Abstract
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide, with a median survival of 6–16 m. Factors responsible for the poor prognosis include late onset diagnosis, underlying cirrhosis and resistance to chemotherapy; 40% of HCCs are clonal and therefore potentially arise from progenitor/stem cells. New insights are provided from several signaling pathways, such as STAT3, NOTCH, hedgehog and transforming growth factor-β (TGFβ), which are involved in stem cell renewal, differentiation, survival, and are commonly deregulated in HCC. Control of stem cell proliferation by the TGFβ, Notch, Wnt and Hedgehog pathways to suppress hepatocellular cancer and to form the endoderm suggest a dual role for this pathway in tumor suppression as well as progression of differentiation from a stem or progenitor stage. This review provides a rationale for detecting and analyzing tumor stem cells as one of the most effective ways to treat cancers such as hepatocellular cancer.
Keywords: TGF, 2-SPECTRIN, stem/progenitor cells, hepatocellular cancer, signal transduction
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide, with a median survival of 6–16 mo. A total of 22,620 new liver and intrahepatic bile duct cancer cases and 18,160 deaths from liver and intrahepatic bile duct cancer are projected to occur in the United States in 2009.1,2 Factors responsible for the poor prognosis include late onset diagnosis, underlying cirrhosis and resistance to chemotherapy; 40% of HCCs are clonal and therefore potentially arise from progenitor/stem cells.3–5
Cancer stem cells (CSCs), while first proposed over 40 y ago, have only recently been identified in hematological malignancies and solid tumors such as liver, breast, prostate, brain and colon.5–9 Stem cells are generally characterized by their capacity for self-renewal through asymmetrical cell division, “multipotency” or the ability to produce progeny in at least two lineages, long-term tissue reconstitution and serial transplantability.10,11 For tumors containing a subpopulation of cancer stem cells, there are at least two proposed mechanisms of CSC origin: oncogenic mutations and/or epigenetic aberrations may inactivate the constraints on normal stem cell expansion, or alternatively, oncogenic mutations and/or epigenetic aberrations in a more differentiated cell generate continually proliferating of cells that no longer enter a post-mitotic differentiated state, thereby creating a pool of self-renewing cells in which further mutations can accumulate.12,13 Potentially, biologically significant pathways that modulate these stem/progenitor cells in cancer tissue could be identified through dual roles in embryonic stem cell development and tumor activation or suppression.4
The plasticity of such cells is reflected by recent studies where pluripotent stem cells from embryonic or adult fibroblasts can be induced by introducing four factors: Oct3/4, Sox2, c-Myc and Klf4, under embryonic stem cell culture conditions.14–16 Conceivably, alterating stem cell/cell cycle activators such as Wnt/β-catenin, Hedgehog, Notch and TGFβ signaling systems or targets such as β-catenin, h-TERT, CDKs, Myb or c-MYC could expedite tumorigenesis or conversely, by specifically targeting these proteins, serve as a powerful cancer-prevention tool. In fact, we have previously demonstrated that disruption of TGFβ signaling by disruption of β2-SPECTRIN (β2SP), a non-pleckstrin homology (PH) domain β-general-spectrin (also known as Embryonic Liver Fodrin isoform, ELF or Spectrin β, non-erythrocytic 1 isoform 2), is critical to the development of hepatocellular cancer.17 Immunohistochemical staining and confocal analysis of HCC revealed cells that label with stem cell markers and have unexpectedly lost TBRII and β2-SPECTRIN. Expression analysis of these tumors revealed marked activation of the IL-6 pathway, suggesting that HCC may develop from IL-6 driven CSCs with disrupted TGFβ signaling. Suppression of IL-6 signaling through the generation of mouse knockouts with a positive IL-6 regulator, ITIH-4, resulted in a reduction of HCC in β2-Spectrin+/− mice, thereby demonstrating a link between two major signaling pathways in the development of cancer and suggesting a possible therapeutic target.5 However, the specific stages that tissue-specific stem cells enter in order to develop into differentiated cells such as hepatocytes and cholangiocytes and the mechanisms of asymmetrical cell division still need to be addressed. The details of the hierarchical biologically functional systems necessary for generating liver stem cell asymmetry will likely come from genetics and in vivo loss-of-function studies.
