Summary
Microbial treatment of environmental contamination by anthropogenic halogenated organic compounds has become popular in recent decades, especially in the subsurface environments. Molecular techniques such as polymerase chain reaction‐based fingerprinting methods have been extensively used to closely monitor the presence and activities of dehalogenating microbes, which also lead to the discovery of new dehalogenating bacteria and novel functional genes. Nowadays, traditional molecular techniques are being further developed and optimized for higher sensitivity, specificity, and accuracy to better fit the contexts of dehalogenation. On the other hand, newly developed high throughput techniques, such as microarray and next‐generation sequencing, provide unsurpassed detection ability, which has enabled large‐scale comparative genomic and whole‐genome transcriptomic analysis. The aim of this review is to summarize applications of various molecular tools in the field of microbially mediated dehalogenation of various halogenated organic compounds. It is expected that traditional molecular techniques and nucleic‐acid‐based biomarkers will still be favoured in the foreseeable future because of relative low costs and high flexibility. Collective analyses of metagenomic sequencing data are still in need of information from individual dehalogenating strains and functional reductive dehalogenase genes in order to draw reliable conclusions.
Introduction
Pollution caused by anthropogenic halogenated organic compounds has been a serious environmental problem since the middle of the 20th century. Halogenated compounds (including chlorinated and brominated) constitute more than 50% of the top hundred species in the 2007 CERCLA Priority List of Hazardous Compounds (http://www.atsdr.cdc.gov/cercla/07list.html). In order to remove halogenated compounds from the anoxic subsurface environments, bacteria that are capable of reductive dehalogenation are promising because they can replace chlorine/bromine with hydrogen and derive energy for growth, i.e. with chlorinated/brominated ethenes, polybrominated diphenyl ethers (PBDEs), polychlorinated biphenyls (PCBs), chlorinated/brominated phenols, chlorobenzenes, and dioxins, through a process called dehalorespiration (Shelton and Tiedje, 1984; Maymó‐Gatell et al., 1997; Boyle et al., 1999; Holliger et al., 1999; Adrian et al., 2000; 2007a; Bunge et al., 2003; May et al., 2008; Ye et al., 2010; L.K. Lee et al., 2011).
Molecular detection and characterization of dehalogenating bacteria have greatly facilitated application of dehalogenating bacteria in bioremediation. Gene expression studies by reverse‐transcript quantitative polymerase chain reaction (RT‐qPCR), RNA‐sequencing (RNA‐seq) and microarray have established links between genes and their dehalogenating activities. Advances in proteomics have initiated the discovery of numerous reductive dehalogenases (RDases) and elucidation of dehalogenation mechanisms. Previously, Northern blotting was used to monitor expression of only a limited number of genes, while microarray technology is able to measure thousands of genes in one chip. For small bacterial genomes, one microarray chip can cover the whole collection of protein‐coding genes, allowing a thorough screening of transcript abundance and gene regulation (West et al., 2008). Therefore, the novel techniques such as microarrays and next‐generation deep genome sequencing further enable the in‐depth study of dehalogenating microbes, in aspects that used to require intensive labour work. Figure 1 depicts various traditional and recently developed molecular techniques that are discussed in this review with their advantages/disadvantages indicated in different scenarios.
Figure 1.
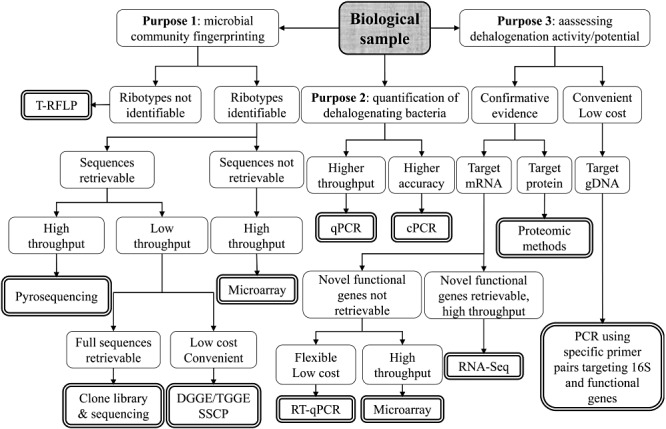
An overview of molecular techniques utilized in the studies of reductive dehalogenation. Double line text boxes indicate various molecular techniques.
This review summarizes molecular techniques that have been utilized or will potentially be applied in studying dehalorespiration of halogenated compounds. It should be noted that there have also been studies on oxygenolytic/hydrolytic dehalogenation as well as co‐metabolic reductive dehalogenation, which will not be covered in this review (Fetzner, 1998; Löffler et al., 2003; Mattes et al., 2010).
Detection and quantification of dehalogenating bacteria
Phylogenetic classification methods
The conventional 16S rRNA gene‐based phylogenetic classification method is still widely used because of the large database available. Genus‐specific primers targeting 16S rRNA genes are available for the detection of various dehalogenating genera, as summarized in Table 1. However, 16S rRNA gene‐based techniques have their disadvantages due to some inherent drawbacks, such as low evolution rates of rRNA gene sequences, and existence of multiple 16S rRNA gene copies in bacterial genomes (Yamamoto and Harayama, 1995; Klappenbach et al., 2001; Acinas et al., 2004). Moreover, the information on 16S rRNA gene alone may not be enough to confirm the phylogeny of a species/strain. Sometimes, microbes sharing very similar 16S rRNA gene sequences (e.g. > 99.5% similarity) are actually different species based on DNA–DNA hybridization results (e.g. only 41 % similarity via DNA–DNA hybridization) (Fox et al., 1992). On the other hand, in the genus of Desulfitobacterium, strains previously thought to belong to different species based on 16S rRNA gene sequences were later found to be in the same species according to > 80% homology in DNA–DNA hybridization (Villemur et al., 2006).
Table 1.
List of genus‐specific primer sets targeting 16S genes of dehalogenating bacteria.
| Group | Primer | Sequence | T | Size | Specificity | Reference |
|---|---|---|---|---|---|---|
| Chloroflexi | Chl348F | GAG GCA GCA GCA AGG AA | 60 | 470 | Chloroflexi | Fagervold et al. (2005) |
| Dehal884R | GGC GGG ACA CTT AAA GCG | |||||
| Acetobacterium | Aceto572f | GGC TCA ACC GGT GAC ATG CA | 59 | 208 | Acetobacterium in KB‐1 | Duhamel and Edwards (2006) |
| Aceto784r | ACT GAG TCT CCC CAA CAC CT | |||||
| Aceto572f | GGC TCA ACC GGT GAC ATG CA | 63 | 219 | Grostern and Edwards (2009) | ||
| Aceto791r | CTG CGG CAC TGA GTC TCC CC | |||||
| Anaeromyxobacter | A60‐86F | (refer to the reference) | Dollhopf et al. (2005) | |||
| A447‐465R | ||||||
| 60F | CGA GAA AGC CCG CAA GGG | 56.5 | 401 | Petrie et al. (2003) | ||
| 461R | ATT CGT CCC TCG CGA CAG T | |||||
| Ade399Fwd | GCA ACG CCG CGT GTG T | 60 | 67 | Thomas et al. (2009) | ||
| Ade466Rev | TCC CTC GCG ACA GTG CTT | |||||
| 2CP444Fwd | TCG CGA GGG ACG AAT AAG G | 60 | 69 | 2CP‐like strains | Thomas et al. (2009) | |
| 2CP513Rev | CGG TGC TTC CTC TCG AGG TA | |||||
| F112 | GTA ATC TGC CCT AGA GTC CGG A | 60 | 115 | A. dehalogenans strain 2CP‐C | Sanford et al. (2007) | |
| R227 | AGA GCG ATA GCT TGT GTA CAG AGG | |||||
| Clostridium | Chis150f | AAA GGR AGA TTA ATA CCG CAT AA | 57 | 540 | Majority of clusters I and II Clostridia | Hung et al. (2008) |
| ClostIr | TTC TTC CTA ATC TCT ACG CA | |||||
| Dehalobacter | Deb179F | TGT ATT GTC CGA GAG GCA | 53 | 828 | Schlötelburg et al. (2002) | |
| Deb1007R | ACT CCC ATA TCT CTA CGG | |||||
| Dre441F | GTT AGG GAA GAA CGG CAT CTG T | 58 | 225 | Smits et al. (2004) | ||
| Dre645R | CCT CTC CTG TCC TCA AGC CAT A | |||||
| Dre441F | GTT AGG GAA GAA CGG CAT CTG T | 58 | 589 | Smits et al. (2004) | ||
| Dre1013R | CGA AGC ACT CCC ATA TCT | |||||
| Dhb477f | GAT TGA CGG TAC CTA ACG AGG | 63 | ∼170 | Grostern and Edwards (2006b) | ||
| Dhb647r | TAC AGT TTC CAA TGC TTT ACG G | |||||
| Dehalobium | 14F | AGA GTT TGA TCC TGG CTC AG | 62 | 1215 | o‐17/DF‐1‐type Chloroflexi | Watts et al. (2005) |
| Dehal1265R | GCT ATT CCT ACC TGC TGT ACC | |||||
| Dehalococcoides | DET730/Dhc730F | GCG GTT TTC TAG GTT GTC | 58 | 620 | Bunge et al. (2003) | |
| DET1350/Dhc1350R | CAC CTT GCT GAT ATG CGG | |||||
| FL2F/Dhc728F/Dco728F | AAG GCG GTT TTC TAG GTT GTC AC | 58 | 436 | Dehalococcoides sp. strain FL2 | Löffler et al. (2000) | |
| FL2R/Dhc1164R | CGT TTC GCG GGG CAG TCT | |||||
| FpDHC1/Dhc1f | GAT GAA CGC TAG CGG CG | 55 | 1377 | Hendrickson et al. (2002)a | ||
| RpDHC1377/1377R | GGT TGG CAC ATC GAC TTC AA | |||||
| FpDHC1/Dhc1f | GAT GAA CGC TAG CGG CG | 59 | ∼260 | Grostern and Edwards (2009) | ||
| Dhc264r | CCT CTC AGA CCA GCT ACC GAT CGA A | |||||
| DHE‐for | AAG GCG GTT TTC TAG GTT | 58 | 443 | Dennis et al. (2003); Yan et al. (2009a) | ||
| DHE‐rev | CGT TTC GCG GGG CAG TCT | |||||
| FpDHC1/Dhc1f | GAT GAA CGC TAG CGG CG | 59 | 258 | Duhamel et al. (2004) | ||
| 259r | CAG ACC AGC TAC CGA TCG AA | |||||
| FpDHC1/Dhc1f | GAT GAA CGC TAG CGG CG | 52 | 1380 | Duhamel et al. (2004) | ||
| 1386r | CCT CCT TGC GGT TGG CAC ATC | |||||
| DeF | GCA ATT AAG ATA GTG GC | 55 | 1373 | Cupples et al. (2003) | ||
| DeR | ACT TCG TCC CAA TTA CC | |||||
| FL2F/Dhc728F/Dco728F | AAG GCG GTT TTC TAG GTT GTC AC | 58 | 216 | Smits et al. (2004) | ||
| Dco944R | CTT CAT GCA TGT CAA AT | |||||
| Dhc193f | GGT TCA YTA AAG CCG YAA GG | 53 | 855 | Dowideit et al. (2010) | ||
| Dhc1048r | CCT GTG CAA RYT CCT GAC T | |||||
| 567F | CGG GAC GTG TCA TTC AAT AC | 55 | 436 | Fennell et al. (2001) | ||
| RpDHC1377/1377R | GGT TGG CAC ATC GAC TTC AA | |||||
| DHC793f | GGG AGT ATC GAC CCT CTC TG | 60 | 153 | Yoshida et al. (2005) | ||
| DHC946r | CGT TYC CCT TTC TGT TCA CT | |||||
| DHC66f | GGT CTT AAG CAA TTA AGA TAG TG | 60 | 114 | Yoshida et al. (2005) | ||
| DHC180r | CAC CAA GCR CCT TRC GGC | |||||
| DhcForward | GGT AAT ACG TAG GAA GCA AGC G | 60 | 98 | ‘D. ethenogenes’ strain 195 | Holmes et al. (2006) | |
| DhcReverse | CCG GTT AAG CCG GGA AAT T | |||||
| Dehalogenimonas | (Thirteen primer sets) | (refer to the reference) | 63 | Yan et al. (2009a) | ||
| Desulfitobacterium | Dd1/ Dsb174F | AAT ACC GNA TAA GCT TAT CCC | 55 | 1199 | El Fantroussi et al. (1997) | |
| Dd2/Dsb1373R | TAG CGA TTC CGA CTT CAT GTT C | |||||
| Dd3/Dsb460F | TCT TCA GGG ACG AAC GGC AG | 55 | 624 | El Fantroussi et al. (1997) | ||
| Dd4/Dsb1084R | CAT GCA CCA CCT GTC TCA T | |||||
| Dsb406F | GTA CGA CGA AGG CCT TCG GGT | 58 | 213 | Smits et al. (2004) | ||
| Dsb619R | CCC AGG GTT GAG CCC TAG GT | |||||
| dsb434f | TAC TGT CTT CAG GGA CGA AC | 60 | 865 | Dowideit et al. (2010) | ||
| dsb1299r | TGA GAC CAG CTT TCT CGG AT | |||||
| Dsb406F | GTA CGA CGA AGG CCT TCG GGT | 58 | 213 | Smits et al. (2004) | ||
| Dsb619R | CCC AGG GTT GAG CCC TAG GT | |||||
| Dsb406F | GTA CGA CGA AGG CCT TCG GGT | 58 | 967 | Smits et al. (2004) | ||
| Dd2/Dsb1373R | TAG CGA TTC CGA CTT CAT GTT C | |||||
| Desulfomonile | Dt1/Dsm59F | CAA GTC GTA CGA GAA ACA TAT C | 55 | 995 | El Fantroussi et al. (1997) | |
| Dt2/Dsm1054R | GAA GAG GAT CGT GTT TCC ACG A | |||||
| Dt3/Dsm205F | GGG TCA AAG TCG GCC TCT CGA CG | 55 | 423 | El Fantroussi et al. (1997) | ||
| Dt4/Dsm628R | GCT TTC ACA TTC GAC TTA TCG | |||||
| DSMON85F | CGG GGT RTG GAG TAA AGT GG | 62 | 1334 | Loy et al. (2004) | ||
| DSMON1419R | CGA CTT CTG GTG CAG TCA RC | |||||
| Desulfovibrio | DSV230 | GRG YCY GCG TYY CAT TAG C | 61 | 610 | Desulfovibrio/Desulfomicrobium | Daly et al. (2000) |
| DSV838 | SYC CGR CAY CTA GYR TYC ATC | |||||
| DSB1180F | CCT AGG GCT ACA CAC GTA CTA A | 61 | 225 | Grostern and Edwards (2006a) | ||
| DSB1405R | CCG GCT TCG GGT AAA ACC AG | |||||
| DSV691‐F | CCG TAG ATA TCT GGA GGA ACA TCA G | 63 | 135 | Fite et al. (2004) | ||
| DSV826‐R | ACA TCT AGC ATC CAT CGT TTA CAG C | |||||
| Desulfuromonas | BB1F/Dsf205F | AAC CTT CGG GTC CTA CTG TC | 58 | 815 | Löffler et al. (2000) | |
| BB1R/Dsf1020R | GCC GAA CTG ACC CCT ATG TT | |||||
| Geobacter | Geo564F | AAG CGT TGT TCG GAW TTA T | 57 | 276 | Geobacteraceae family | Cummings et al. (2003); Sanford et al. (2007)b |
| Geo840R | GGC ACT GCA GGG GTC AAT A | |||||
| Geo196F | GAA TAT GCT CCT GAT TC | 53 | 820 | Geobacter sp. strain SZ | Sung (2005) | |
| Geo999R | ACC CTC TAC TTT CAT AG | |||||
| Geo73f | CTT GCT CTT TCA TTT AGT GG | 59 | 412 | Geobacter sp. strain SZ | Duhamel and Edwards (2006) | |
| Geo485r | AAG AAA ACC GGG TAT TAA CC | |||||
| Geo196F | GAA TAT GCT CCT GAT TC | 50 | 357 | Geobacter sp. strain SZ | Amos et al. (2007) | |
| Geo535R | TAA ATC CGA ACA ACG CTT | |||||
| Geo63F | CAG GCC TAA CAC ATG CAA GT | 62 | 1443 | Geobacteraceae family | Dennis et al. (2003) | |
| Geo418R | CCG ACC ATT CCT TAG GAC | |||||
| Sulfurospirillum | Sulfuro114f | GCT AAC CTG CCC TTT AGT GG | 59 | 307 | Sulfurospirillum in culture KB‐1 | Löffler et al. (2005); Duhamel and Edwards (2006) |
| Sulfuro421r | GTT TAC ACA CCG AAA TGC GT |
This table contains most of the primer sets for dehalogenating bacteria, but should not be considered all inclusive. Primers are genus‐specific unless specified according to statements in the references. For some primers, more than one primer names are listed, separated by ‘/’.
