Summary
Methane‐oxidizing bacteria (MOB) have a large potential as a microbial sink for the greenhouse gas methane as well as for biotechnological purposes. However, their application in biotechnology has so far been hampered, in part due to the relative slow growth rate of the available strains. To enable the availability of novel strains, this study compares the isolation of MOB by conventional dilution plating with miniaturized extinction culturing, both performed after an initial enrichment step. The extinction approach rendered 22 MOB isolates from four environmental samples, while no MOB could be isolated by plating. In most cases, extinction culturing immediately yielded MOB monocultures making laborious purification redundant. Both type I (Methylomonas spp.) and type II (Methylosinus sp.) MOB were isolated. The isolated methanotrophic diversity represented at least 11 different strains and several novel species based on 16S rRNA gene sequence dissimilarity. These strains possessed the particulate (100%) and soluble (64%) methane monooxygenase gene. Also, 73% of the strains could be linked to a highly active fast‐growing mixed MOB community. In conclusion, miniaturized extinction culturing was more efficient in rapidly isolating numerous MOB requiring little effort and fewer materials, compared with the more widely applied plating procedure. This miniaturized approach allowed straightforward isolation and could be very useful for subsequent screening of desired characteristics, in view of their future biotechnological potential.
Introduction
Next to carbon dioxide, methane is the second most important greenhouse gas contributing to one‐fifth of global warming (Dalal and Allen, 2008). Yearly, 30 Tg of atmospheric methane is removed through oxidation by aerobic methane‐oxidizing bacteria (MOB), a unique group of bacteria capable of utilizing methane as their sole carbon and energy source (Trotsenko and Murrell, 2008). The key enzyme of these microorganisms, particulate or soluble methane monooxygenase (pMMO and sMMO), is remarkable in its broad substrate specificity (Semrau et al., 2010). To date, MOB have shown potential in three fields of industrial biotechnology, namely (i) geoengineering the climate through assimilation of methane and thus mitigating greenhouse effects (Scheutz et al., 2009), (ii) bioremediation of pollutants via co‐metabolism by MMOs (Wendlandt et al., 2010), and (iii) production of commercially relevant metabolites (Zhang et al., 2008; Jiang et al., 2010). Despite the multifunctional potential of these bacteria, there are still several factors limiting large‐scale applicability in industrial processes, which are mostly related to the MOB themselves, such as slow growth rates and low substrate affinity (Jiang et al., 2010). In order to achieve the full potential of MOB and communities for such biotechnological applications, a straightforward isolation technique is of high value, as it allows the optimization of the growth conditions of the MOB of interest. However, their cultivation is still laborious (Bowman, 2006). To date, only few MOB have been studied and examined thoroughly for further biotechnological applications (Jiang et al., 2010). The examined cultures were often not selected for their optimal use in these bioprocesses but were just the only available methanotrophic cultures at the time.
The first MOB were isolated by Söhngen (1906), but it was not until 1970 that Whittenbury and co‐workers established a successful procedure to isolate and characterize methane‐oxidizing bacteria by plating on nitrate‐ or ammonium mineral salts (NMS/AMS) medium (Whittenbury et al., 1970). Since then, novel research on MOB mainly encompassed exploring new environments and using cultivation conditions adapted to the corresponding environment (Dedysh et al., 1998a; Wise et al., 1999; Svenning et al., 2003; Bussmann et al., 2004). Most of the methanotrophic diversity thus obtained could be phylogenetically positioned within the Alphaproteobacteria[such as Methylosinus (Type II)] and the Gammaproteobacteria[such as Methylomonas (Type Ia) and Methylococcus (Type Ib)] (Dedysh, 2009). Since the recognition of ‘The Great Anomaly’ (Staley and Konopka, 1985), alternative ways to increase the general cultivability of the microbial diversity were explored, for example by application of growth conditions that closely mimic the natural environment (Dedysh et al., 1998a), prolonged incubation at low temperatures (Song et al., 2009) or the use of alternative gelling agents replacing agar (Janssen et al., 2002; Dedysh et al., 2007; Stott et al., 2008; Tamaki et al., 2009). Another approach was extinction culturing by diluting a sample to the point of extinction and thereby purifying to a less complex sample containing only one or a few organisms (Button et al., 1993; Schut et al., 1993), which was further optimized by the development of high‐throughput culturing methods (Connon and Giovannoni, 2002; Rappe et al., 2002; Stingl et al., 2007). To date, MOB are still mostly isolated via plate methods, whereby one or several liquid enrichment steps are followed by a serial dilution onto plates (Dunfield et al., 2003; Dedysh et al., 2004; Heyer et al., 2005; Tsubota et al., 2005; Wartiainen et al., 2006). However, these procedures are very laborious and almost always require elaborate purification.