Human Liver Progenitor Cells and Cancer Stem Cells
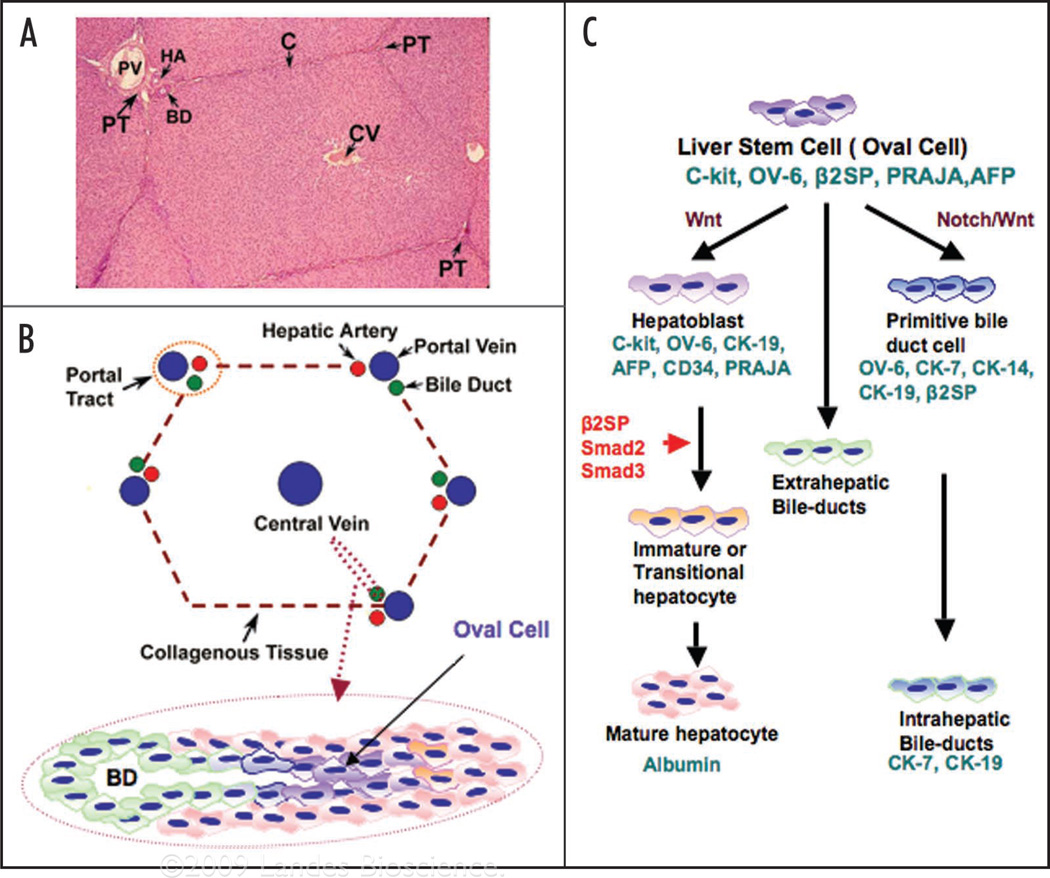
Hepatocytes and cholangiocytes are believed to arise from a liver progenitor cell (called an oval cell). The oval cell, a small cell with an oval nucleus and scanty cytoplasm, is derived from a liver stem cell niche located close to the portal space and is considered to be a progenitor cell with the ability to differentiate into hepatocytes or cholangiocytes (Fig. 1). The existence of a hepatic stem cell compartment gives rise to expectations regarding the practical applications of such research. Understanding and identifying the bipotential hepatic stem cells may hold promise for new therapeutic treatments for a wide range of liver pathologies ranging from congenital metabolic diseases, end-stage liver cirrhosis and hepatocarcinogenesis. Human hepatic stem cells most likely can give rise to hepatocellular carcinoma as well as cholangiocarcinomas.18,19 In these models, a periportal population of small “primitive” oval epithelial cells proliferate either in association with or before hepatocyte multiplication. Several studies have shown a progenitor cell phenotype in a substantial number of hepatocellular carcinomas (HCCs). Detailed immunophenotyping of HCCs revealed that 28–50% of HCCs express markers of progenitor cells such as CK7 and CK19. These tumors also consist of cells that have an intermediate phenotype between progenitors and mature hepatocytes. In fact, HCCs that express hepatocyte and biliary cell markers such as albumin, CK7 and CK19 carry a significantly poorer prognosis and higher recurrence after surgical resection and liver transplantation.20 Nevertheless, the question remains whether this immature intermediate phenotype represents progenitor cell differentiation arrest or dedifferentiation of mature hepatocytes. Histological and immunophenotyping studies favor the progenitor cell differentiation arrest model. Fifty-five percent of small dysplastic foci (less than 1 mm in size), which represent the earliest premalignant lesions, are comprised of progenitor cells and intermediate hepatocytes.21 Moreover, a side population of cells, with characteristics of both hepatocytic and cholangiocytic lineages, sorted from the human HCC cell lines huh7 and PLC/PRF/5 cells, was found to give rise to persistently aggressive tumors upon serial transplantation in immunodeficient NOD/SCID mice.22 Furthermore, mice homozygous for β2-Spectrin (β2SP−/−) undergo mid-gestational death with hypoplastic livers.17 Heterozygous β2SP+/− mice, however, spontaneously developed tumors of the liver with an incidence of over 40%. The liver lesions included early centrilobular steatosis, dysplasia in most sections, with nuclear disarray and stratification, mitosis and apoptosis, proceeding to poorly differentiated carcinoma. Thus interruption of the TGFβ pathway could result in hepatocellular carcinoma through disruption of a normal pattern of cellular differentiation by hepatic progenitor/stem cells (Fig. 2).
Figure 1.
Liver stem cell and TGFβ signaling via Smad proteins in liver development. (A) H&E staining for hepatic lobule; (B) Schematic representation of hepatic lobule and oval cell (liver progenitor cell); (C) Schematic diagram of TGFβ signaling in hepatocyte precursors directs development and differentiation. Smad 2, 3 and β2-SPECTRIN expression increases through development. Liver progenitor cells (LPCs) and rodent oval cells express phenotypic markers of both (immature) hepatocytes (α-fetoprotein and albumin) and bile duct cells (such as biliary-type CKs, oval cell marker 6, CK7, CK14 and CK19) as well as hematopoietic markers (such as the receptor for stem cell factor, c-kit and CD34). Another TGFβ adaptor protein, PRAJA, and c-kit expression decrease through development.
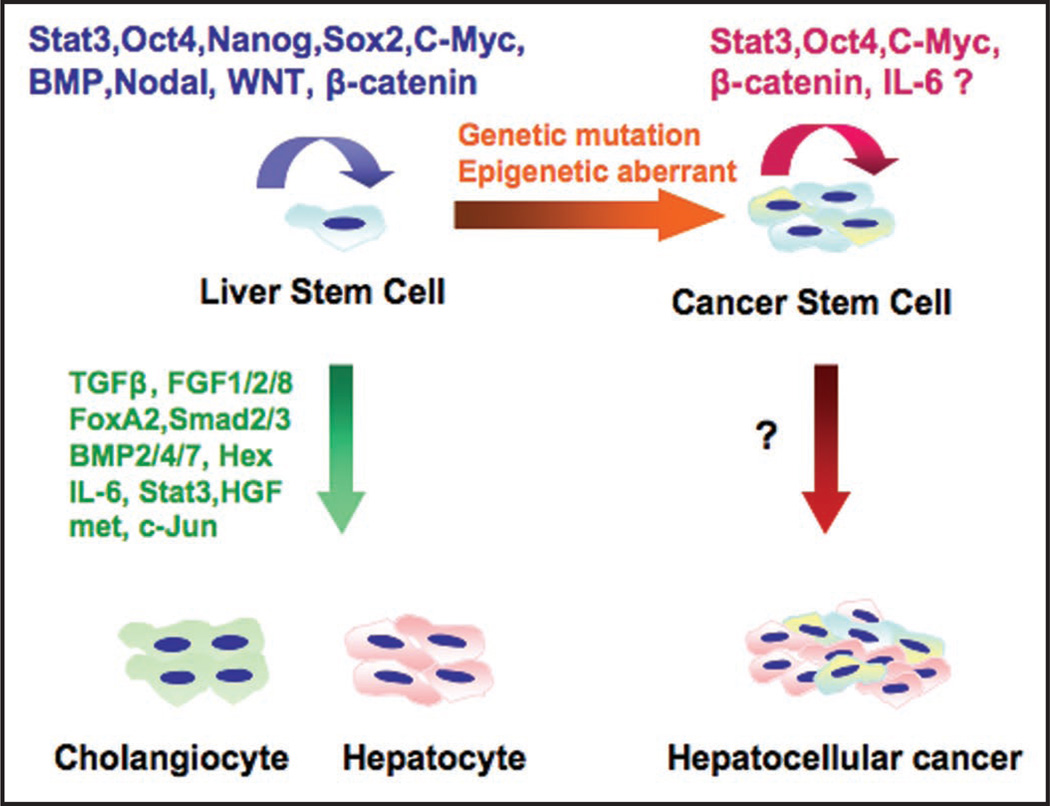
Figure 2.