There are in total seven primer sets in the study by Hendrickson and colleagues (2002), among which three sets need to raise their annealing temperatures to ensure specificity on Dehalococcoides according to Yan and colleagues (2009a).
This primer set also amplifies Anaeromyxobacter 16S rRNA genes according to Bedard and colleagues (2007).
According to rrnDB as of June 2011 (Z.M.P. Lee et al., 2009), among the 1074 entries of Bacteria and Archaea, only 20.2% genomes contain single‐copy of 16S rRNA gene operons and the average number of 16S rRNA operons is 3.94 copies per genome (refer to Table 2 for 16S rRNA gene copies in genomes of common dehalogenating bacteria). Multiple and sometimes heterogeneous 16S rRNA gene copies in a single genome have notable inconvenience when analysing the phylogenetic sequence (Tourova, 2003) and also when querying the abundance of organisms (Fogel et al., 1999). For example, in order to query methanogen abundance in dechlorinating consortia, 16S gene copy number per genome in Methanococcus maripaludis has be to be estimated in order to convert measured gene copy numbers into actual cell numbers (Daprato et al., 2007).
Table 2.
Numbers of SSU rRNA gene copies per genome in common dehalogenating bacteria (Villemur et al., 2006; Z.M.P. Lee et al., 2009).
| Genus | Species | Strain | 16S | ITS | 23S | 5S | tRNA |
|---|---|---|---|---|---|---|---|
| Dehalococcoides | ethenogenes | 195 | 1 | 0 | 1 | 1 | 46 |
| Dehalococcoides | sp. | BAV1 | 1 | 0 | 1 | 1 | 46 |
| Dehalococcoides | sp. | CBDB1 | 1 | 0 | 1 | 1 | 47 |
| Desulfovibrio | vulgaris | DP4 | 5 | 5 | 5 | 6 | 68 |
| Desulfovibrio | desulfuricans | G20 | 4 | 4 | 4 | 4 | 66 |
| Desulfovibrio | vulgaris | Hildenborough | 5 | 5 | 5 | 6 | 68 |
| Desulfovibrio | vulgaris | Miyazaki F | 4 | 4 | 4 | 4 | 64 |
| Desulfitobacterium | hafniense | Y51 | 6 | 6 | 6 | 6 | 59 |
| Desulfitobacterium | hafniense | DCB‐2 | 6 | – | – | – | – |
| Clostridium | (Multiple) | (Multiple) | 9 | 8.72 | 9 | 8.81 | 79.16 |
| Anaeromyxobacter | dehalogenans | 2CP‐C | 2 | 2 | 2 | 2 | 49 |
| Anaeromyxobacter | sp. | Fw109‐5 | 2 | 2 | 2 | 2 | 49 |
| Anaeromyxobacter | sp. | K | 2 | 2 | 2 | 2 | 49 |
| Enterobacter | sp. | 638 | 7 | 7 | 7 | 8 | 84 |
| Enterobacter | sakazakii | ATCC BAA‐894 | 7 | 7 | 7 | 8 | 80 |
All data are presented as of 8Jun2011 from rrnDB. SSU rRNA gene copy numbers for Clostridium are average of 27 Clostridium strains.
To tackle these problems, the use of the rpoB gene (Dahllöf et al., 2000), the fast‐evolving gene gyrB and the internal transcribed spacer (ITS) (Yamamoto and Harayama, 1995; Dauga, 2002; Brown and Fuhrman, 2005; Stingl et al., 2007) have been proposed to complement the 16S rRNA gene‐based classification method (at least for some phylogenetic groups). The gyrB‐ and ITS‐based phylogenetic analysis might serve as a promising way to determine culture's purity and suggest phylogenetic distances among strains, especially for bacterial groups sharing very similar 16S gene sequences, e.g. ‘Dehalococcoides’ (Cheng and He, 2009).
Biomarker based‐stable isotope probing (SIP)
Dependence on bacteria's cultivability limits discovery of some difficult‐to‐cultivate bacterial species that degrade environmental pollutants. This challenge can be circumvented by biomarker based‐SIP, which uses stable isotope as a tracer and analyses biomarkers after cells incorporate the isotope‐containing substrate into biomass (Radajewski et al., 2000; Manefield et al., 2002). The available biomarkers include DNA, rRNA, and phospholipid‐derived fatty acid (Neufeld et al., 2007) as well as mRNA and protein (Jehmlich et al., 2010; Dumont et al., 2011). SIP works well when the bacteria in query are able to break down targeted substrates and incorporate the labelled atoms into biomass, examples including benzene (Herrmann et al., 2010), phenol (Manefield et al., 2002), biphenyl (Leigh et al., 2007; Sul et al., 2009) and nitrotoluenes (Gallagher et al., 2010).
However, SIP encounters problems with reductive dehalogenation where assimilation of atoms in the halogenated substrates does not usually take place (Holliger et al., 1999). An alternate way is adding 13C‐labelled carbon source (usually 13C‐acetate) together with unlabelled halogenated compounds to the bacterial consortia, as proposed by Kittelmann and Friedrich (2008a) in a study of microbial community in pristine river sediment. The underlying principle is that acetate‐utilizing bacteria should also be actively involved in the dehalogenation process when halogenated compounds are supplied as the major electron acceptors (Kittelmann and Friedrich, 2008a). Following this strategy, several novel bacteria were identified, which played important roles in tetrachloroethene (PCE) dechlorination, such as bacterial cluster LC from river sediments and Dehalobium from tidal flat sediments (Kittelmann and Friedrich, 2008a,b). Notably, all the dechlorinators identified in the study of Kittelmann and Friedrich belong to the Chloroflexi phylum. It is possible that some dehalogenating bacteria capable of fermentation were missed out by SIP since they may utilize other fatty acids instead of acetate as a carbon source. Therefore, the results of SIP in dehalogenation application are of importance, but may not be considered comprehensive in terms of its coverage of potential dehalogenating bacteria.
Quantification techniques
Quantitative real‐time PCR (qPCR) and competitive PCR (cPCR) are two powerful PCR‐based nucleic acid quantification techniques. Difference in their mechanisms is that qPCR quantifies fluorescence intensities during the amplification process while cPCR measures signals at the amplification endpoint. In addition, applying molecular fingerprinting techniques on serially diluted samples may also provide quantitative estimates of operational taxonomic units, a strategy called ‘qfingerprinting’ (Ramette, 2009).
qPCR finds its extremely versatile usage in quantification of dehalogenating species due to its huge merits in producing precise and fast results (Cupples, 2008) and offering high sensitivity compared with terminal restriction fragment length polymorphism (T‐RFLP) and RFLP plus clone sequencing (Freeborn et al., 2005; Rahm et al., 2006a). A fast approach of synthesizing DNA standards and controls using long oligonucleotide hybridization has made the setup of qPCR even more convenient (David et al., 2008). The wide applications of qPCR in dehalogenation studies include: (i) establishing relationship between species and dehalogenating activities (Lendvay et al., 2003; Yoshida et al., 2005; Taşet al., 2009; 2010a); (ii) examining interactions between dehalogenating bacteria and other species (Duhamel and Edwards, 2006; 2007; Cheng et al., 2010); (iii) demonstrating growth‐linked dehalorespiration (He et al., 2003; Bedard et al., 2007; Grostern and Edwards, 2009; Yan et al., 2009b; L.K. Lee et al., 2011) and assessing culture purity (Sung et al., 2006); (iv) assessing spatial and temporal distributions of dehalogenating bacteria (Amos et al., 2009); and (v) analysing effects of growth factors on dehalogenating bacteria (He et al., 2007). As a standardized method, qPCR is sometimes used as a validation for other quantification methods (Adrian et al., 2007a). However, the accuracy and precision of qPCR is prone to interference such as PCR amplification inhibition and differences in PCR amplification/DNA extraction efficiency (Cupples, 2008). Holmes and colleagues (2006) successfully applied a four‐gene plasmid standard to lower down the discrepancy between the 16S rRNA gene and RDase gene copy number. However, this methodology lacks flexibility, and can only increase precision of qPCR measurement but not accuracy.
cPCR is more accurate than qPCR in quantifying nucleic acids, showing good reproducibility when detecting very small variations of nucleic acid concentrations (Cupples et al., 2003; Zentilin and Giacca, 2007). Recent modification of cPCR (namely, alternately binding probe competitive PCR) allows good fitting standard curve (R = 0.999) and lower detection limit (10 copies µl−1 template DNA), specifically for Dehalococcoides (Miyata et al., 2010). cPCR has been used in enumeration of Dehalococcoides (Cupples et al., 2003; 2004), Desulfitobacterium (Lévesque et al., 1998), Dehalobium (May et al., 2008), and the Chloroflexi bacteria group (Fagervold et al., 2007). Although cPCR is both accurate and reliable, its limitations are obvious, which mainly lie in the construction of competitor standards that need to be as close as possible to the targeted template and in the cumbersome post‐PCR electrophoresis‐based detection and analysis step (Zentilin and Giacca, 2007). Its throughput is limited as multiple reactions are needed to quantify one single nucleic acid fragment.
Traditional fingerprinting techniques
Various molecular fingerprinting techniques are available aiming at retrieving microbial community structure information. Cloning and sequencing reveal nearly full‐length sequences of 16S rRNA genes and thus allow discrimination based on subtle differences in the gene sequences. However, cloning and sequencing are performed at the expense of tedious work and high sequencing costs especially when a large number of clones are needed. Denaturing or temperature gradient gel electrophoresis (DGGE/TGGE) can separate DNA sequences differing only by one base pair (Myers et al., 1987; Muyzer and Smalla, 1998), which can be useful in the detection of Dehalococcoides strains with high identity of 16S rRNA genes (Hendrickson et al., 2002). Due to its low cost, fast results, high sensitivity (as low as 1% of total population), semi‐quantitative ability, and good resolution, DGGE has been widely used in characterizing dehalogenating communities (Duhamel et al., 2002; 2004; May et al., 2008; Narihiro et al., 2010). However, DGGE/TGGE has multiple limitations (e.g. relatively short sequences of only 200–400 bp) as described by Muyzer and Smalla (1998). The choice of hypervariable regions (e.g. ‘V1, V9 and V3 – the most variable regions’ versus ‘V1 and V4 – the most heterogeneous regions in terms of melting temperature’) in 16S rRNA genes has significant impact on the resolving power of DGGE and thus the diversity implicated for the microbial community (Yu and Morrison, 2004). To further improve DGGE's separation resolution towards complex microbial communities, Wang and He (2011) developed a new method T‐RFs‐2D that separates terminal restriction fragments (T‐RFs) of 16S rRNA genes on a two‐dimensional gel electrophoresis. When characterizing a microbial community in a complex river‐sediment that dechlorinates PCBs, T‐RFs‐2D separated 63 DNA fragments, while traditional DGGE detected only 41 DNA fragments in the same sample.