The main objective of this study was to combine and optimize several of the above‐mentioned cultivation approaches for the specific isolation of methane‐oxidizing bacteria, and to compare the isolation efficiency of the resulting two‐step liquid isolation procedure, consisting of an initial prolonged enrichment and subsequent miniaturized extinction culturing, with conventional dilution plating. Initial enrichment followed by extinction culturing greatly simplified purification procedures and easily rendered novel methanotrophic strains, which were further characterized by sequence analysis of the 16S rRNA gene, repetitive element sequence based PCR fingerprinting (rep‐PCR), pmoA and mmoX gene amplification (encoding for pMMO and sMMO respectively) as well as sMMO activity assays. Most of the strains were traced back to highly active and fast‐growing methanotrophic communities through denaturing gradient gel electrophoresis (DGGE) targeting the pmoA gene.
Results
Isolation of fast‐growing MOB
Four environmental samples (wastewater treatment plant, wetland, biofilter and slurry pit) were serially diluted and enriched, while monitoring methane and oxygen consumption and carbon dioxide production. During the first four weeks, higher dilutions were gradually found positive with each subsequent measurement, stabilizing by the fifth week (Fig. S1). These data were used to calculate the abundance of cultivable MOB under given growth conditions (MPN g−1 expressed as MPN index [95% confidence]): cultivable MOB were most abundant in the wastewater treatment plant (4.3 × 104 MOB g−1[9.0 × 103 to 1.8 × 105 MOB g−1]), followed by wetland (4.3 × 103 MOB g−1[9.0 × 102 to 1.8 × 104 MOB g−1]) and then the slurry pit and biofilter samples (both 2.3 × 102 MOB g−1[4.6 × 101 to 9.4 × 102 MOB g−1]). After 5 weeks of incubation at 20°C, 23 dilutions were found positive for methane oxidation and were used further for MOB isolation.
The extinction culturing procedure was performed in duplicate to isolate MOB from these enrichment cultures, resulting in 46 extinction series, of which only four did not show growth after two weeks of incubation. The highest dilutions of the remaining 42 series were transferred to gas‐tight vials and diluted nitrate mineral salts (dNMS) medium solidified with gellan gum for confirmation of methane consumption and purity check respectively. Methane oxidation was observed in 27 series, of which 14 were immediately monocultures. Seven methane‐oxidizing cultures consisted of an MOB in co‐culture with a non‐methanotrophic bacterium (identified as a member of Nocardioides with 16S rRNA gene sequence analysis) forming a distinct colony morphology surrounding the methanotrophic colonies. From the remaining six methane‐oxidizing cultures, several different colony morphologies were found upon plating. Subsequent purification of these MOB was achieved after a maximum of three sub‐cultivation steps. One dilution series resulted in a monoculture (identified as Ancylobacter with 16S rRNA gene sequence analysis), but did not oxidize CH4. Fourteen dilutions were discarded since these were not able to oxidize CH4 and did not result in a pure culture upon plating.
Dilution plating was performed in parallel. The 23 initial methane‐oxidizing enrichments were diluted, plated on dNMS (solidified with gellan gum) and incubated under atmospheric conditions supplemented with CH4. Randomly, 200 colonies were picked up and purified. Almost all purified isolates (197 out of 200) showed heterotrophic growth on diluted TSA without CH4 and were therefore not considered as potential MOB. The three remaining isolates, not able to grow on diluted TSA, also failed to grow on solid dNMS with or without CH4 added to the headspace. To confirm these results, eight randomly selected isolates obtained via dilution plating were identified to the genus level through 16S rRNA gene sequence analysis. These sequences were affiliated with Nocardioides, Zoogloea, Rhizobium, Pseudomonas, Polaromonas, Rhodobacter and Enterobacter, genera not harbouring known MOB.