Directed stem cell renewal and liver differentiation by multiple signaling pathways. Proteins that direct self-renewal of embryonic stem cells and then differentiation of stem cells into mature hepatocytes and cholangiocytes. The mechanisms directing transformation of stem cells to cancer stem cells and hepatocellular cancer, however, remain to be elucidated.
The Notch and Wnt Pathway and Cancer Stem Cells
Multiple signaling pathways control stem cell proliferation. The Notch signaling pathway plays an important role in stem cell self-renewal and differentiation.7,23–26 Other signaling pathways influence whether Notch functions as a oncogene or tumor suppressor in a particular tissue.27 The activated intracellular form of Notch3, as well as the Notch ligand Jagged, are highly expressed in HCC.28–30 Notch-dependent transformation is associated with ERK activation downstream of the Ras pathway, which increases Notch mRNA stability and is required for transcription of the Notch target gene, Hes-1.31–33 Conversely, Notch-1 has been reported to function as a tumor suppressor and participate in cross-talk with other signaling pathways such as Ras/Raf/MEK/ERK through the regulation of the PTEN tumor suppressor.34 Recent evidence indicates that activation of Notch1 signaling will increase death receptor 5 (DR5) expression with enhancement of TRAIL-induced apoptosis in HCC cells; overexpressing Notch1 in HCC cell lines results in inhibiting HCC growth and inducing apoptosis in vitro and in vivo.35,36
The Wnt signaling pathway also plays a crucial role in regulating stem and progenitor cell expansion.8,37 Among the 19 members of the Wnt family, Wnt-1, -2, -3, -3a and 10b are ligands for the canonical Wnt pathway, which activates β-catenin/TCF-mediated transcription and induces transformation of mammary epithelial cells.38,39 Wnt-1 was originally identified as the Int-1 gene activated by retroviral insertion of MMTV in the mammary gland.40 The Wnt pathway stabilizes β-catenin, a coactivator of the TCF transcription factor family, in its monomeric form.41 Activation of the Wnt receptor, Frizzled, in association with its co-receptor LRP-5/6, activates disheveled, which results in dissociation of the tetrameric GSK3β/β-catenin/APC/axin complex, reduction of β-catenin phosphorylation, β-catenin stabilization, and its entry into the nucleus where it associates with and coactivates TCF. Stabilization of β-catenin can result from direct inhibition of GSK3β by PKC and ERK, and transformation of mammary epithelial cells resulting from increased β-catenin/TCF signaling has been linked to PKCα activation downstream of PDK1.42–47 Conversely, GSK3β activation phosphorylates β-catenin and axin to target β-catenin for ubiquitination and proteasomal degradation. In differentiated cells, β-catenin binds to cadherins, as well as other tight-junction proteins, to form adherens junctions that are important for lumen integrity and secretory epithelial cell function.39
Interplay between the Wnt, Hedgehog, BMP and Notch pathways often determines whether stem cells self-renew or differentiate in epithelial tissues. For instance, Wnt signaling is activated in the colonic crypts and maintains cells in a proliferative state. Increased activity of the Wnt pathway leads to enlarged crypts and intestinal tumors whereas, Wnt inhibition results in loss of the stem cell compartment altogether. Wnt signaling is essential for maintenance of this stem cell compartment and regulates cellular differentiation.48 This effect of Wnt signaling can be mimicked by stabilizing cytoplasmic/nuclear β-catenin alone, and the effects of Wnt signaling on stem cells are modulated through association with other signaling pathways including Notch, SHH and TGFβ signaling.4,49,50 Understanding of the regulation of this stem cell compartment has come mostly from examining skin, intestinal and hematopoietic cells. Mutations and deletions of AXIN1 and AXIN2 and overexpression of β-catenin and FZD-7 have been described in 17–40% of human HCCs analyzed.51–56 Not all studies, however, demonstrate a correlation between elevated nuclear β-catenin and expression of its transcriptional targets in HCC, suggesting that the expression of these target genes is also regulated by alternative signaling pathways.57–59