T‐RFLP is a sensitive and high‐throughput molecular fingerprinting method (Marsh, 1999). It was claimed that T‐RFLP detected more ‘ribotypes’ and was considerably more sensitive than DGGE (Marsh et al., 1998). However, T‐RFLP has often to be combined with clone library and sequencing to identify each fragment (Bunge et al., 2008; Kittelmann and Friedrich, 2008a,b) or with in silico analysis based on database sequences (Sung et al., 2006). Moreover, because of its non‐confirmative results and the emergence of other new high‐throughput fingerprinting techniques such as microarray and 16S‐pyrosequencing, T‐RFLP has become less frequently used except in initial tentative profiling of microbial community structures and changes.
There are other fingerprinting techniques such as single‐strand conformation polymorphism (SSCP), amplified ribosomal DNA restriction analysis (ARDRA), and ribosomal intergenic spacer analysis (RISA), which were summarized and compared in a review by Justé and colleagues (2008). They were also occasionally applied in the studies of microbial reductive dehalogenation, albeit at a lower frequency. It should be noted that sometimes there is discrepancy observed in ribotype identities obtained using different 16S‐rRNA‐gene‐based techniques, which may be caused by biases in sample preparations of different techniques. For example, in a study of microbial community in a biofilm sample that aerobically degraded PCBs, although SSCP and clone library/sequencing detected bacteria species belonging to same genera, none of the sequences obtained by SSCP was identical to the sequences of clones obtained by PCR of 16S rRNA genes or RT‐PCR of 16S rRNA (Tillmann et al., 2005).
High‐throughput fingerprinting techniques
The next‐generation sequencing and microarray techniques are developed that could overcome the limitations of traditional fingerprinting techniques, i.e. only limited number of DNA fragments can be displayed on the DGGE/TGGE gel or on the T‐RFLP profile.
Next‐generation sequencing techniques (Shendure and Ji, 2008) pushed forward genome sequencing by providing a low‐cost and ultra‐fast sequencing technique, which does not require cloning of sample DNA fragments. One of the sequencing techniques, pyrosequencing, was later applied in high‐throughput sequencing of 16S rRNA gene fragments amplified from genomic DNA for microbial community analysis (Roesch et al., 2007). Multiplex barcoded pyrosequencing has further enhanced efficiency by pooling together primer barcoded DNA from multiple samples in a single run (Parameswaran et al., 2007; Smith et al., 2010). Zhang and colleagues (2010) successfully applied massively parallel pyrosequencing of a hypervariable region of the 16S rRNA genes on microbial samples from biofilm reactors with dechlorination activities. Dehalococcoides was found to thrive on the biofilm via dechlorinating trichloroethene (TCE), while a more diverse microbial community was observed in the biofilm fed with multiple chlorinated compounds, including sulfate‐reducing bacteria (Desulfovibrio) and nitrate‐reducing bacteria (Geothrix and Pseudomonas). J. Lee and colleagues (2011) retrieved over 10 000 sequences by using pyrosequencing on tidal flat microbial communities, and found Desulfuromonas and Desulfovibrio as potential PCE dechlorinators while Dehalococcoides was not detected. The pyrosequencing technique possesses a much higher resolution than conventional clone‐library based approach.
Phylogenetic oligonucleotide arrays (POAs) (e.g. the PhyloChips) can detect the presence and abundance of Bacteria and Archaea by hybridization between matched DNA fragments and probes designed to target prokaryotic 16S rRNA genes (Brodie et al., 2006). A recent application of PhyloChip revealed a significantly altered community structure when monitoring microbial community prior to and after the oil spillage in the Gulf of Mexico (Hazen et al., 2010). In another TCE‐contaminated site, PhyloChip measurement exhibited that TCE‐respiring Dehalococcoides decreased, but methane‐oxidizing organisms capable of TCE co‐metabolism increased in wells distant from electron donor injection location (Conrad et al., 2010). The above observation indicates that electron donor addition that aimed at enhancing reductive dechlorination might also stimulate co‐metabolism of TCE. Another POA designed by Sanguin and colleagues (2006a,b) showed that microbial community structure was significantly affected by as low as 1 p.p.m. TCE in soil and the most affected microorganisms from TCE treatment were identified (Nemir et al., 2010). However, unlike the PhyloChip which can distinguish bacterial phylogeny down to subfamily level, this POA has only 742 probes and thus possesses a much lower phylogenetic resolution.
The genome‐probing microarray (GPM) is another type of microarray that spots bacterial genome DNAs instead of oligonucleotides onto glass slides to query sample DNA (Bae et al., 2005). Without the aid of PCR amplification, the detection limit of GPM was 2.5 ng of sample genomic DNA even in the presence of non‐target DNA, which was added to test its effect on hybridization and detection sensitivity. The detection sensitivity of GPM was 0.25% of total microbial community. GPM avoided bias caused by PCR amplification and achieved a species‐specific detection; however, genomic DNA needs to be prepared for each bacterial strain thus preventing large‐scale production of arrays and application of this technique. Another limitation of GPM is that uncultured microorganisms cannot be used to establish genome probes on array chips. However, this was later solved by Chang and colleagues (2008a) using digital multiple displacement amplification to amply genomes from uncultured single bacterial cells. Despite these drawbacks, GPM is believed to have advantage over traditional DNA–DNA hybridization by having higher reproducibility, a lower background, and a less time‐consuming procedure (Chang et al., 2008b).
Besides the above‐mentioned POA and GPM, latter sections of this review will discuss other types of microarrays, i.e. functional gene arrays (FGAs) and whole‐genome arrays (WGAs).
Assessing culture purity
Obtaining pure dehalogenating cultures is important for in‐depth study of their physiological properties and dehalogenation mechanisms. Culture purity can be indicated by microscopic observation of uniform cell morphology, or by observing a single 16S rRNA fragment as detected by fingerprinting techniques (Yan et al., 2009b). In addition to molecular fingerprinting methods as mentioned above to assess culture purity, nested‐PCR using genus‐specific primer sets (Table 1) would be a recommended approach to detect other populations possibly existing in extreme minor populations. This is due to the fact that in some seemingly pure dehalogenating cultures, there might be another strain that is actually responsible for dehalogenating but could only grow to a very low cell density caused by the rather low concentrations of halogenated compounds such as PCBs, PBDEs and dioxins. Colony picking from solid phase medium is an important isolation method but should not be relied on as evidence for purity of culture, since other possible taxa could be carried over during the colony picking process such as the case for coculture DPH‐1 (Chang et al., 2000; Fletcher et al., 2008).
Even though all 16S rRNA gene‐based techniques indicate a single 16S rRNA gene pattern in a culture, it may still be possible that multiple strains with the same 16S rRNA gene sequence exist, especially for Dehalococcoides. This is perfectly demonstrated in the isolation process of Dehalococcoides sp. strain GT (Sung et al., 2006). To cope with this challenge, RDases have been quantified together with 16S rRNA genes by qPCR based on the fact that common RDase genes such as tceA and bvcA are single copy genes in the Dehalococcoides genome (Krajmalnik‐Brown et al., 2004; Sung et al., 2006). Pure culture identification must be performed carefully with appropriate molecular tools, or conclusions can be questionable. In a study of a PCE‐to‐ethene dechlorinating culture originated from Bitterfeld (Germany), one single Dehalococcoides strain in the culture was claimed to be responsible for all dechlorination steps from PCE to ethene, based on substrate specificity and 16S rRNA gene‐based DGGE tests (Cichocka et al., 2010). However, the purity of Dehalococcoides in the culture was still questionable (unless a simultaneous quantification of RDase genes were performed), although the author attributed the random variations in 16S rRNA gene sequences of Dehalococcoides clones to method‐introduced errors.
Investigation on dehalogenating activity
Identification of novel functional genes
Techniques available. Identification of novel functional genes responsible for dehalogenation is crucial in elucidating mechanisms of catalytic dehalogenation and in optimizing dehalogenation rates. In particular, dehalogenating bacteria possess RDases that catalyse the terminal electron transfer in the dehalorespiration process (Holliger et al., 1999). Ni and colleagues (1995) successfully identified the first RDase from Desulfomonile tiedjei strain DCB‐1 by using chromatography separation. This RDase catalyses dechlorination of 3‐chlorobenzoate to benzoate in an energy‐yielding process. Up till now, more than 20 RDases have been linked to specific dehalogenation activities (Table 3), although PCB/PBDE/dioxin‐related RDases remain largely undiscovered (Sakaki and Munetsuna, 2010) except for a few tentative cases (Zanaroli et al., 2010). Earlier identification of RDases was achieved by protein separation combined with in vitro activity test, and subsequent N‐terminal sequencing of the enzyme. Later, after more homologue sequences were obtained, degenerate PCR primers based on conserved regions of RDases became popular in pulling out putative RDases. The rapid development of next‐generation sequencing and microarray techniques also greatly aided novel RDase identification. Figure 2 depicts the common workflow of RDase gene identification.
Table 3.
List of identified reductive dehalogenases and approaches employed.
| RDase | Microorganism | Main substrate | Identification technique | Reference |
|---|---|---|---|---|
| 3‐ClBA–RD | Desulfomonile tiedjei strain DCB‐1 | 3‐chlorobenzoate | LC + in vitro | Ni et al. (1995) |
| CprA | Desulfitobacterium chlororespirans strain Co23 | 3‐chloro‐4‐hydroxybenzoate, chlorinated phenols | LC + in vitro + Amino | Löffler et al. (1996); Krasotkina et al. (2001) |
| PceA | Sulfurospirillum multivorans | PCE, TCE, cis‐/trans‐DBE | LC + in vitro + Amino | Neumann et al. (1996; 1998); Ye et al. (2010) |
| PceA | Dehalobacter restrictus | PCE, TCE | LC + in vitro + Amino | Schumacher et al. (1997); Maillard et al. (2003) |
| PceA | Dehalococcoides ethenogenes strain 195 | PCE | LC + in vitro + PAGE | Magnuson et al. (1998) |
| TceA | Dehalococcoides ethenogenes strain 195 | TCE | LC + in vitro + PAGE | Magnuson et al. (1998; 2000) |
| PceA | Desulfitobacterium hafniense strain PCE‐S | PCE, TCE, cis‐/trans‐DBE, VB | LC + in vitro + Amino | Miller et al. (1998); Ye et al. (2010) |
| CprA | Desulfitobacterium hafniense strain DCB‐2 | 3‐chloro‐4‐hydroxyphenylacetate | LC + in vitro + Amino | Christiansen et al. (1998) |
| CprA | Desulfitobacterium dehalogenans | A number of ortho‐chlorinated phenols | LC + in vitro + Amino | van de Pas et al. (1999) |
| PceA | Desulfitobacterium sp. strain PCE1 | PCE | LC + in vitro + Amino | van de Pas et al. (2001) |
| CprA | Desulfitobacterium sp. strain PCE1 | Cl‐OH‐phenylacetate | LC + in vitro + Amino | van de Pas et al. (2001) |
| PceA | Desulfitobacterium hafniense strain TCE1 | PCE, TCE | LC + in vitro + Amino | van de Pas et al. (2001) |
| PceC | Coculture DPH‐1 (containing Desulfitobacterium hafniense strain JH1) | PCE, TCE | LC + in vitro + Amino | Okeke et al. (2001) |
| PceA | Desulfitobacterium sp. strain Y51 | PCE, TCE | LC + in vitro + Amino | Suyama et al. (2002) |
| CrdA | Desulfitobacterium hafniense strain PCP‐1 | 2,4,6‐TCP, PCP | LC + in vitro + Amino + Genome | Boyer et al. (2003) |
| CprA5 | Desulfitobacterium hafniense strain PCP‐1 | 3,5‐DCP | LC + in vitro + MS + Genome | Thibodeau et al. (2004) |
| VcrA | Dehalococcoides sp. strain VS | VC, cis‐/trans‐/1,1‐DCE | LC + in vitro + Amino | Müller et al. (2004) |
| BvcA | Dehalococcoides sp. strain BAV1 | VC | Dege + qPCR | Krajmalnik‐Brown et al. (2004) |
| CbrA | Dehalococcoides sp. strain CBDB1 | Chlorinated benzenes | PAGE + in vitro + MS + Dege + T‐RFLP + Genome | Adrian et al. (2007b; Wagner et al. (2009) |
| DcaA | Desulfitobacterium dichloroeliminans strain DCA1 | 1,2‐DCA | Dege + qPCR | Marzorati et al. (2007) |
| CBDBA1453 | Dehalococcoides sp. strain CBDB1 | 1,2,3‐TCB | Dege + T‐RFLP + Genome | Wagner et al. (2009) |
| CBDBA187 | Dehalococcoides sp. strain CBDB1 | 1,2,3‐TCB | Dege + T‐RFLP + Genome | Wagner et al. (2009) |
| CBDBA1624 | Dehalococcoides sp. strain CBDB1 | 1,2,4‐TCB | Dege + T‐RFLP + Genome | Wagner et al. (2009) |
| WL RdhA1 | Dehalobacter sp. | 1,2‐DCA | Dege + qPCR | Grostern and Edwards (2009) |
| (eight RdhAs) | Dehalococcoides culture TUT2264 | Chloroethenes | Dege + qPCR | Futamata et al. (2009) |
| MbrA | Dehalococcoides sp. strain MB | TCE | Dege + qPCR | Chow et al. (2010) |
| CprA3 | Desulfitobacterium hafniense strain PCP‐1 | PCP, TeCP, TCP | LC + in vitro + MS + Genome | Bisaillon et al. (2010) |
Techniques: LC, chromatography separation; Amino, amino acid sequencing; PAGE, PAGE gel separation; Dege, degenerate primer detection; in vitro, in vitro activity test of RDases; Genome, sequenced genome of the targeted strain; MS, mass spectrometry detection of peptides; qPCR, transcriptional analysis by qPCR; T‐RFLP, transcriptional analysis by T‐RFLP.
