeneous PDX1(+) pancreatic progenitors from human ES cell-derived endoderm cells. J Mol Cell Biol. 2010;2:50–60. doi: 10.1093/jmcb/mjp037. [DOI] [PubMed] [Google Scholar]
- 50.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 51.Fujimoto K, Polonsky KS. Pdx1 and other factors that regulate pancreatic beta-cell survival. Diabetes Obes Metab. 2009;11(Suppl 4):30–37. doi: 10.1111/j.1463-1326.2009.01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalgin G, Ward AB, Haole T, Beattie CE, Nechiporuk A, Prince VE. Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development. 2011;138:4597–4608. doi: 10.1242/dev.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vetere A, Marsich E, Di Piazza M, Koncan R, Micali F, Paoletti S. Neurogenin3 triggers beta-cell differentiation of retinoic acid-derived endoderm cells. Biochem J. 2003;371:831–841. doi: 10.1042/BJ20021524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dohrmann C, Gruss P, Lemaire L. Pax genes and the differentiation of hormone-producing endocrine cells in the pancreas. Mech Dev. 2000;92:47–54. doi: 10.1016/s0925-4773(99)00324-x. [DOI] [PubMed] [Google Scholar]
- 55.Chen C, Zhang Y, Sheng X, Huang C, Zang YQ. Differentiation of embryonic stem cells towards pancreatic progenitor cells and their transplantation into streptozotocin-induced diabetic mice. Cell Biol Int. 2008;32:456–461. doi: 10.1016/j.cellbi.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 56.Sirchia SM, Ren M, Pili R, Sironi E, Somenzi G, Ghidoni R, Toma S, Nicolo G, Sacchi N. Endogenous reactivation of the RARbeta2 tumor suppressor gene epigenetically silenced in breast cancer. Cancer Res. 2002;62:2455–2461. [PubMed] [Google Scholar]
- 57.Youssef EM, Estecio MR, Issa JP. Methylation and regulation of expression of different retinoic acid receptor beta isoforms in human colon cancer. Cancer Biol Ther. 2004;3:82–86. doi: 10.4161/cbt.3.1.591. [DOI] [PubMed] [Google Scholar]
- 58.House MG, Herman JG, Guo MZ, Hooker CM, Schulick RD, Lillemoe KD, Cameron JL, Hruban RH, Maitra A, Yeo CJ. Aberrant hypermethylation of tumor suppressor genes in pancreatic endocrine neoplasms. Ann Surg. 2003;238:423–431. doi: 10.1097/01.sla.0000086659.49569.9e. discussion 431–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato N, Fukushima N, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2008;21:238–244. doi: 10.1038/modpathol.3800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, Calonne E, Volkmar U, Igoillo-Esteve M, Naamane N, Del Guerra S, Masini M, Bugliani M, Marchetti P, Cnop M, Eizirik DL, Fuks F. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.