In addition, Wnt signaling can promote stem cell renewal and rescue of the hematopoietic stem cell (HSC) compartment after irradiation either through the canonical (Wnt-3a) or possibly also through the non-canonical (Wnt-5a) pathway.60,61 In fact, repopulation of a HSC compartment from CD34+ cells was augmented more than three fold through intraperitoneal injection of Wnt-5a alone. These effects could be mimicked through activation of the Wnt signaling pathway by a degradation-resistant β-catenin, while HSC compartment reconstitution can be inhibited by overexpression of Axin.60 How the Wnt pathway promotes stem cell renewal and is involved in liver cancer stem cells, however, is still unclear. Meanwhile, Notch acts jointly with Wnt to sustain stem cell proliferation, and is essential for the differentiation of specific cell types. Moreover, Hedgehog signaling promotes differentiation and restricts crypt formation that is mediated through its effect on BMP signaling.62
Liver Cancer Stem Cells and the TGFβ Signaling Pathway
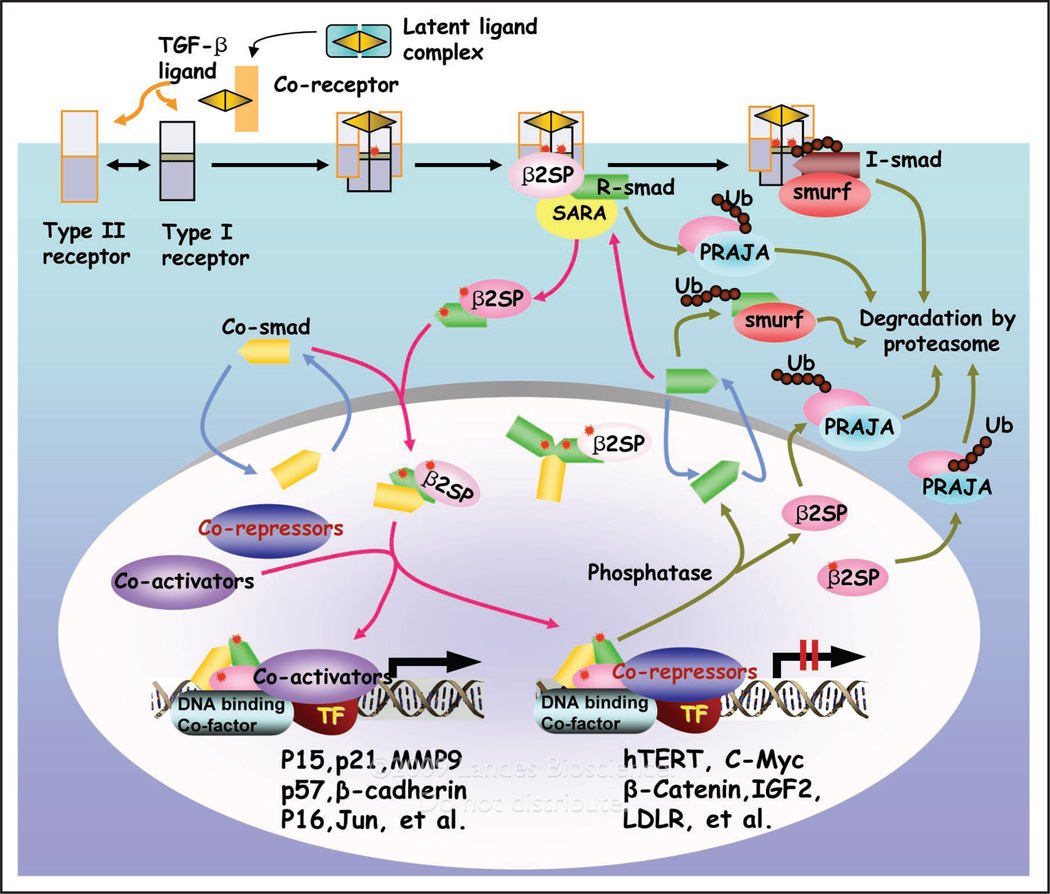
Recent evidence also suggests a key role for the TGFβ signaling pathway in both foregut cancer suppression and normal gut endoderm development, suggesting a dual role in liver tumor suppression as well as the transition of stem cells to a progenitor and fully differentiated phenotype.4,63 The TGFβ signaling pathway appears to be most prominent at the interface between development and cancer in liver and gut epithelial cells.4 Smad signaling has been shown to be pivotal for embryonic hepatocyte proliferation, as well as in the formation of GI cancers.64–66 Smad activation is modulated by various receptor- or Smad-interacting proteins that include ubiquitin and SUMO (small ubiquitin-related modifier) ligases, as well as multiple adaptor proteins that include SARA (Smad anchor for receptor activation), Filamin and β2-SPECTRIN. β2-SPECTRIN, a β-spectrin first isolated from foregut endodermal stem cell librairies, is crucial for the propagation of TGFβ signaling.67 Specifically, β2-SPECTRIN associates with Smad3, presenting it to the cytoplasmic domain of the TGFβ Type I receptor complex; followed by heteromeric complex association with Smad4, nuclear translocation and target gene activation (Fig. 3).17 Disruption of β2-SPECTRIN in mice leads to disruption of TGFβ signaling, resulting in a phenotype similar to Smad2+/−/Smad3+/− mutant mice with mid-gestational death due to gastrointestinal, liver, neural and heart defects, and loss of intrahepatic bile ducts. Interestingly, while the liver lineage is established, hepatocytes are poorly formed and liver architecture is lost with an absence of primitive bile ducts as in the Smad2+/−/Smad3+/− mutants. Moreover, bile duct formation can be induced in liver explant cultures treated with TGFβ. In this model of haploinsufficiency, the addition of TGFβ appears sufficient to drive up Smad2 and Smad3 levels and the formation of a limiting plate and bile ducts. Moreover, a TGFβ/BMP regulated protein PRAJA is expressed in hepatoblasts and modulates β2SP and Smad3, adding to the complexity of TGFβ signaling in the liver.68
Figure 3.
TGF ligands signal through distinct receptors and Smads that are modulated by adaptor proteins and ubiquitinators. TGF binds to serine threonine kinase receptor complexes that phosphorylate R-Smads (Smad2/Smad3) as well as adaptor proteins such as 2-spectrin (2SP). R-Smads, 2-spectrin and Smad4 form a heteromeric complex, and translocate to the nucleus, activating target genes. At all levels, Smad modulation occurs through adaptor proteins as well as E3 ligases such as PRAJA and Smurfs, generating the diverse and complex signals. Arrows indicate signal flow and are color coded: black for ligand and receptor activation, red for Smad and receptor activation, olive green for Smad and receptor inactivation and formation of a transcriptional complex, and blue for Smad nucleocytoplasmic shuttling. Phosphate groups and ubiquitin (Ub) are represented by red stars and dark red circles, respectively.
Thus, emerging new data indicate that TGFβ is important for a stem cell to transition to a progenitor cell phenotype and ultimately its conversion to a fully differentiated phenotype in the liver and biliary system. Ultimately, the common mediators Smad4 and β2SP heterozygotes survive to adulthood only to succumb to a gastro-intestinal cancers. β2SP+/− heterozygous mice develop HCC.21,69–71 Moreover, inactivation of at least one of the TGFβ signaling components occurs in almost all gastrointestinal tumors.4,69 Thus, it is conceivable that tumors arise in organs lacking crucial differentiating factors such as Smad2 with Smad3 at the progenitor to transitional stages or at the stages when stem cells divide into progenitor cells, which then develop into immature epithelial cells. The data support the idea that the absence of TGFβ driven epithelial differentiation favors carcinogenesis. Impaired TGFβ signaling may distinguish cancer stem cells from normal stem cells and give clues toward identifying a human progenitor cell pool and other functional pathways that become activated in such “cancer stem/progenitor cells.”