Compounds: PCP, pentachlorophenol; TeCP, tetrachlorophenol; TCP, trichlorophenol; DCP, dichlorophenol; PCE, tetrachloroethene; TCE, trichloroethene; DCE, dichloroethene VC, vinyl chloride; DCA: dichloroethane; TCB, trichlorobenzene; DBE, dibromoethene; VB, vinyl bromide.
Figure 2.
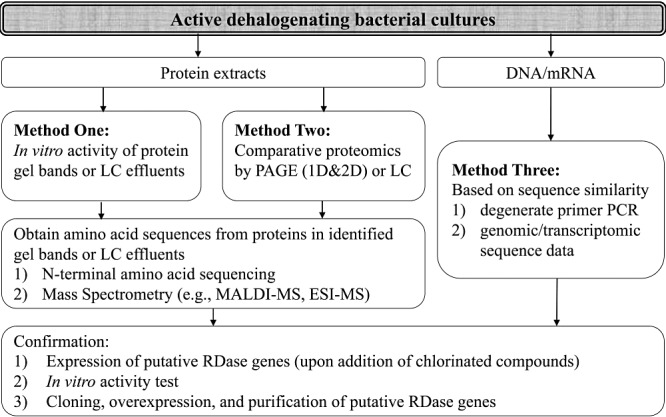
Common work flow of reductive dehalogenase gene identification.
Proteomic methods can identify RDases with no prior knowledge of the RDase gene sequences. Separation of whole cell proteins is achieved by either liquid chromatography (LC) (Magnuson et al., 1998) or polyacrylamide gel electrophoresis (PAGE) (Adrian et al., 2007b). Differential abundance of proteins in cultures with or without the targeted halogenated compounds may also give a hint of some possible candidates, since most RDase genes are inducible rather than constitutive (Cole et al., 1995; Lee et al., 2006). However, when using comparative proteomics technique to pick out potential RDases, one needs to bear in mind that differentially expressed genes might also result from the response of bacteria towards toxic substances and thus cannot guarantee a positive identification. After separation of proteins, native PAGE gel bands or LC effluent fractions containing RDase activities are collected for further analysis, e.g. SDS‐PAGE (Ni et al., 1995; Magnuson et al., 1998; Maillard et al., 2003), N‐terminal amino acid sequencing (Miller et al., 1998), or mass spectrometry (Thibodeau et al., 2004; Adrian et al., 2007b).
For certain bacterial species such as those within the Chloroflexi phylum, the commonly encountered problem in identification of RDases is extremely low biomass, which may be due to low energy yield and growth rate under anaerobic conditions, and may also be due to low solubility of some chloroaromatic compounds. Low biomass severely limits the application of proteomic techniques that usually require a large protein amount in order to ensure successful detection, either in gel or by chromatography (Müller et al., 2004; Adrian et al., 2007b). With accumulating RDase sequences identified (confirmed or putative) in recent years, using degenerate PCR primers to probe unknown cultures has become more popular (Table 4) (Krajmalnik‐Brown et al., 2004; Regeard et al., 2004; Chow et al., 2010). To increase the chances of finding the most expressed RDases by degenerate primers, construction of clone library based on complementary DNA (cDNA) rather than on genomic DNA is helpful (Lee et al., 2008). It is impractical to cover all possible RDase sequences using one degenerate primer set as demonstrated by Wagner and colleagues (2009), who designed 13 primer sets to cover 32 RDases in Dehalococcoides sp. strain CBDB1. It is perceivable that clone libraries with multiple degenerate primer sets may retrieve more putative RDases, but work load is significantly higher.
Table 4.
List of degenerate primer sets for reductive dehalogenase gene identification.
| Primer pair | Primer | Targeted region | Size | Detected RDase genes | Reference |
|---|---|---|---|---|---|
| 1 | RRF2 | Twin arginine motif in strain 195 | 1500 ∼ 1700 | 7 RDase genes in BAV1 including bvcA | Krajmalnik‐Brown et al. (2004) |
| 7 RDases genes in MB including mbrA (together with RDH F1C/R1C) | Chow et al. (2010) | ||||
| 13 RDase genes in CBDB1, 14 RDases in FL2 | Hölscher et al. (2004) | ||||
| B1R | ‘WYEW’ motif in B genes in strain 195 | ||||
| 8 RDase genes in culture TUT2264 | Futamata et al. (2009) | ||||
| 4 RDase genes in environmental samples, including two novel RDases | Lee et al. (2008) | ||||
| 2 | RDH F1C | Twin arginine motif | 1200 | 7RDases genes in MB including mbrA (together with RRF1 and B1R) | Chow et al. (2010) |
| RDH R1C | ‘PIDD’ motif | ||||
| 3 | mern2 | Upstream of ISB region of orfA in strain CBDB1 | 1000 | One RDase gene in CBDB1 | Hölscher et al. (2004) |
| mern5 | |||||
| 4 | fdehal | Upstream of ISB region of orfA in strain CBDB1 | 500 | Two RDase genes in CBDB1 | Hölscher et al. (2004) |
| rdehal | |||||
| 5 | ceRD2L/ceRD2S | Conserved sequence: ‘AARLFGA(D/S)(L/S)VG’ | 750 ∼ 900 | Two known pceA, one new RDase gene in Desulfitobacterium sp. | Regeard et al. (2004) |
| dcaA in Desulfitobacterium dichloroeliminans strain DCA1 | Marzorati et al. (2006; 2007) | ||||
| RD7r | Conserved sequence: ‘C(V/E)AVCP’ | ||||
| 7 RDase genes (together with RRF2/RD7r) | Grostern and Edwards (2009) | ||||
| 6 | ceRD2L/ceRD2S | Conserved sequence: ‘AARLFGA(D/S)(L/S)VG’ | 550 | Two new RDase genes in D. restrictus, one new RDase in S. multivorans | Regeard et al. (2004) |
| RD5r | Conserved sequence: ‘P(D/T)KPI(D/K)(A/F)G’ | ||||
| 7 | RD4f/RD4r/RD5f | (refer to the reference) | No amplicons | Regeard et al. (2004) | |
| 8 | RRF2 | Twin arginine motif in strain 195 | 1000 | 7RDase genes (together with ceRD2L/S&RD7r) | Grostern and Edwards (2009) |
| RD7r | Conserved sequence: ‘C(V/E)AVCP’ | ||||
| 9 | Dhu1080f | highly conserved ISB region | 450 | Two RDase genes in a 2‐bromophenol‐degrading consortium | Rhee et al. (2003) |
| Dhu1350r | |||||
| 10 | Dhar1000f | highly conserved iron–sulfur cluster binding motifs | 350 | One RDase gene in a 2‐bromophenol‐degrading consortium | Rhee et al. (2003) |
| Dhu1350r | No amplicons | Kittelmann and Friedrich (2008b) | |||
| 11 | (unnamed primer sets) | Conserved regions in several known pceA genes | 330 | Two pceA genes | Kimoto et al. (2010) |
Metagenomic sequencing and whole‐genome sequencing extract huge amounts of sequence information from bacterial genomes, and thus pave the way for rapid identification of novel putative RDases. For example, the complete genome sequence of Dehalococcoides sp. strain CBDB1 revealed 32 putative RDases, implying the enormous dehalogenating potential of this microbe (Kube et al., 2005). Recently released complete genome sequence of the novel Chloroflexi microbe Dehalobium chlorocoercia strain DF‐1 by J. Craig Venter Institute revealed at least 35 putative RDases, which may be responsible for DF‐1's ability to dechlorinate PCB congeners as well as chlorinated ethenes (http://www.jcvi.org). Chan and colleagues (2010) verified activities of putative hydrolytic dehalogenases identified from five sequenced microbial genomes by expressing them in E. coli. The strategy of cloning, overexpression and purification of selected proteins as adopted in this study proved to be effective in screening potential functional genes from genome sequencing data.
Similar to the degenerate primers method, putative RDase genes in genome sequences can only be identified if they share a significant sequence similarity with identified RDases, thus certain novel RDases may be missed out if they are only distantly related with existing RDases. To tackle this, sequencing of bacterial transcriptome is a promising way to select possible candidates among the most abundant transcripts (Mao et al., 2008; Ansorge, 2009). In such scenarios, there will be more positive identifications because transcripts with either small sizes or low BLAST scores in public databases will still be identified as long as they are highly expressed upon the addition of halogenated compounds.
Understanding RDase structures. So far, several consensus sequences in RDases have been identified to be related to reductive dehalogenation, such as the iron–sulfur cluster binding motifs, cobalamin binding motifs, and twin‐arginine signal sequence (Hölscher et al., 2004), as well as some conserved amino acid residues (e.g. tryptophan and histidine) that are potentially involved in catalysis of chloroethenes (Smidt et al., 2000). It is known that critical changes of amino acids in active sites may cause significant shift in catalytic activities as demonstrated in studies on hydrolytic dehalogenases (Pavlova et al., 2009; Beloqui et al., 2010). Obtaining such information with RDases will help in modification of RDases to achieve higher catalytic rates and broader substrate ranges.
Up to now, what we know about RDase catalysing mechanisms is still limited to the above‐mentioned conserved regions. While next‐generation gene sequencing has yielded billions of base pairs of gene sequences from either isolates or environmental samples, interestingly, the boosting RDase gene pools have not brought in revolutionary insights into structure–function analysis of the RDases. One reason is that expressing RDase genes in host cells is difficult due to their instability after purification and absence of activity after overexpression (Neumann et al., 1998; Sakaki and Munetsuna, 2010). More needs to be done to overcome such difficulties when trying to produce active RDases since this is the prerequisite for site‐specific mutagenesis and further identification of RDase active sites. One possible solution is to create genetically modified strains using natural dehalogenating bacteria, either by modifying genes on chromosomes or introducing expression vectors containing RDase gene sequences. This strategy takes advantage of the natural transcription/translation in these bacteria and may circumvent the difficulties in constructing a suitable expression system in the commonly used Escherichia coli host strains.
On the other hand, deficiency in analysis of metagenomic data hinders novel enzyme identification (Fernández‐Arrojo et al., 2010). It should be noted that sequencing data only provide an inventory of genes rather than proofs in functionality. A significant portion (5%) of open reading frames in the newly sequenced genomic data have little homology with genes of known functions, implying for many previously undescribed genes (Harrington et al., 2007). Also, miss‐annotation exists in gene databases, especially in those without manual curation (Schnoes et al., 2009). To facilitate protein identification, semi‐rational protein design that utilizes computational tools has become popular recently (Beloqui et al., 2010; Lutz, 2010). By preselecting promising target sites and limiting amino acid diversity, semi‐rational protein design greatly reduces library sizes, which are usually large in directed evolution of proteins. Thus, it holds promise for identification and modification of novel RDases in future.
Monitoring dehalogenating activities
Biomarkers indicating dehalogenating activities. Traditionally, the most evident sign for dehalogenation activity is direct monitoring of microbial degradation of substrates in situ (Kjellerup et al., 2008). However, biomarker‐based techniques (e.g. DNA, mRNA, protein and phospholipid) are mainstream detection methods due to their high sensitivity (White et al., 2005; Lee et al., 2008; Futamata et al., 2009; Lu et al., 2009; Werner et al., 2009). DNA fragments such as 16S rRNA genes (Lu et al., 2009) or functional genes of dehalogenating bacteria indicate dehalogenating potential but are only indirectly related to dehalogenating activity because: (i) quantification of cell numbers (16S rRNA gene copies) often does not reflect the actual physiological state of the microbial community (Röling, 2007), as shown by the discrepancy between dehalogenating bacteria cell counts and in situ activity (Freeborn et al., 2005; Ritalahti et al., 2010); and (ii) functional genes may be present but not expressed at all, or targeted functional genes do not cover the entire group of genes with similar functions, since the current RDase database is far from complete (Ritalahti et al., 2010).
In view of the limitation of gene copy numbers, it is recommended to monitor gene expression (mRNA abundance) in order to assess in situ dehalogenation activity, using techniques such as RT‐qPCR and microarrays. To account for mRNA loss during sample preparation, the addition of exogenous internal reference mRNA substantially improved the quantification accuracy for laboratory cultures (Johnson et al., 2005; Futamata et al., 2009). Transcripts of key functional genes such as RDase genes were found to correlate with active dechlorination of chlorinated ethenes (Lee et al., 2006; Futamata et al., 2009). For example, Wagner and colleagues (2009) adopted an innovative T‐RFLP method to monitor the expression of all 32 RDases in strain CBDB1 genome, which is less labour‐intensive and more cost‐effective. However, because of primer degeneracy, certain low level transcripts were not successfully amplified. Moreover, primer degeneracy also leads to biased PCR amplification among different RDase transcripts, making this method only semi‐quantitative. Nevertheless, T‐RFLP seems promising for simultaneous monitoring of gene homologues other than 16S rRNA genes as long as suitable primer sets are available. Besides tracking the RDase transcripts, some other key genes (e.g. hydrogenase genes) in the respiratory chain may also be monitored for assessing microbial activities (Rahm et al., 2006b; Rahm and Richardson, 2008a,b; Rowe et al., 2008). It is noteworthy to point out that under certain stress conditions (e.g. elevated temperature and presence of oxygen), expression of functional genes may be upregulated but the corresponding microbial activity does not elevate simultaneously (Amos et al., 2008; Fletcher et al., 2011).
Proteins translated from mRNA are more confirmative evidence for dehalogenating activities because specific RDases directly catalyse the transformation of halogenated compounds. However, protein biomarkers are less utilized compared with nucleic acids because of lack of convenient and sensitive method for their detection and identification. Based on available Dehalococcoides genome annotation, mass spectrometry can identify specific peptides matching several respiratory enzymes (e.g. hydrogenases, formate dehydrogenase and several strain‐specific RDases) present in active dechlorinating cultures, which may be used as biomarkers in environmental samples (Morris et al., 2006; 2007; Fung et al., 2007). However, conventional mass‐spectrometry‐based proteomic analyses are susceptible to contaminating proteins, and can only be carried out in less complex systems, or in membrane‐associated cell fractions (Morris et al., 2007). Werner and colleagues (2009) introduced a highly selective and sensitive protein identification method in the detection of specific proteins in a complex environment. This method, referred to as multiple‐reaction monitoring mass spectrometry, is able to quantify as low as 5 fmol peptide and requires protein from merely 1.4 × 106Dehalococcoides cells for analysis.