Epigenetic Regulation of DNA Replication in Liver Cancer Stem Cells and HCC
Although the initiation of DNA replication is considered to be highly coordinated through multiple protein-DNA and protein-protein interactions, it is not well understood how particular locations within the eukaryotic chromosome are selected as origins of DNA replication. Chromatin organization depends on epigenetic information encoded in postsynthetic modifications of histones and of DNA itself rather than on particular nucleotide sequences.72 The role of epigenetic control in regulating the initiation of DNA replication is poorly understood.
Accurate duplication of genetic material is central to cell proliferation. In eukaryotes, S phase is tightly controlled during development. For instance, initiation events occur at random sites in early embryos but later appear restricted to preferred DNA regions. Epigenetic changes dependent on chromatin organization and/or the availability of specific factors likely control origin choice. The use of the dynamic molecular combing technology coupled with specific labeling of neo-synthesized DNA and FISH (fluorescent in situ hybridization) demonstrated that the efficiency and spacing of initiation sites are still flexible in mammalian somatic cells, and strongly rely on nucleotide availability. In all conditions, initiation events appear confined to short AT-rich sequences previously identified as matrix attachment regions, which suggests a direct involvement of these features in origin specification.73,74
Histones are subject to several post-translational modifications that occur mainly in their N-termini including methylation, acetylation, phosphorylation, sumoylation and ubiquitination. Histone acetylation75 and Histone 3 methylation76,77 appear to be important molecular events during the initiation of DNA replication, but these processes are neither sufficient nor necessary to determine the initiation of DNA replication. Furthermore, they are also not sufficient to dictate the spatial and temporal regulation of chromosomal domains.78 Therefore, it is likely that multiple histone modifications collaborate to specify the replication program in vivo.
In mammals, DNA methylation occurs by covalent modification at the fifth carbon (C5) of the pyrimidine ring of approximately 4% of all cytosines and is mostly located in CpG dinucleotides. A family of methylated DNA binding proteins (MDBs) binds to methylated CpGs that, in general, contribute to transcriptional silencing by interacting with histone deacetylases and transcriptional corepressors.79 The effect of DNA methylation on ORI activity has been addressed in mouse and human inactive X chromosomes, where most CpG islands are methylated and transcriptionally silent as opposed to their nonmethylated and expressed status in the active homologues. The results showed that ORIs at active nonmethylated CpG islands replicated earlier than their inactive methylated alleles during the S phase.80 This evidence indicates that CpG island methylation does not prevent initiation of DNA replication but completely abolished transcription.81,82
The function of CpG dinucleotide methylation is to maintain the stability of chromosome structures. Satellite DNA in the heterochromatin, in particular, is more heavily methylated at the CpG islands than the rest of the genomic DNA. It has been postulated that under-methylated satellite sequences confer abnormal chromatin structures and predispose to chromosomal instability by enhancing chromosome recombinations. A high frequency of chromosome rearrangements with peri-centromeric or heterochromatic breakpoints have been observed in many tumors, which may suggest a relationship between satellite hypomethylation, chromosome instability and carcinogenesis. Wong et al. demonstrated the role of DNA hypomethylation in 1q copy gain in HCC by examining the methylation status of chromosome 1 heterochromatin DNA (band 1q12). Thirty-six histologically confirmed samples of HCC were studied (24 samples consisted of paired tumor and adjacent nontumorous liver tissues while 12 samples were of the tumor only) and suggest a role for Satellite 2 DNA demethylation in the early stages of the stepwise progression of liver carcinogenesis.83
Cell division cycle 6 (CDC6) is an essential regulator of DNA replication in eukaryotic cells. Its best-characterized function is the assembly of prereplicative complexes at origins of replication during the G1 phase of the cell division cycle. However, CDC6 also plays important roles in the activation and maintenance of the checkpoint mechanisms that coordinate S phase and mitosis, and recent studies have unveiled its proto-oncogenic activity. CDC6 overexpression interferes with the expression of INK4/ARF tumor suppressor genes through a mechanism involving the epigenetic modification of chromatin at the INK4/ARF locus. In addition, CDC6 overexpression in primary cells may promote DNA hyper-replication and induce a senescence response similar to that caused by oncogene activation. These findings indicate that deregulation of CDC6 expression in human cells poses a serious risk of carcinogenesis. Overexpression of CDC6 has been described in 32 HCC patients with or without lymph node metastasis by high-throughput microarray.84 CpG island hypermethylation of CDC6 gene was shown in human prostate cancer and benign prostate hyperplasias,85 but a role for CDC6 in HCC has not been explored.
Epigenetic Regulation of Gene Expression in Liver Cancer Stem Cells and HCC
The pattern of DNA methylation is often altered significantly in liver progenitor cells, cancer stem cells and HCC. One key component of the cancer epigenome is an altered DNA methylation pattern composed of global demethylation but promoter localized hypermethylation compared to normal cells, these changes basically contribute to an altered structure and function of DNA, potentially involving the regulation of gene transcription or expression.86 The methyl-CpG-binding protein MeCP2 associates with histone deacetylases (HDACs) and a deacetylation-dependent co-repressor (mSin3) specifically interacts with methylated DNA and mediates transcriptional repression.87 A growing body of evidence suggests that aberrant DNA methylation of CpG islands around promoter regions can have the same effects as mutations in the coding regions on the inactivation of tumor-suppressor genes.88
p16 is one of tumor suppressor genes most intensively studied. The gene encoding the p16 protein is located on 9p21 with a total length of 8.5 kb, including three exons and two introns. p16 can inhibit the activity of the cell cycle-dependent protein kinases CDK4 and CDK6 and reduce the phosphorylation level of the Rb protein, which thereby inhibits cell proliferation. The most common epigentic alteration of the p16 gene in HCC was de novo methylation, which was detected in 48% of the cases. p16 gene allele loss was observed at a lower frequency (21%). These results indicate that alterations of the p16 gene plays an important role in the genesis of HCC.89 Yoshikawa et al. identified a correlation between the aberrant methylation in the CpG island of SOCS-1 and its transcription silencing in HCC cell lines.90 An incidence of 65% of hypermethylation of SOCS-1 gene was detected in 26 human primary HCC tumor samples.90 The restoration of SOCS-1 function in cells with methylation-silenced SOCS-1 and constitutively activated JAK2 activity retards the anchorage-independent growth of cells.90 Moreover, recent evidence shows that Drosophila melanogaster sal-like gene 3 (SALL3) is a novel inhibitory factor for DNA methyltransferase 3 α (DNMT3A), which acts by a direct interaction between the double zinc finger motif of SALL3 and the PWWP domain of DNMT3A. SALL3 expression reduces DNMT3A-mediated CpG island methylation in cell culture and in vitro. SALL3 is inducible by BMP-4 and silenced by associated DNA methylation in hepatocellular carcinoma.91 Furthermore, by analyzing 106 tissues (64 HCC samples and 42 samples without liver disorders) and seven hepatocarcinoma cell lines (HepG2, HepG3B2, C3A, SNU-182, SNU-398, SBU-449 and SNU-475), Iliopoulos et al. demonstrated that telomerase reverse transcriptase (hTERT) expression levels were inversely correlated with DNA methylation levels in HCC and normal tissues. They also found that hTERT expression was regulated by DNA methylation and histone H3-K9 modifications that effect the ability of c-myc binding to the E-box 1 site in hTERT promoter.92
Interestingly, DNA methylation and histone methylation/acetylation serve to modulate gene expression and represent a powerful mechanism that contributes to tumor growth by the altered expression of genes involved in tumor cell behavior. However, it is not entirely clear why many differentiation/apoptosis-related gene promoters in cancer cells contain hypermethylated (or aberrantly methylated) CpG islands. We propose that genetic damage caused by chromosomal instability may impair epigenetic modifications (or inheritance) of proliferation/differentiation/apaptosis-specific genes, such as oncogenes, tumor suppressor genes, and DNA repair genes or other genes. Although the mechanism focuses mainly on methylated DNA modifications and the perturbation of histone methylation/acetylation, other as yet unknown epigenetic mechanisms that are involved in tumoriogenesis may also be causative.