Transcriptomic analysis by microarray and next‐generation sequencing. The construction of microarray and next‐generation sequencing techniques has turned high‐throughput transcriptomic analysis into reality (Schena et al., 1995; Wang et al., 2009). Metabolism of key dechlorinators such as ‘Dehalococcoides ethenogenes’ strain 195 is of constant interest to researchers. A WGA was designed to cover > 99% of the predicted protein‐coding sequences for strain 195, based on which a series of studies were performed to query its metabolic pathways (Johnson et al., 2008; 2009; West et al., 2008; Tang et al., 2009). Using this array, changes in the strain 195 transcriptome were captured and linked to availability of growth factors such as corrinoid cofactor, electron acceptor, electron donor, carbon source and nitrogen source (Johnson et al., 2008; 2009; P.K.H. Lee et al., 2009). When targeting on genomic DNA rather than mRNA, comparative genomics by using WGAs have yielded interesting results in analysing intraspecies genome mutations among Dehalococcoides (West et al., 2008; P.K.H. Lee et al., 2011). The above studies show that although Dehalococcoides strains are similar to each other in genomes, they differ in genes located in integrated elements or high‐plasticity regions where RDase genes usually locate.
Functional gene arrays target genes involved in key metabolic processes and are used to study microbially mediated geochemical, ecological and environmental processes, such as E‐FGA (McGrath et al., 2010) and GeoChip (He et al., 2010). In the GeoChip 3.0, the number of probes was increased to 27 812, covering 56 990 functional genes for carbon, nitrogen, phosphorus and sulfur cycles, energy metabolism, and notably, degradation of organic contaminants including chlorinated compounds (He et al., 2010). Both genomic DNA and cDNA from reverse transcribed RNA can be detected by GeoChip, since its probes were designed based on protein‐coding gene sequences (He et al., 2010). GeoChip found its versatile usage in tracking functional microbial communities in bioremediation sites (Leigh et al., 2007; Taşet al., 2009; Van Nostrand et al., 2009). An example was the study of dechlorinating community in soil samples from Ebro River (Taşet al., 2009), in which new probes were designed and added to the array in order to cover all RDases in public databases. Results showed that Dehalococcoides activity varied significantly at different locations.
In recent years, sequencing of cDNA library using next‐generation sequencing techniques, termed as RNA‐Seq, has gained enormous attention in the study of transcriptomics (Wang et al., 2009). Comparisons of microarray and RNA‐Seq are frequently made, usually in favour of RNA‐Seq in view of inherent limitations of microarray techniques (Shendure, 2008; Wang et al., 2009). Nevertheless, microarray is still frequently used because of shorter time to retrieve results and reasonable cost (Agarwal et al., 2010). It is expected that with reducing sequencing costs and further improvement of protocols, RNA‐Seq will gradually replace microarray in most bacterial genome‐wide transcriptomic analyses (Croucher and Thomson, 2010).
Future perspective
Rapid development of molecular techniques has revolutionized the study of dehalogenation in many ways, and some basic issues need to be re‐evaluated, such as choices of biomarkers, evolution of mainstream techniques and overall analysis strategies.
From nucleic acids to peptide fragments as targeted biomarkers?
It is interesting to know whether characteristic peptides will surpass DNA/RNA and become the most frequent bioremediation biomarkers in the future. There are predictions of expecting the rise of proteomics in functional microbial ecology, judging from recent research trends that incorporate more data from shotgun proteomics (Maron et al., 2007; Desai et al., 2010). However, unlike nucleic acids which can be conveniently amplified and targeted, protein detection lacks suitable amplification methods, and is limited by sophisticated instrumentations. Therefore, before fast and high‐throughput protein sequencing techniques become available, genomics/transcriptomics are still the most welcome approaches we can rely on to obtain an overall picture of microbial status in situ.
From traditional low‐throughput techniques to novel high‐throughput techniques?
Current PCR‐based quantification/fingerprinting techniques are continuously being optimized to better suit to characterize dehalogenating microbial communities, to improve coverage, specificity and sensitivity in the detection of dehalogenating bacteria. On the other hand, emerging sequencing and microarray techniques allow analysis of very complex microbial community and thorough screening of gene expression in the microbial genome, which greatly facilitate the identification of functional genes and their regulation mechanisms with decreasing costs of genome/metagenome sequencing and tailor‐made microarrays. Nevertheless, relative low costs and high flexibility will still keep traditional low‐throughput molecular tools important in monitoring key dehalogenating bacteria and functional genes. The two sets of techniques are perfectly complementary to each other rather than replacing one for another.
From specific microbes and genes to an integrated network?
Taş and colleagues (2010b) argued an end of the so‐called reductionist approaches in the studies of Dehalococcoides, which are confined to only a few selective biomarkers. Instead, they contended for a switch to a strategy aiming at the entire bioremediation system. Similar points were also raised out by Vieites and colleagues (2009) and Frias‐Lopez and colleagues (2008). Although it is absolutely necessary to view the behaviour of microbes and functional genes collectively, it is still too early to move our focus from key dehalogenating bacteria and biomarkers to the holistic approaches. For example, many uncertainties and contributing factors exist in the analysis of metagenomic/metatranscriptomic sequencing data, such as miss‐annotation of genes and lack of information on protein function analyses. More information is needed on functional genes and their regulation mechanisms before we can expect reliable inferences of relationship between genes and activities from next‐generation sequencing data.
References
- Acinas S.G., Marcelino L.A., Klepac‐Ceraj V., Polz M.F. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L., Szewzyk U., Wecke J., Görisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- Adrian L., Hansen S.K., Fung J.M., Görisch H., Zinder S.H. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ Sci Technol. 2007a;41:2318–2323. doi: 10.1021/es062076m. [DOI] [PubMed] [Google Scholar]
- Adrian L., Rahnenführer J., Gobom J., Hölscher T. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol. 2007b;73:7717–7724. doi: 10.1128/AEM.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Koppstein D., Rozowsky J., Sboner A., Habegger L., Hillier L.W. Comparison and calibration of transcriptome data from RNA‐Seq and tiling arrays. BMC Genomics. 2010;11:383–398. doi: 10.1186/1471-2164-11-383. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos B.K., Sung Y., Fletcher K.E., Gentry T.J., Wu W.M., Criddle C.S. Detection and quantification of Geobacter lovleyi strain SZ: implications for bioremediation at tetrachloroethene‐ and uranium‐impacted sites. Appl Environ Microbiol. 2007;73:6898–6904. doi: 10.1128/AEM.01218-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos B.K., Ritalahti K.M., Cruz‐Garcia C., Padilla‐Crespo E., Löffler F.E. Oxygen effect on Dehalococcoides viability and biomarker quantification. Environ Sci Technol. 2008;42:5718–5726. doi: 10.1021/es703227g. [DOI] [PubMed] [Google Scholar]
- Amos B.K., Suchomel E.J., Pennell K.D., Löffler F.E. Spatial and temporal distributions of Geobacter lovleyi and Dehalococcoides spp. during bioenhanced PCE‐NAPL dissolution. Environ Sci Technol. 2009;43:1977–1985. doi: 10.1021/es8027692. [DOI] [PubMed] [Google Scholar]
- Ansorge W.J. Next‐generation DNA sequencing techniques. New Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Bae J.‐W., Rhee S.‐K., Park J.R., Chung W.‐H., Nam Y.‐D., Lee I. Development and evaluation of genome‐probing microarrays for monitoring lactic acid bacteria. Appl Environ Microbiol. 2005;71:8825–8835. doi: 10.1128/AEM.71.12.8825-8835.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard D.L., Ritalahti K.A., Löffler F.E. The Dehalococcoides population in sediment‐free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol. 2007;73:2513–2521. doi: 10.1128/AEM.02909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloqui A., Polaina J., Vieites J.M., Reyes‐Duarte D., Torres R., Golyshina O.V. Novel hybrid esterase‐haloacid dehalogenase enzyme. ChemBioChem. 2010;11:1975–1978. doi: 10.1002/cbic.201000258. et al. [DOI] [PubMed] [Google Scholar]
- Bisaillon A., Beaudet R., Lépine F., Déziel E., Villemur R. Identification and characterization of a novel CprA reductive dehalogenase specific to highly chlorinated phenols from Desulfitobacterium hafniense strain PCP‐1. Appl Environ Microbiol. 2010;76:7536–7540. doi: 10.1128/AEM.01362-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer A., Pagé‐Bélanger R., Saucier M., Villemur R., Lépine F., Juteau P. Purification, cloning and sequencing of an enzyme mediating the reductive dechlorination of 2,4,6‐trichlorophenol from Desulfitobacterium frappieri PCP‐1. Biochem J. 2003;373:297–303. doi: 10.1042/BJ20021837. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A.W., Phelps C.D., Young L.Y. Isolation from estuarine sediments of a Desulfovibrio strain which can grow on lactate coupled to the reductive dehalogenation of 2,4,6‐tribromophenol. Appl Environ Microbiol. 1999;65:1133–1140. doi: 10.1128/aem.65.3.1133-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie E.L., DeSantis T.Z., Joyner D.C., Baek S.M., Larsen J.T., Andersen G.L. Application of a high‐density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.V., Fuhrman J.A. Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat Microb Ecol. 2005;41:15–23. [Google Scholar]
- Bunge M., Adrian L., Kraus A., Opel M., Lorenz W.G., Andreesen J.R. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature. 2003;421:357–360. doi: 10.1038/nature01237. et al. [DOI] [PubMed] [Google Scholar]
- Bunge M., Wagner A., Fischer M., Andreesen J.R., Lechner U. Enrichment of a dioxin‐dehalogenating Dehalococcoides species in two‐liquid phase cultures. Environ Microbiol. 2008;10:2670–2683. doi: 10.1111/j.1462-2920.2008.01688.x. [DOI] [PubMed] [Google Scholar]
- Chan W.Y., Wong M., Guthrie J., Savchenko A.V., Yakunin A.F., Pai E.F. Sequence‐ and activity‐based screening of microbial genomes for novel dehalogenases. Microb Biotech. 2010;3:107–120. doi: 10.1111/j.1751-7915.2009.00155.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.‐W., Sung Y., Kim K.‐H., Nam Y.‐D., Roh S.W., Kim M.‐S. Development of microbial genome‐probing microarrays using digital multiple displacement amplification of uncultivated microbial single cells. Environ Sci Technol. 2008a;42:6058–6064. doi: 10.1021/es8006029. et al. [DOI] [PubMed] [Google Scholar]
- Chang H.W., Nam Y.D., Jung M.Y., Kim K.H., Roh S.W., Kim M.S. Statistical superiority of genome‐probing microarrays as genomic DNA–DNA hybridization in revealing the bacterial phylogenetic relationship compared to conventional methods. J Microbiol Methods. 2008b;75:523–530. doi: 10.1016/j.mimet.2008.08.003. et al. [DOI] [PubMed] [Google Scholar]
- Chang Y.C., Hatsu M., Jung K., Yoo Y.S., Takamizawa K. Isolation and characterization of a tetrachloroethylene dechlorinating bacterium, Clostridium bifermentans DPH‐1. J Biosci Bioeng. 2000;89:489–491. doi: 10.1016/s1389-1723(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Cheng D., He J. Isolation and characterization of ‘Dehalococcoides’ sp. strain MB, which dechlorinates tetrachloroethene to trans‐1,2‐dichloroethene. Appl Environ Microbiol. 2009;75:5910–5918. doi: 10.1128/AEM.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D., Chow W.L., He J. A Dehalococcoides‐containing co‐culture that dechlorinates tetrachloroethene to trans‐1,2‐dichloroethene. ISME J. 2010;4:88–97. doi: 10.1038/ismej.2009.90. [DOI] [PubMed] [Google Scholar]
- Chow W.L., Cheng D., Wang S., He J. Identification and transcriptional analysis of trans‐DCE‐producing reductive dehalogenases in Dehalococcoides species. ISME J. 2010;4:1020–1030. doi: 10.1038/ismej.2010.27. [DOI] [PubMed] [Google Scholar]
- Christiansen N., Ahring B.K., Wohlfarth G., Diekert G. Purification and characterization of the 3‐chloro‐4‐hydroxy‐phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 1998;436:159–162. doi: 10.1016/s0014-5793(98)01114-4. [DOI] [PubMed] [Google Scholar]
- Cichocka D., Nikolausz M., Haest P.J., Nijenhuis I. Tetrachloroethene conversion to ethene by a Dehalococcoides‐containing enrichment culture from Bitterfeld. FEMS Microbiol Ecol. 2010;72:297–310. doi: 10.1111/j.1574-6941.2010.00845.x. [DOI] [PubMed] [Google Scholar]
- Cole J.R., Fathepure B.Z., Tiedje J.M. Tetrachloroethene and 3‐chlorobenzoate dechlorination activities are co‐induced in Desulfomonile tiedjei DCB‐1. Biodegradation. 1995;6:167–172. doi: 10.1007/BF00695347. [DOI] [PubMed] [Google Scholar]
- Conrad M.E., Brodie E.L., Radtke C.W., Bill M., Delwiche M.E., Lee M.H. Field evidence for co‐metabolism of trichloroethene stimulated by addition of electron donor to groundwater. Environ Sci Technol. 2010;44:4697–4704. doi: 10.1021/es903535j. et al. [DOI] [PubMed] [Google Scholar]