Therapeutic Strategies and Perspective
Activation of individual oncogenes or the loss of individual tumour suppressor genes is not sufficient to induce tumorigenesis. Therefore, it is likely that multiple genetic and epigenetic aberrations occur in tumoriogenesis. Understanding the mechanisms operating in liver progenitor cells and cancer stem cells is essential to innovate novel therapeutic approaches.
Epigenetic inheritance of DNA methylation and histone methylation/acetylation is believed to be essential for maintaining genome stability and transcriptional patterns during liver development and liver stem cell differentiation. Aberrant DNA methylation and histone methylation/acetylation has also been connected to silencing of tumor suppressor genes in cancer, and the enzymes regulating DNA methylation and histone methylation/acetylation are therefore important candidate targets for the development of anti-cancer drugs.
Differences in post-transcriptional and post-translational modifications in normal and cancerous cells reveal that different splicing variants of the adhesion receptor, CD44, are expressed differently in each. Hence, future therapeutic strategies can focus on targeting this cell surface marker with specified antibodies.93,94 Differentiation of cancer cells into less aggressive, more differentiated cells has also been shown to be a successful strategy, particularly in treatment of acute promyleocytic leukemia. All-trans retinoic acid (ATRA) after normal chemotherapy resulted in a 90% remission and 70% cure rate.95 However, the response of a cancer stem cell to a differentiating agent such as retinoic acid may not be predictable and may result in inappropriate differentiation. For example, some breast cancer cells trans-differentiate into cells with the genotypic and phenotypic characteristics of blood vessels when treated with RA.96 Differentiation therapy in HCC is an attractive strategy, but will require a major effort to define the liver cancer stem cell and its differentiation pathways.
Targeting the stem cell niche is another promising therapeutic strategy. The specified microenvironment in which stem cells reside often dictates self-renewal and reproduction.97 Alteration of components of the stem cell niche can effectively change stem cell fate, as in the case of experimental parathyroid hormone (PTH) induction.97–99 Furthermore, human-ES-cell-derived fibroblast-like cells (hdFs) provide a supportive environment for stem cells through insulin-like growth factor 2 (IGF-II).100 Targeting IGF-II, therefore, can manipulate the stem cell microenvironment. How this therapy may be efficacious in cancers of the liver, however, remains to be elucidated.
A clear and precise delineation of markers specific to stages of CSC formation, from cell surface markers such as CD90, CD45 and CD133 to stem cell markers such as Nanog, Oct3/4, STAT3, as well as the precise tumor suppressive role of the TGFβ/Smad/B2SP pathway, remain crucial for developing therapeutic strategies targeting CSCs in HCC.
Abbreviations
- CSCs
cancer stem cells
- HCC
hepatocellular cancer
- LPCs
liver progenitor cells
References
- 1.Lin L, Amin R, Gallicano GI, Glasgow E, Jogunoori W, Jessup JM, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGFbeta signaling. Oncogene. 2009;28:961–972. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Mishra L, Banker T, Murray J, Byers S, Thenappan A, He AR, et al. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra L, Derynck R, Mishra B. Transforming growth factor-beta signaling in stem cells and cancer. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGFbeta and IL-6 signaling. Proc Natl Acad Sci USA. 2008;105:2445–2450. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 8.Tsai RY. A molecular view of stem cell and cancer cell self-renewal. Int J Biochem Cell Biol. 2004;36:684–694. doi: 10.1016/j.biocel.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 11.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 13.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 16.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELFbeta-spectrin-deficient mice. Science. 2003;299:574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 18.Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 19.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 20.Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 21.Weinstein M, Monga SP, Liu Y, Brodie SG, Tang Y, Li C, et al. Smad proteins and hepatocyte growth factor control parallel regulatory pathways that converge on beta1-integrin to promote normal liver development. Mol Cell Biol. 2001;21:5122–5131. doi: 10.1128/MCB.21.15.5122-5131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 23.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 24.Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 25.Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 26.Tien AC, Rajan A, Bellen HJ. A Notch updated. J Cell Biol. 2009;184:621–629. doi: 10.1083/jcb.200811141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev. 2004;14:48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Giovannini C, Lacchini M, Gramantieri L, Chieco P, Bolondi L. Notch3 intracellular domain accumulates in HepG2 cell line. Anticancer Res. 2006;26:2123–2127. [PubMed] [Google Scholar]
- 29.Gramantieri L, Giovannini C, Lanzi A, Chieco P, Ravaioli M, Venturi A, et al. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int. 2007;27:997–1007. doi: 10.1111/j.1478-3231.2007.01544.x. [DOI] [PubMed] [Google Scholar]
- 30.Gao J, Chen C, Hong L, Wang J, Du Y, Song J, et al. Expression of Jagged1 and its association with hepatitis B virus X protein in hepatocellular carcinoma. Biochem Biophys Res Commun. 2007;356:341–347. doi: 10.1016/j.bbrc.2007.02.130. [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald K, Harrington A, Leder P. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene. 2000;19:4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- 32.Gonsalves FC, Weisblat DA. MAPK regulation of maternal and zygotic Notch transcript stability in early development. Proc Natl Acad Sci USA. 2007;104:531–536. doi: 10.1073/pnas.0609851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockhausen MT, Sjolund J, Axelson H. Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res. 2005;310:218–228. doi: 10.1016/j.yexcr.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Chappell WH, Green TD, Spengeman JD, McCubrey JA, Akula SM, Bertrand FE. Increased protein expression of the PTEN tumor suppressor in the presence of constitutively active Notch-1. Cell Cycle. 2005;4:1389–1395. doi: 10.4161/cc.4.10.2028. [DOI] [PubMed] [Google Scholar]
- 35.Qi R, An H, Yu Y, Zhang M, Liu S, Xu H, et al. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 2003;63:8323–8329. [PubMed] [Google Scholar]
- 36.Wang C, Qi R, Li N, Wang Z, An H, Zhang Q, et al. Notch1 signaling sensitizes tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human hepatocellular carcinoma cells by inhibiting Akt/Hdm2-mediated p53 degradation and upregulating p53-dependent DR5 expression. J Biol Chem. 2009;284:16183–16190. doi: 10.1074/jbc.M109.002105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu H, Julius MA, Giarre M, Zheng Z, Brown AM, Kitajewski J. Transformation by Wnt family proteins correlates with regulation of beta-catenin. Cell Growth Differ. 1997;8:1349–1358. [PubMed] [Google Scholar]
- 39.Hatsell S, Rowlands T, Hiremath M, Cowin P. Beta-catenin and Tcfs in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2003;8:145–158. doi: 10.1023/a:1025944723047. [DOI] [PubMed] [Google Scholar]
- 40.Nusse R, van Ooyen A, Cox D, Fung YK, Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984;307:131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 41.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 42.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 43.Cook D, Fry MJ, Hughes K, Sumathipala R, Woodgett JR, Dale TC. Wingless inactivates glycogen synthase kinase-3 via an intracellular signalling pathway which involves a protein kinase C. EMBO J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]
- 44.Chen RH, Ding WV, McCormick F. Wnt signaling to beta-catenin involves two interactive components. Glycogen synthase kinase-3beta inhibition and activation of protein kinase C. J Biol Chem. 2000;275:17894–17899. doi: 10.1074/jbc.M905336199. [DOI] [PubMed] [Google Scholar]
- 45.Kim D, Rath O, Kolch W, Cho KH. A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK pathways. Oncogene. 2007;26:4571–4579. doi: 10.1038/sj.onc.1210230. [DOI] [PubMed] [Google Scholar]
- 46.Xie Z, Yuan H, Yin Y, Zeng X, Bai R, Glazer RI. 3-phosphoinositide-dependent protein kinase-1 (PDK1) promotes invasion and activation of matrix metalloproteinases. BMC Cancer. 2006;6:77. doi: 10.1186/1471-2407-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng X, Xu H, Glazer RI. Transformation of mammary epithelial cells by 3-phosphoinositide-dependent protein kinase-1 (PDK1) is associated with the induction of protein kinase Calpha. Cancer Res. 2002;62:3538–3543. [PubMed] [Google Scholar]
- 48.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 50.Clarke AR. Wnt signalling in the mouse intestine. Oncogene. 2006;25:7512–7521. doi: 10.1038/sj.onc.1210065. [DOI] [PubMed] [Google Scholar]
- 51.Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, Binot F, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120:1763–1773. doi: 10.1053/gast.2001.24798. [DOI] [PubMed] [Google Scholar]
- 52.Taniguchi K, Roberts LR, Aderca IN, Dong X, Qian C, Murphy LM, et al. Mutational spectrum of beta-catenin, AXIN1 and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene. 2002;21:4863–4871. doi: 10.1038/sj.onc.1205591. [DOI] [PubMed] [Google Scholar]
- 53.Ishizaki Y, Ikeda S, Fujimori M, Shimizu Y, Kurihara T, Itamoto T, et al. Immunohistochemical analysis and mutational analyses of beta-catenin, Axin family and APC genes in hepatocellular carcinomas. Int J Oncol. 2004;24:1077–1083. [PubMed] [Google Scholar]
- 54.Park JY, Park WS, Nam SW, Kim SY, Lee SH, Yoo NJ, et al. Mutations of beta-catenin and AXIN I genes are a late event in human hepatocellular carcinogenesis. Liver Int. 2005;25:70–76. doi: 10.1111/j.1478-3231.2004.0995.x. [DOI] [PubMed] [Google Scholar]
- 55.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 56.Merle P, de la Monte S, Kim M, Herrmann M, Tanaka S, Von Dem Bussche A, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 57.Prange W, Breuhahn K, Fischer F, Zilkens C, Pietsch T, Petmecky K, et al. Beta-catenin accumulation in the progression of human hepatocarcinogenesis correlates with loss of E-cadherin and accumulation of p53, but not with expression of conventional WNT-1 target genes. J Pathol. 2003;201:250–259. doi: 10.1002/path.1448. [DOI] [PubMed] [Google Scholar]
- 58.Inagawa S, Itabashi M, Adachi S, Kawamoto T, Hori M, Shimazaki J, et al. Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival. Clin Cancer Res. 2002;8:450–456. [PubMed] [Google Scholar]
- 59.Joo M, Lee HK, Kang YK. Expression of beta-catenin in hepatocellular carcinoma in relation to tumor cell proliferation and cyclin D1 expression. J Korean Med Sci. 2003;18:211–217. doi: 10.3346/jkms.2003.18.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 61.Murdoch B, Chadwick K, Martin M, Shojaei F, Shah KV, Gallacher L, et al. Wnt-5A augments repopulating capacity and primitive hematopoietic development of human blood stem cells in vivo. Proc Natl Acad Sci USA. 2003;100:3422–3427. doi: 10.1073/pnas.0130233100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pitsouli C, Perrimon N. Developmental biology: Our fly cousins’ gut. Nature. 2008;454:592–593. doi: 10.1038/454592a. [DOI] [PubMed] [Google Scholar]