- Croucher N.J., Thomson N.R. Studying bacterial transcriptomes using RNA‐seq. Curr Opin Microbiol. 2010;13:619–624. doi: 10.1016/j.mib.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings D., Snoeyenbos‐West O., Newby D., Niggemyer A., Lovley D., Achenbach L. Diversity of Geobacteraceae species inhabiting metal‐polluted freshwater lake sediments ascertained by 16S rDNA analyses. Microb Ecol. 2003;46:257–269. doi: 10.1007/s00248-005-8002-3. et al. [DOI] [PubMed] [Google Scholar]
- Cupples A.M. Real‐time PCR quantification of Dehalococcoides populations: methods and applications. J Microbiol Methods. 2008;72:1–11. doi: 10.1016/j.mimet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Cupples A.M., Spormann A.M., McCarty P.L. Growth of a Dehalococcoides‐like microorganism on vinyl chloride and cis‐dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol. 2003;69:953–959. doi: 10.1128/AEM.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples A.M., Spormann A.M., McCarty P.L. Comparative evaluation of chloroethene dechlorination to ethene by Dehalococcoides‐like microorganisms. Environ Sci Technol. 2004;38:4768–4774. doi: 10.1021/es049965z. [DOI] [PubMed] [Google Scholar]
- Dahllöf I., Baillie H., Kjelleberg S. rpoB‐based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl Environ Microbiol. 2000;66:3376–3380. doi: 10.1128/aem.66.8.3376-3380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly K., Sharp R.J., McCarthy A.J. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate‐reducing bacteria. Microbiology-UK. 2000;146:1693–1705. doi: 10.1099/00221287-146-7-1693. [DOI] [PubMed] [Google Scholar]
- Daprato R.C., Löffler F.E., Hughes J.B. Comparative analysis of three tetrachloroethene to ethene halorespiring consortia suggests functional redundancy. Environ Sci Technol. 2007;41:2261–2269. doi: 10.1021/es061544p. [DOI] [PubMed] [Google Scholar]
- Dauga C. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. Int J Syst Evol Microbiol. 2002;52:531–547. doi: 10.1099/00207713-52-2-531. [DOI] [PubMed] [Google Scholar]
- David M.M., Sapkota A.R., Simonet P., Vogel T.M. A novel and rapid method for synthesizing positive controls and standards for quantitative PCR. J Microbiol Methods. 2008;73:73–77. doi: 10.1016/j.mimet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Dennis P.C., Sleep B.E., Fulthorpe R.R., Liss S.N. Phylogenetic analysis of bacterial populations in an anaerobic microbial consortium capable of degrading saturation concentrations of tetrachloroethylene. Can J Microbiol. 2003;49:15–27. doi: 10.1139/w03-008. [DOI] [PubMed] [Google Scholar]
- Desai C., Pathak H., Madamwar D. Advances in molecular and ‘‐omics’ technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresour Technol. 2010;101:1558–1569. doi: 10.1016/j.biortech.2009.10.080. [DOI] [PubMed] [Google Scholar]
- Dollhopf S.L., Hyun J.‐H., Smith A.C., Adams H.J., O'Brien S., Kostka J.E. Quantification of ammonia‐oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl Environ Microbiol. 2005;71:240–246. doi: 10.1128/AEM.71.1.240-246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowideit K., Scholz‐Muramatsu H., Miethling‐Graff R., Vigelahn L., Freygang M., Dohrmann A.B. Spatial heterogeneity of dechlorinating bacteria and limiting factors for in situ trichloroethene dechlorination revealed by analyses of sediment cores from a polluted field site. FEMS Microbiol Ecol. 2010;71:444–459. doi: 10.1111/j.1574-6941.2009.00820.x. et al. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Edwards E.A. Microbial composition of chlorinated ethene‐degrading cultures dominated by Dehalococcoides. FEMS Microbiol Ecol. 2006;58:538–549. doi: 10.1111/j.1574-6941.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Edwards E.A. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2‐dichloroethane. Environ Sci Technol. 2007;41:2303–2310. doi: 10.1021/es062010r. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Wehr S.D., Yu L., Rizvi H., Seepersad D., Dworatzek S. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis‐dichloroethene and vinyl chloride. Water Res. 2002;36:4193–4202. doi: 10.1016/s0043-1354(02)00151-3. et al. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Mo K., Edwards E.A. Characterization of a highly enriched Dehalococcoides‐containing culture that grows on vinyl chloride and trichloroethene. Appl Environ Microbiol. 2004;70:5538–5545. doi: 10.1128/AEM.70.9.5538-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M.G., Pommerenke B., Casper P., Conrad R. DNA‐, rRNA‐ and mRNA‐based stable isotope probing of aerobic methanotrophs in lake sediment. Environ Microbiol. 2011;13:1153–1167. doi: 10.1111/j.1462-2920.2010.02415.x. [DOI] [PubMed] [Google Scholar]
- El Fantroussi S., Mahillon J., Naveau H., Agathos S. Introduction of anaerobic dechlorinating bacteria into soil slurry microcosms and nested‐PCR monitoring. Appl Environ Microbiol. 1997;63:806–811. doi: 10.1128/aem.63.2.806-811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagervold S.K., Watts J.E.M., May H.D., Sowers K.R. Sequential reductive dechlorination of meta‐chlorinated polychlorinated biphenyl congeners in sediment microcosms by two different Chloroflexi phylotypes. Appl Environ Microbiol. 2005;71:8085–8090. doi: 10.1128/AEM.71.12.8085-8090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagervold S.K., May H.D., Sowers K.R. Microbial reductive dechlorination of Aroclor 1260 in Baltimore Harbor sediment microcosms is catalyzed by three phylotypes within the phylum Chloroflexi. Appl Environ Microbiol. 2007;73:3009–3018. doi: 10.1128/AEM.02958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell D.E., Carroll A.B., Gossett J.M., Zinder S.H. Assessment of indigenous reductive dechlorinating potential at a TCE‐contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ Sci Technol. 2001;35:1830–1839. doi: 10.1021/es0016203. [DOI] [PubMed] [Google Scholar]
- Fernández‐Arrojo L., Guazzaroni M.‐E., López‐Cortés N., Beloqui A., Ferrer M. Metagenomic era for biocatalyst identification. Curr Opin Biotechnol. 2010;21:725–733. doi: 10.1016/j.copbio.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Fetzner S. Bacterial dehalogenation. Appl Microbiol Biotechnol. 1998;50:633–657. doi: 10.1007/s002530051346. [DOI] [PubMed] [Google Scholar]
- Fite A., Macfarlane G.T., Cummings J.H., Hopkins M.J., Kong S.C., Furrie E. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53:523–529. doi: 10.1136/gut.2003.031245. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K.E., Ritalahti K.M., Pennell K.D., Takamizawa K., Löffler F.E. Resolution of culture Clostridium bifermentans DPH‐1 into two populations, a Clostridium sp. and tetrachloroethene‐dechlorinating Desulfitobacterium hafniense strain JH1. Appl Environ Microbiol. 2008;74:6141–6143. doi: 10.1128/AEM.00994-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K.E., Costanza J., Cruz‐Garcia C., Ramaswamy N.S., Pennell K.D., Löffler F.E. Effects of elevated temperature on Dehalococcoides dechlorination performance and DNA and RNA biomarker abundance. Environ Sci Technol. 2011;45:712–718. doi: 10.1021/es1023477. [DOI] [PubMed] [Google Scholar]
- Fogel G.B., Collins C.R., Li J., Brunk C.F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- Fox G.E., Wisotzkey J.D., Jurtshuk P. How close is close – 16S ribosomal RNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- Freeborn R.A., West K.A., Bhupathiraju V.K., Chauhan S., Rahm B.G., Richardson R.E. Phylogenetic analysis of TCE‐dechlorinating consortia enriched on a variety of electron donors. Environ Sci Technol. 2005;39:8358–8368. doi: 10.1021/es048003p. et al. [DOI] [PubMed] [Google Scholar]
- Frias‐Lopez J., Shi Y., Tyson G.W., Coleman M.L., Schuster S.C., Chisholm S.W. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung J.M., Morris R.M., Adrian L., Zinder S.H. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3‐dichlorophenol. Appl Environ Microbiol. 2007;73:4439–4445. doi: 10.1128/AEM.00215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futamata H., Kaiya S., Sugawara M., Hiraishi A. Phylogenetic and transcriptional analyses of a tetrachloroethene‐dechlorinating ‘Dehalococcoides’ enrichment culture TUT2264 and its reductive‐dehalogenase genes. Microbes Environ. 2009;24:330–337. doi: 10.1264/jsme2.me09133. [DOI] [PubMed] [Google Scholar]
- Gallagher E.M., Young L.Y., McGuinness L.M., Kerkhof L.J. Detection of 2,4,6‐trinitrotoluene‐utilizing anaerobic bacteria by 15N and 13C incorporation. Appl Environ Microbiol. 2010;76:1695–1698. doi: 10.1128/AEM.02274-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grostern A., Edwards E.A. A 1,1,1‐trichloroethane‐degrading anaerobic mixed microbial culture enhances biotransformation of mixtures of chlorinated ethenes and ethanes. Appl Environ Microbiol. 2006a;72:7849–7856. doi: 10.1128/AEM.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grostern A., Edwards E.A. Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl Environ Microbiol. 2006b;72:428–436. doi: 10.1128/AEM.72.1.428-436.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grostern A., Edwards E.A. Characterization of a Dehalobacter coculture that dechlorinates 1,2‐dichloroethane to ethene and identification of the putative reductive dehalogenase gene. Appl Environ Microbiol. 2009;75:2684–2693. doi: 10.1128/AEM.02037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington E.D., Singh A.H., Doerks T., Letunic I., von Mering C., Jensen L.J. Quantitative assessment of protein function prediction from metagenomics shotgun sequences. Proc Natl Acad Sci USA. 2007;104:13913–13918. doi: 10.1073/pnas.0702636104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen T.C., Dubinsky E.A., DeSantis T.Z., Andersen G.L., Piceno Y.M., Singh N. Deep‐sea oil plume enriches indigenous oil‐degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. et al. [DOI] [PubMed] [Google Scholar]
- He J., Ritalahti K.M., Yang K.L., Koenigsberg S.S., Löffler F.E. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424:62–65. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- He J., Holmes V.F., Lee P.K.H., Alvarez‐Cohen L. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol. 2007;73:2847–2853. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z.L., Deng Y., Van Nostrand J.D., Tu Q.C., Xu M.Y., Hemme C.L. GeoChip 3.0 as a high‐throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010;4:1167–1179. doi: 10.1038/ismej.2010.46. et al. [DOI] [PubMed] [Google Scholar]
- Hendrickson E.R., Payne J.A., Young R.M., Starr M.G., Perry M.P., Fahnestock S. Molecular analysis of Dehalococcoides16S ribosomal DNA from chloroethene‐contaminated sites throughout North America and Europe. Appl Environ Microbiol. 2002;68:485–495. doi: 10.1128/AEM.68.2.485-495.2002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann S., Kleinsteuber S., Chatzinotas A., Kuppardt S., Lueders T., Richnow H.‐H. Functional characterization of an anaerobic benzene‐degrading enrichment culture by DNA stable isotope probing. Environ Microbiol. 2010;12:401–411. doi: 10.1111/j.1462-2920.2009.02077.x. et al. [DOI] [PubMed] [Google Scholar]
- Holliger C., Wohlfarth G., Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- Holmes V.F., He J., Lee P.K.H., Alvarez‐Cohen L. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl Environ Microbiol. 2006;72:5877–5883. doi: 10.1128/AEM.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher T., Krajmalnik‐Brown R., Ritalahti K.M., von Wintzingerode F., Görisch H., Löffler F.E. Multiple nonidentical reductive‐dehalogenase‐homologous genes are common in Dehalococcoides. Appl Environ Microbiol. 2004;70:5290–5297. doi: 10.1128/AEM.70.9.5290-5297.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C.H., Cheng C.H., Cheng L.H., Liang C.M., Lin C.Y. Application of Clostridium‐specific PCR primers on the analysis of dark fermentation hydrogen‐producing bacterial community. Int J Hydrogen Energy. 2008;33:1586–1592. [Google Scholar]
- Jehmlich N., Schmidt F., Taubert M., Seifert J., Bastida F., von Bergen M. Protein‐based stable isotope probing. Nat Protocols. 2010;5:1957–1966. doi: 10.1038/nprot.2010.166. et al. [DOI] [PubMed] [Google Scholar]
- Johnson D.R., Lee P.K.H., Holmes V.F., Alvarez‐Cohen L. An internal reference technique for accurately quantifying specific mRNAs by real‐time PCR with application to the tceA reductive dehalogenase gene. Appl Environ Microbiol. 2005;71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.R., Brodie E.L., Hubbard A.E., Andersen G.L., Zinder S.H., Alvarez‐Cohen L. Temporal transcriptomic microarray analysis of ‘Dehalococcoides ethenogenes’ strain 195 during the transition into stationary phase. Appl Environ Microbiol. 2008;74:2864–2872. doi: 10.1128/AEM.02208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.R., Nemir A., Andersen G.L., Zinder S.H., Alvarez‐Cohen L. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol Lett. 2009;294:198–206. doi: 10.1111/j.1574-6968.2009.01569.x. [DOI] [PubMed] [Google Scholar]
- Justé A., Thomma B., Lievens B. Recent advances in molecular techniques to study microbial communities in food‐associated matrices and processes. Food Microbiol. 2008;25:745–761. doi: 10.1016/j.fm.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kimoto H., Suye S., Makishima H., Arai J., Yamaguchi S., Fujii Y. Cloning of a novel dehalogenase from environmental DNA. Biosci Biotechnol Biochem. 2010;74:1290–1292. doi: 10.1271/bbb.100027. et al. [DOI] [PubMed] [Google Scholar]