- 63.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 65.Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- 66.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 67.Mishra L, Cai T, Yu P, Monga SP, Mishra B. β2-Spectrin3 encodes a novel 200-kD beta-spectrin: role in liver development. Oncogene. 1999;18:353–364. doi: 10.1038/sj.onc.1202313. [DOI] [PubMed] [Google Scholar]
- 68.Monga SP, Tang Y, Candotti F, Rashid A, Wildner O, Mishra B, et al. Expansion of hepatic and hematopoietic stem cells utilizing mouse embryonic liver explants. Cell Transplant. 2001;10:81–89. [PubMed] [Google Scholar]
- 69.Blake T, Ganesan N, Shetty K, Mishra L. TGFbeta Signaling in Gastrointestinal Stem Cells. Sci STKE. (Connections Map, as seen September 2005; http://stke.sciencemag.org/cgi/cm/stkecm;CMP_17699). [Google Scholar]
- 70.Feng XH, Derynck R. Specificity and versatility in TGFbeta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 71.Roberts A, Mishra L. Role of TGFbeta in stem cells and cancer. Oncogene. 2005;24:5667. [Google Scholar]
- 72.Antequera F. Genomic specification and epigenetic regulation of eukaryotic DNA replication origins. EMBO J. 2004;23:4365–4370. doi: 10.1038/sj.emboj.7600450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McNairn AJ, Gilbert DM. Epigenomic replication: linking epigenetics to DNA replication. Bioessays. 2003;25:647–656. doi: 10.1002/bies.10305. [DOI] [PubMed] [Google Scholar]
- 74.Méchali M. DNA replication origins: from sequence specificity to epigenetics. Nat Rev Genet. 2001;2:640–645. doi: 10.1038/35084598. [DOI] [PubMed] [Google Scholar]
- 75.Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 2002;16:2479–2484. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stedman W, Deng Z, Lu F, Lieberman PM. ORC MCM, and histone hyperacetylation at the Kaposi’s sarcoma-associated herpesvirus latent replication origin. J Virol. 2004;78:12566–12575. doi: 10.1128/JVI.78.22.12566-12575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, Schepers A, et al. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 2005;24:1406–1417. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 80.Cohen SM, Brylawski BP, Cordeiro-Stone M, Kaufman DG. Same origins of DNA replication function on the active and inactive human × chromosomes. J Cell Biochem. 2003;88:923–931. doi: 10.1002/jcb.10429. [DOI] [PubMed] [Google Scholar]
- 81.Gómez M, Brockdorff N. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc Natl Acad Sci USA. 2004;101:6923–7298. doi: 10.1073/pnas.0401854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goto Y, Gomez M, Brockdorff N, Feil R. Differential patterns of histone methylation and acetylation distinguish active and repressed alleles at X-linked genes. Cytogenet Genome Res. 2002;99:66–74. doi: 10.1159/000071576. [DOI] [PubMed] [Google Scholar]
- 83.Wong N, Lam WC, Lai PB, Pang E, Lau WY, Johnson PJ. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am J Pathol. 2001;159:465–671. doi: 10.1016/S0002-9440(10)61718-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee CF, Ling ZQ, Zhao T, Fang SH, Chang WC, Lee SC, et al. Genomic-wide analysis of lymphatic metastasis-associated genes in human hepatocellular carcinoma. World J Gastroenterol. 2009;15:356–365. doi: 10.3748/wjg.15.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bastian PJ, Ellinger J, Heukamp LC, Kahl P, Müller SC, von Rücker A. Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing radical prostatectomy. Eur Urol. 2007;51:665–674. doi: 10.1016/j.eururo.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 86.Ting AH, et al. The cancer epigenome-components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 87.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 88.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chaubert P, Guillou L, Kurt AM, Bertholet MM, Metthez G, Leisinger HJ, et al. Frequent p16INK4 (MTS1) gene inactivation in testicular germ cell tumors. Am J Pathol. 1997;151:859–865. [PMC free article] [PubMed] [Google Scholar]
- 90.Yoshikawa H, Matsubara K, Qian GS, et al. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 91.Shikauchi Y, Saiura A, Kubo T, Niwa Y, Yamamoto J, Murase Y, et al. SALL3 interacts with DNMT3A and shows the ability to inhibit CpG island methylation in hepatocellular carcinoma. Mol Cell Biol. 2009;29:1944–1958. doi: 10.1128/MCB.00840-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iliopoulos D, Satra M, Drakaki A, Poultsides GA, Tsezou A. Epigenetic regulation of hTERT promoter in hepatocellular carcinomas. Int J Oncol. 2009;34:391–399. [PubMed] [Google Scholar]
- 93.Spira AI, Carducci MA. Differentiation therapy. Curr Opin Pharmacol. 2003;3:338–343. doi: 10.1016/s1471-4892(03)00081-x. [DOI] [PubMed] [Google Scholar]
- 94.Miletti-Gonzalez KE, Chen S, Muthukumaran N, Saglimbeni GN, Wu X, Yang J, et al. The CD44 receptor interacts with P-glycoprotein to promote cell migration and invasion in cancer. Cancer Res. 2005;65:6660–6667. doi: 10.1158/0008-5472.CAN-04-3478. [DOI] [PubMed] [Google Scholar]
- 95.Massard C, Deutsch E, Soria JC. Tumour stem cell-targeted treatment: elimination or differentiation. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2006;17:1620. doi: 10.1093/annonc/mdl074. [DOI] [PubMed] [Google Scholar]
- 96.Endo Y, Deonauth K, Prahalad P, Hoxter B, Zhu Y, Byers SW. Role of Sox-9, ER81 and VE-cadherin in retinoic acid-mediated trans-differentiation of breast cancer cells. PLoS ONE. 2008;3:2714. doi: 10.1371/journal.pone.0002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adams GB, Martin RP, Alley IR, Chabner KT, Cohen KS, Calvi LM, et al. Therapeutic targeting of a stem cell niche. Nature Biotech. 2007;25:238. doi: 10.1038/nbt1281. [DOI] [PubMed] [Google Scholar]
- 98.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 99.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 100.Martinez-Iglesias O, Ruiz-Llorente L, Sanchez-Martinez R, Garcia L, Zambrano A, Aranda A. Histone deacetylase inhibitors: mechanism of action and therapeutic use in cancer. Clin Transl Oncol. 2008;10:395–398. doi: 10.1007/s12094-008-0221-x. [DOI] [PubMed] [Google Scholar]