- Kittelmann S., Friedrich M.W. Identification of novel perchloroethene‐respiring microorganisms in anoxic river sediment by RNA‐based stable isotope probing. Environ Microbiol. 2008a;10:31–46. doi: 10.1111/j.1462-2920.2007.01427.x. [DOI] [PubMed] [Google Scholar]
- Kittelmann S., Friedrich M.W. Novel uncultured Chloroflexi dechlorinate perchloroethene to trans‐dichloroethene in tidal flat sediments. Environ Microbiol. 2008b;10:1557–1570. doi: 10.1111/j.1462-2920.2008.01571.x. [DOI] [PubMed] [Google Scholar]
- Kjellerup B.V., Sun X.L., Ghosh U., May H.D., Sowers K.R. Site‐specific microbial communities in three PCB‐impacted sediments are associated with different in situ dechlorinating activities. Environ Microbiol. 2008;10:1296–1309. doi: 10.1111/j.1462-2920.2007.01543.x. [DOI] [PubMed] [Google Scholar]
- Klappenbach J.A., Saxman P.R., Cole J.R., Schmidt T.M. rrnDB: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajmalnik‐Brown R., Hölscher T., Thomson I.N., Saunders F.M., Ritalahti K.M., Löffler F.E. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70:6347–6351. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasotkina J., Walters T., Maruya K.A., Ragsdale S.W. Characterization of the B12‐ and iron–sulfur‐containing reductive dehalogenase from Desulfitobacterium chlororespirans. J Biol Chem. 2001;276:40991–40997. doi: 10.1074/jbc.M106217200. [DOI] [PubMed] [Google Scholar]
- Kube M., Beck A., Zinder S.H., Kuhl H., Reinhardt R., Adrian L. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol. 2005;23:1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- Lee L.K., Ding C., Yang K.L., He J. Complete debromination of tetra‐ and penta‐ brominated diphenyl ethers by a coculture consisting of Dehalococcoides and Desulfovibrio species. Environ Sci Technol. 2011;45:8475–8482. doi: 10.1021/es201559g. [DOI] [PubMed] [Google Scholar]
- Lee J., Lee T.K., Löffler F.E., Park J. Characterization of microbial community structure and population dynamics of tetrachloroethene‐dechlorinating tidal mudflat communities. Biodegradation. 2011;22:687–698. doi: 10.1007/s10532-010-9429-x. [DOI] [PubMed] [Google Scholar]
- Lee P.K.H., Johnson D.R., Holmes V.F., He J., Alvarez‐Cohen L. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl Environ Microbiol. 2006;72:6161–6168. doi: 10.1128/AEM.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.K.H., Macbeth T.W., Sorenson K.S., Deeb R.A., Alvarez‐Cohen L. Quantifying genes and transcripts to assess the in situ physiology of ‘Dehalococcoides’ spp. in a trichloroethene‐contaminated groundwater site. Appl Environ Microbiol. 2008;74:2728–2739. doi: 10.1128/AEM.02199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.K.H., He J., Zinder S.H., Alvarez‐Cohen L. Evidence for nitrogen fixation by ‘Dehalococcoides ethenogenes’ strain 195. Appl Environ Microbiol. 2009;75:7551–7555. doi: 10.1128/AEM.01886-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.K.H., Cheng D., Hu P., West K.A., Dick G.J., Brodie E.L. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 2011;5:1014–1024. doi: 10.1038/ismej.2010.202. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Z.M.P., Bussema C., Schmidt T.M. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh M.B., Pellizari V.H., Uhlík O., Sutka R., Rodrigues J., Ostrom N.E. Biphenyl‐utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs) ISME J. 2007;1:134–148. doi: 10.1038/ismej.2007.26. et al. [DOI] [PubMed] [Google Scholar]
- Lendvay J.M., Löffler F.E., Dollhopf M., Aiello M.R., Daniels G., Fathepure B.Z. Bioreactive barriers: bioaugmentation and biostimulation for chlorinated solvent remediation. Environ Sci Technol. 2003;37:1422–1431. et al. [Google Scholar]
- Lévesque M.J., Beaudet R., Bisaillon J.G., Villemur R. Quantification of Desulfitobacterium frappieri strain PCP‐1 and Clostridium‐like strain 6 in mixed bacterial populations by competitive polymerase chain reaction. J Microbiol Methods. 1998;32:263–271. [Google Scholar]
- Löffler F.E., Sanford R.A., Tiedje J.M. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl Environ Microbiol. 1996;62:3809–3813. doi: 10.1128/aem.62.10.3809-3813.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler F.E., Sun Q., Li J.R., Tiedje J.M. 16S rRNA gene‐based detection of tetrachloroethene‐dechlorinating Desulfuromonas and Dehalococcoides species. Appl Environ Microbiol. 2000;66:1369–1374. doi: 10.1128/aem.66.4.1369-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler F.E., Cole J.R., Ritalahti K.M., Tiedje J.M. Diversity of dechlorinating bacteria. In: Häggblom M.M., Bossert I.D., editors. Kluwer Academic Press; 2003. pp. 53–87. [Google Scholar]
- Löffler F.E., Sanford R.A., Ritalahti K.M. Enrichment, cultivation, and detection of reductively dechlorinating bacteria. Methods Enzymol. 2005;397:77–111. doi: 10.1016/S0076-6879(05)97005-5. [DOI] [PubMed] [Google Scholar]
- Loy A., Kusel K., Lehner A., Drake H.L., Wagner M. Microarray and functional gene analyses of sulfate‐reducing prokaryotes in low‐sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl Environ Microbiol. 2004;70:6998–7009. doi: 10.1128/AEM.70.12.6998-7009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wilson J., Kampbell D. Comparison of an assay for Dehalococcoides DNA and a microcosm study in predicting reductive dechlorination of chlorinated ethenes in the field. Environ Pollut. 2009;157:809–815. doi: 10.1016/j.envpol.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Lutz S. Beyond directed evolution–semi‐rational protein engineering and design. Curr Opin Biotechnol. 2010;21:734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K.C., Mondav R., Sintrajaya R., Slattery B., Schmidt S., Schenk P.M. Development of an environmental functional gene microarray for soil microbial communities. Appl Environ Microbiol. 2010;76:7161–7170. doi: 10.1128/AEM.03108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson J.K., Stern R.V., Gossett J.M., Zinder S.H., Burris D.R. Reductive dechlorination of tetrachloroethene to ethene by two‐component enzyme pathway. Appl Environ Microbiol. 1998;64:1270–1275. doi: 10.1128/aem.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson J.K., Romine M.F., Burris D.R., Kingsley M.T. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl Environ Microbiol. 2000;66:5141–5147. doi: 10.1128/aem.66.12.5141-5147.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard J., Schumacher W., Vazquez F., Regeard C., Hagen W.R., Holliger C. Characterization of the corrinoid iron–sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl Environ Microbiol. 2003;69:4628–4638. doi: 10.1128/AEM.69.8.4628-4638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M., Whiteley A.S., Griffiths R.I., Bailey M.J. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C.H., Evans C., Jensen R.V., Sobral B.W.S. Identification of new genes in Sinorhizobium meliloti using the genome sequencer FLX system. BMC Microbiol. 2008;8:72. doi: 10.1186/1471-2180-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron P.‐A., Ranjard L., Mougel C., Lemanceau P. Metaproteomics: a new approach for studying functional microbial ecology. Microb Ecol. 2007;53:486–493. doi: 10.1007/s00248-006-9196-8. [DOI] [PubMed] [Google Scholar]
- Marsh T.L. Terminal restriction fragment length polymorphism (T‐RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr Opin Microbiol. 1999;2:323–327. doi: 10.1016/S1369-5274(99)80056-3. [DOI] [PubMed] [Google Scholar]
- Marsh T.L., Liu W.T., Forney L.J., Cheng H. Beginning a molecular analysis of the eukaryal community in activated sludge. Water Sci Technol. 1998;37:455–460. [Google Scholar]
- Marzorati M., Borin S., Brusetti L., Daffonchio D., Marsilli C., Carpani G. Response of 1,2‐dichloroethane‐adapted microbial communities to ex‐situ biostimulation of polluted groundwater. Biodegradation. 2006;17:41–56. doi: 10.1007/s10532-005-9004-z. et al. [DOI] [PubMed] [Google Scholar]
- Marzorati M., de Ferra F., Van Raemdonck H., Borin S., Allifranchini E., Carpani G. A novel reductive dehalogenase, identified in a contaminated groundwater enrichment culture and in Desulfitobacterium dichloroeliminans strain DCA1 is linked to dehalogenation of 1,2‐dichloroethane. Appl Environ Microbiol. 2007;73:2990–2999. doi: 10.1128/AEM.02748-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes T.E., Alexander A.K., Coleman N.V. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Rev. 2010;34:445–475. doi: 10.1111/j.1574-6976.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- May H.D., Miller G.S., Kjellerup B.V., Sowers K.R. Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl Environ Microbiol. 2008;74:2089–2094. doi: 10.1128/AEM.01450-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymó‐Gatell X., Chien Y.T., Gossett J.M., Zinder S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- Miller E., Wohlfarth G., Diekert G. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE‐S. Arch Microbiol. 1998;169:497–502. doi: 10.1007/s002030050602. [DOI] [PubMed] [Google Scholar]
- Miyata R., Adachi K., Tani H., Kurata S., Nakamura K., Tsuneda S. Quantitative detection of chloroethene‐reductive bacteria Dehalococcoides spp. using alternately binding probe competitive polymerase chain reaction. Mol Cell Probes. 2010;24:131–137. doi: 10.1016/j.mcp.2009.11.005. et al. [DOI] [PubMed] [Google Scholar]
- Morris R.M., Sowell S., Barofsky D., Zinder S., Richardson R. Transcription and mass‐spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ Microbiol. 2006;8:1499–1509. doi: 10.1111/j.1462-2920.2006.01090.x. [DOI] [PubMed] [Google Scholar]
- Morris R.M., Fung J.M., Rahm B.G., Zhang S., Freedman D.L., Zinder S.H. Comparative proteomics of Dehalococcoides spp. reveals strain‐specific peptides associated with activity. Appl Environ Microbiol. 2007;73:320–326. doi: 10.1128/AEM.02129-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.A., Rosner B.M., von Abendroth G., Meshulam‐Simon G., McCarty P.L., Spormann A.M. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70:4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G., Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek Int J Gen Mol Microbiol. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- Myers R.M., Maniatis T., Lerman L.S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- Narihiro T., Kaiya S., Futamata H., Hiraishi A. Removal of polychlorinated dioxins by semi‐aerobic fed‐batch composting with biostimulation of ‘Dehalococcoides’. J Biosci Bioeng. 2010;109:249–256. doi: 10.1016/j.jbiosc.2009.08.498. [DOI] [PubMed] [Google Scholar]
- Nemir A., David M.M., Perrussel R., Sapkota A., Simonet P., Monier J.‐M. Comparative phylogenetic microarray analysis of microbial communities in TCE‐contaminated soils. Chemosphere. 2010;80:600–607. doi: 10.1016/j.chemosphere.2010.03.036. et al. [DOI] [PubMed] [Google Scholar]
- Neufeld J.D., Wagner M., Murrell J.C. Who eats what, where and when? Isotope‐labelling experiments are coming of age. ISME J. 2007;1:103–110. doi: 10.1038/ismej.2007.30. [DOI] [PubMed] [Google Scholar]
- Neumann A., Wohlfarth G., Diekert G. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J Biol Chem. 1996;271:16515–16519. doi: 10.1074/jbc.271.28.16515. [DOI] [PubMed] [Google Scholar]
- Neumann A., Wohlfarth G., Diekert G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J Bacteriol. 1998;180:4140–4145. doi: 10.1128/jb.180.16.4140-4145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni S.S., Fredrickson J.K., Xun L.Y. Purification and characterization of a novel 3‐chlorobenzoate reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB‐1. J Bacteriol. 1995;177:5135–5139. doi: 10.1128/jb.177.17.5135-5139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke B.C., Chang Y.C., Hatsu M., Suzuki T., Takamizawa K. Purification, cloning, and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium bifermentans DPH‐1. Can J Microbiol. 2001;47:448–456. [PubMed] [Google Scholar]
- Parameswaran P., Jalili R., Tao L., Shokralla S., Gharizadeh B., Ronaghi M. A pyrosequencing‐tailored nucleotide barcode design unveils opportunities for large‐scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pas B.A., Smidt H., Hagen W.R., van der Oost J., Schraa G., Stams A.J.M. Purification and molecular characterization of ortho‐chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem. 1999;274:20287–20292. doi: 10.1074/jbc.274.29.20287. et al. [DOI] [PubMed] [Google Scholar]
- van de Pas B.A., Gerritse J., de Vos W.M., Schraa G., Stams A.J.M. Two distinct enzyme systems are responsible for tetrachloroethene and chlorophenol reductive dehalogenation in Desulfitobacterium strain PCE1. Arch Microbiol. 2001;176:165–169. doi: 10.1007/s002030100316. [DOI] [PubMed] [Google Scholar]
- Pavlova M., Klvana M., Prokop Z., Chaloupkova R., Banas P., Otyepka M. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat Chem Biol. 2009;5:727–733. doi: 10.1038/nchembio.205. et al. [DOI] [PubMed] [Google Scholar]
- Petrie L., North N.N., Dollhopf S.L., Balkwill D.L., Kostka J.E. Enumeration and characterization of iron(III)‐reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI) Appl Environ Microbiol. 2003;69:7467–7479. doi: 10.1128/AEM.69.12.7467-7479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radajewski S., Ineson P., Parekh N.R., Murrell J.C. Stable‐isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- Rahm B.G., Richardson R.E. Correlation of respiratory gene expression levels and pseudo‐steady‐state PCE respiration rates in Dehalococcoides ethenogenes. Environ Sci Technol. 2008a;42:416–421. doi: 10.1021/es071455s. [DOI] [PubMed] [Google Scholar]
- Rahm B.G., Richardson R.E. Dehalococcoides' gene transcripts as quantitative bioindicators of tetrachloroethene, trichloroethene, and cis‐1,2‐dichloroethene dehalorespiration rates. Environ Sci Technol. 2008b;42:5099–5105. doi: 10.1021/es702912t. [DOI] [PubMed] [Google Scholar]
- Rahm B.G., Chauhan S., Holmes V.F., Macbeth T.W., Sorenson K.S.J., Alvarez‐Cohen L. Molecular characterization of microbial populations at two sites with differing reductive dechlorination abilities. Biodegradation. 2006a;17:523–534. doi: 10.1007/s10532-005-9023-9. [DOI] [PubMed] [Google Scholar]
- Rahm B.G., Morris R.M., Richardson R.E. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl Environ Microbiol. 2006b;72:5486–5491. doi: 10.1128/AEM.00855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl Environ Microbiol. 2009;75:2495–2505. doi: 10.1128/AEM.02409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regeard C., Maillard J., Holliger C. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J Microbiol Methods. 2004;56:107–118. doi: 10.1016/j.mimet.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Rhee S.K., Fennell D.E., Häggblom M.M., Kerkhof L.J. Detection by PCR of reductive dehalogenase motifs in a sulfidogenic 2‐bromophenol‐degrading consortium enriched from estuarine sediment. FEMS Microbiol Ecol. 2003;43:317–324. doi: 10.1111/j.1574-6941.2003.tb01072.x. [DOI] [PubMed] [Google Scholar]
- Ritalahti K.M., Hatt J.K., Lugmayr V., Henn L., Petrovskis E.A., Ogles D.M. Comparing on‐site to off‐site biomass collection for Dehalococcoides biomarker gene quantification to predict in situ chlorinated ethene detoxification potential. Environ Sci Technol. 2010;44:5127–5133. doi: 10.1021/es100408r. et al. [DOI] [PubMed] [Google Scholar]
- Roesch L.F., Fulthorpe R.R., Riva A., Casella G., Hadwin A.K.M., Kent A.D. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röling W.F.M. Do microbial numbers count? Quantifying the regulation of biogeochemical fluxes by population size and cellular activity. FEMS Microbiol Ecol. 2007;62:202–210. doi: 10.1111/j.1574-6941.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- Rowe A.R., Lazar B.J., Morris R.M., Richardson R.E. Characterization of the community structure of a dechlorinating mixed culture and comparisons of gene expression in planktonic and biofloc‐associated ‘Dehalococcoides’ and Methanospirillum species. Appl Environ Microbiol. 2008;74:6709–6719. doi: 10.1128/AEM.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T., Munetsuna E. Enzyme systems for biodegradation of polychlorinated dibenzo‐p‐dioxins. Appl Microbiol Biotechnol. 2010;88:23–30. doi: 10.1007/s00253-010-2765-2. [DOI] [PubMed] [Google Scholar]
- Sanford R.A., Wu Q., Sung Y., Thomas S.H., Amos B.K., Prince E.K. Hexavalent uranium supports growth of Anaeromyxobacter dehalogenans and Geobacter spp. with lower than predicted biomass yields. Environ Microbiol. 2007;9:2885–2893. doi: 10.1111/j.1462-2920.2007.01405.x. et al. [DOI] [PubMed] [Google Scholar]
- Sanguin H., Herrera A., Oger‐Desfeux C., Dechesne A., Simonet P., Navarro E. Development and validation of a prototype 16S rRNA‐based taxonomic microarray for Alphaproteobacteria. Environ Microbiol. 2006a;8:289–307. doi: 10.1111/j.1462-2920.2005.00895.x. et al. [DOI] [PubMed] [Google Scholar]
- Sanguin H., Remenant B., Dechesne A., Thioulouse J., Vogel T.M., Nesme X. Potential of a 16S rRNA‐based taxonomic microarray for analyzing the rhizosphere effects of maize on Agrobacterium spp. and bacterial communities. Appl Environ Microbiol. 2006b;72:4302–4312. doi: 10.1128/AEM.02686-05. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schlötelburg C., von Wintzingerode C., Hauck R., von Wintzingerode F., Hegemann W., Göbel U.B. Microbial structure of an anaerobic bioreactor population that continuously dechlorinates 1,2‐dichloropropane. FEMS Microbiol Ecol. 2002;39:229–237. doi: 10.1111/j.1574-6941.2002.tb00925.x. [DOI] [PubMed] [Google Scholar]
- Schnoes A.M., Brown S.D., Dodevski I., Babbitt P.C. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher W., Holliger C., Zehnder A.J.B., Hagen W.R. Redox chemistry of cobalamin and iron–sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 1997;409:421–425. doi: 10.1016/s0014-5793(97)00520-6. [DOI] [PubMed] [Google Scholar]
- Shelton D.R., Tiedje J.M. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3‐chlorobenzoic acid. Appl Environ Microbiol. 1984;48:840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- Shendure J., Ji H.L. Next‐generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Smidt H., Akkermans A.D.L., van der Oost J., de Vos W.M. Halorespiring bacteria‐molecular characterization and detection. Enzyme Microb Technol. 2000;27:812–820. doi: 10.1016/s0141-0229(00)00316-1. [DOI] [PubMed] [Google Scholar]
- Smith A.M., Heisler L.E., St. Onge R.P., Farias‐Hesson E., Wallace I.M., Bodeau J. Highly‐multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res. 2010;38:e142. doi: 10.1093/nar/gkq368. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits T.H.M., Devenoges C., Szynalski K., Maillard J., Holliger C. Development of a real‐time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J Microbiol Methods. 2004;57:369–378. doi: 10.1016/j.mimet.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Stingl U., Tripp H.J., Giovannoni S.J. Improvements of high‐throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J. 2007;1:361–371. doi: 10.1038/ismej.2007.49. [DOI] [PubMed] [Google Scholar]
- Sul W.J., Park J., Quensen J.F., Rodrigues J.L.M., Seliger L., Tsoi T.V. DNA‐stable isotope probing integrated with metagenomics for retrieval of biphenyl dioxygenase genes from polychlorinated biphenyl‐contaminated river sediment. Appl Environ Microbiol. 2009;75:5501–5506. doi: 10.1128/AEM.00121-09. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y. School of Civil and Environmental Engineering, Georgia Institute of Technology; 2005. [Google Scholar]
- Sung Y., Ritalahti K.M., Apkarian R.P., Löffler F.E. Quantitative PCR confirms purity of strain GT, a novel trichloroethene‐to‐ethene‐respiring Dehalococcoides isolate. Appl Environ Microbiol. 2006;72:1980–1987. doi: 10.1128/AEM.72.3.1980-1987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama A., Yamashita M., Yoshino S., Furukawa K. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J Bacteriol. 2002;184:3419–3425. doi: 10.1128/JB.184.13.3419-3428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.J.J., Yi S., Zhuang W.Q., Zinder S.H., Keasling J.D., Alvarez‐Cohen L. Investigation of carbon metabolism in ‘Dehalococcoides ethenogenes’ strain 195 by use of isotopomer and transcriptomic analyses. J Bacteriol. 2009;191:5224–5231. doi: 10.1128/JB.00085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taş N., van Eekert M.H.A., Schraa G., Zhou J.Z., de Vos W.M., Smidt H. Tracking functional guilds: ‘Dehalococcoides’ spp. in European river basins contaminated with hexachlorobenzene. Appl Environ Microbiol. 2009;75:4696–4704. doi: 10.1128/AEM.02829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taş N., Heilig H., van Eekert M.H.A., Schraa G., de Vos W.M., Smidt H. Concurrent hexachlorobenzene and chloroethene transformation by endogenous dechlorinating microorganisms in the Ebro River sediment. FEMS Microbiol Ecol. 2010a;74:682–692. doi: 10.1111/j.1574-6941.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- Taş N., Van Eekert M.H.A., De Vos W.M., Smidt H. The little bacteria that can – diversity, genomics and ecophysiology of ‘Dehalococcoides’ spp. in contaminated environments. Microb Biotechnol. 2010b;3:389–402. doi: 10.1111/j.1751-7915.2009.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau J., Gauthier A., Duguay M., Villemur R., Lépine F., Juteau P. Purification, cloning, and sequencing of a 3,5‐dichlorophenol reductive dehalogenase from Desulfitobacterium frappieri PCP‐1. Appl Environ Microbiol. 2004;70:4532–4537. doi: 10.1128/AEM.70.8.4532-4537.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.H., Padilla‐Crespo E., Jardine P.M., Sanford R.A., Löffler F.E. Diversity and distribution of Anaeromyxobacter strains in a uranium‐contaminated subsurface environment with a nonuniform groundwater flow. Appl Environ Microbiol. 2009;75:3679–3687. doi: 10.1128/AEM.02473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann S., Strömpl C., Timmis K.N., Abraham W.‐R. Stable isotope probing reveals the dominant role of Burkholderia species in aerobic degradation of PCBs. FEMS Microbiol Ecol. 2005;52:207–217. doi: 10.1016/j.femsec.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Tourova T.P. Copy number of ribosomal operons in prokaryotes and its effect on phylogenetic analyses. Microbiology. 2003;72:389–402. [PubMed] [Google Scholar]
- Van Nostrand J.D., Wu W.M., Wu L.Y., Deng Y., Carley J., Carroll S. GeoChip‐based analysis of functional microbial communities during the reoxidation of a bioreduced uranium‐contaminated aquifer. Environ Microbiol. 2009;11:2611–2626. doi: 10.1111/j.1462-2920.2009.01986.x. et al. [DOI] [PubMed] [Google Scholar]
- Vieites J.M., Guazzaroni M.‐E., Beloqui A., Golyshin P.N., Ferrer M. Metagenomics approaches in systems microbiology. FEMS Microbiol Rev. 2009;33:236–255. doi: 10.1111/j.1574-6976.2008.00152.x. [DOI] [PubMed] [Google Scholar]
- Villemur R., Lanthier M., Beaudet R., Lépine F. The Desulfitobacterium genus. FEMS Microbiol Rev. 2006;30:706–733. doi: 10.1111/j.1574-6976.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- Wagner A., Adrian L., Kleinsteuber S., Andreesen J.R., Lechner U. Transcription analysis of genes encoding homologues of reductive dehalogenases in ‘Dehalococcoides’ sp. strain CBDB1 by using terminal restriction fragment length polymorphism and quantitative PCR. Appl Environ Microbiol. 2009;75:1876–1884. doi: 10.1128/AEM.01042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., He J. Separation of fluorescence‐labelled terminal restriction fragment DNA on a two‐dimensional gel (T‐RFs‐2D) – an efficient approach for microbial consortium characterization. Environ Microbiol. 2011;13:2565–2575. doi: 10.1111/j.1462-2920.2011.02527.x. [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. RNA‐Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J.E.M., Fagervold S.K., May H.D., Sowers K.R. A PCR‐based specific assay reveals a population of bacteria within the Chloroflexi associated with the reductive dehalogenation of polychlorinated biphenyls. Microbiology. 2005;151:2039–2046. doi: 10.1099/mic.0.27819-0. [DOI] [PubMed] [Google Scholar]
- Werner J.J., Ptak A.C., Rahm B.G., Zhang S., Richardson R.E. Absolute quantification of Dehalococcoides proteins: enzyme bioindicators of chlorinated ethene dehalorespiration. Environ Microbiol. 2009;11:2687–2697. doi: 10.1111/j.1462-2920.2009.01996.x. [DOI] [PubMed] [Google Scholar]
- West K.A., Johnson D.R., Hu P., DeSantis T.Z., Brodie E.L., Lee P.K.H. Comparative genomics of ‘Dehalococcoides ethenogenes’ 195 and an enrichment culture containing unsequenced ‘Dehalococcoides’ strains. Appl Environ Microbiol. 2008;74:3533–3540. doi: 10.1128/AEM.01835-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D.C., Geyer R., Peacock A.D., Hedrick D.B., Koenigsberg S.S., Sung Y. Phospholipid furan fatty acids and ubiquinone‐8: Lipid biomarkers that may protect Dehalococcoides strains from free radicals. Appl Environ Microbiol. 2005;71:8426–8433. doi: 10.1128/AEM.71.12.8426-8433.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol. 1995;61:1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Rash B.A., Rainey F.A., Moe W.M. Detection and quantification of Dehalogenimonas and ‘Dehalococcoides’ populations via PCR‐based protocols targeting 16S rRNA genes. Appl Environ Microbiol. 2009a;75:7560–7564. doi: 10.1128/AEM.01938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Rash B.A., Rainey F.A., Moe W.M. Isolation of novel bacteria within the Chloroflexi capable of reductive dechlorination of 1,2,3‐trichloropropane. Environ Microbiol. 2009b;11:833–843. doi: 10.1111/j.1462-2920.2008.01804.x. [DOI] [PubMed] [Google Scholar]
- Ye L.D., Schilhabel A., Bartram S., Boland W., Diekert G. Reductive dehalogenation of brominated ethenes by Sulfurospirillum multivorans and Desulfitobacterium hafniense PCE‐S. Environ Microbiol. 2010;12:501–509. doi: 10.1111/j.1462-2920.2009.02093.x. [DOI] [PubMed] [Google Scholar]
- Yoshida N., Takahashi N., Hiraishi A. Phylogenetic characterization of a polychlorinated‐dioxin‐dechlorinating microbial community by use of microcosm studies. Appl Environ Microbiol. 2005;71:4325–4334. doi: 10.1128/AEM.71.8.4325-4334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Morrison M. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR‐denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2004;70:4800–4806. doi: 10.1128/AEM.70.8.4800-4806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanaroli G., Balloi A., Negroni A., Daffonchio D., Fava F. A Dehaloccoides‐like bacterium and a new reductive dehalogenase are responsible for PCB dechlorination in marine sediments under in situ biogeochemical conditions. J Biotechnol. 2010;150:S270–S270. [Google Scholar]
- Zentilin L., Giacca M. Competitive PCR for precise nucleic acid quantification. Nat Protoc. 2007;2:2092–2104. doi: 10.1038/nprot.2007.299. [DOI] [PubMed] [Google Scholar]
- Zhang H.S., Ziv‐El M., Rittmann B.E., Krajmalnik‐Brown R. Effect of dechlorination and sulfate reduction on the microbial community structure in denitrifying membrane‐biofilm reactors. Environ Sci Technol. 2010;44:5159–5164. doi: 10.1021/es100695n. [DOI] [PubMed] [Google Scholar]