Identification and characterization of MOB isolates
In total, 22 purified MOB isolates were retrieved from four different samples (Table 1). Dereplication with rep‐PCR fingerprint analysis grouped the isolates into 11 distinct clusters, representing 11 unique strains (Fig. 1). Randomly chosen representatives of each strain were further identified to the genus level with 16S rRNA gene sequence analysis. Ten out of 11 methanotrophic strains were assigned to the gammaproteobacterial genus Methylomonas, while the remaining strain was assigned to the alphaproteobacterial genus Methylosinus (Table 1). Pairwise comparisons of the 16S rRNA gene sequences of the 10 newly isolated strains with the type strains of all species of Methylomonas suggested that representatives of potentially novel Methylomonas species were isolated from all four environments, with 16S rRNA gene sequence similarities below 98% (Stackebrandt and Ebers, 2006). The 11 reference strains contained the pmoA gene, seven of which also harboured the mmoX gene and showed sMMO activity (Table 1): six Methylomonas strains isolated from the WWTP sample as well as the Methylosinus sp. strain.
Table 1.
Genus assignment of 11 representative MOB strains (rep‐PCR; Fig. 1) based on 16S rRNA gene sequence analysis (> 1400 bp).
| Sample | Representative Strain | sMMO & mmoX | Genus identification | Type strain with highest 16S rRNA gene sequence similarity to query sequence | |||
|---|---|---|---|---|---|---|---|
| Species name | Strain number | Sequence similarity | Accession number | ||||
| WWTP | D1 | Yes | Methylomonas | Methylomonas methanica | NCIMB 11130T | 98.5% | AF304196 |
| C1 | Yes | Methylomonas | Methylomonas methanica | NCIMB 11130T | 98.4% | AF304196 | |
| G1 | Yes | Methylomonas | Methylomonas methanica | NCIMB 11130T | 98.6% | AF304196 | |
| E1 | Yes | Methylomonas | Methylomonas methanica | NCIMB 11130T | 98.6% | AF304196 | |
| H1 | Yes | Methylomonas | Methylomonas methanica | NCIMB 11130T | 98.6% | AF304196 | |
| K1 | Yes | Methylomonas | Methylomonas methanica | NCIMB 11130T | 98.3% | AF304196 | |
| B1 | No | Methylomonas | Methylomonas scandinavica | SR5T | 97.5% | AJ131369 | |
| Slurry Pit | I1 | No | Methylomonas | Methylomonas scandinavica | SR5T | 97.5% | AJ131369 |
| Wetland | A6a | No | Methylomonas | Methylomonas fodinarum | ACM 3268T | 96.2% | X72778 |
| J1 | No | Methylomonas | Methylomonas fodinarum | ACM 3268T | 95.6% | X72778 | |
| F1 | Yes | Methylosinus | Methylosinus sporium | NCIMB 11126T | 98.9% | Y18946 | |
| Biofilter | A6a | No | Methylomonas | Methylomonas fodinarum | ACM 3268T | 96.2% | X72778 |
Five wetland and two biofilter isolates clustered together according to rep‐PCR, wetland isolate A6 was randomly selected as representative strain.
Similarity values of 16S rRNA gene sequence to closest type strain, origin of the strains and results of naphthalene oxidation assay and mmoX gene amplification are given.
Figure 1.
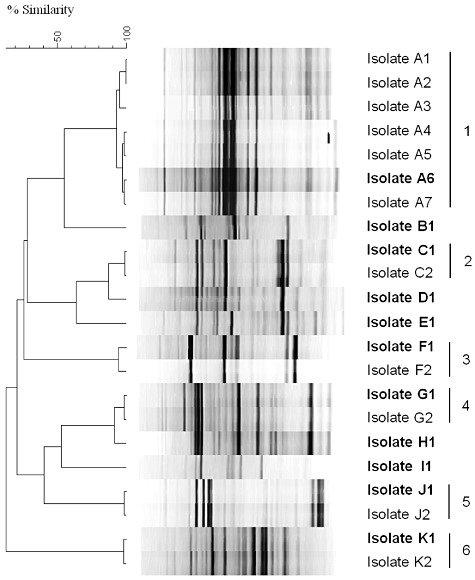
GTG5 rep‐PCR fingerprinting using Pearson product moment correlation coefficient and UPGMA. MOB isolates were divided into 11 groups: 5 isolates showing unique profiles (B1, D1, E1, H1 and I2) and 6 groups of isolates (1–6) showing identical profiles. F1 and F2 isolates belonged to the genus Methylosinus, all the other isolates were identified as members of Methylomonas (Table 1). A representative of each cluster was selected randomly (indicated in bold) for further characterization.
Detection of the isolated MOB in fast‐growing methanotrophic communities
Sequence batch enrichments from the original samples were set up in parallel with the dilution series enrichments used for MOB isolation, with the same cultivation conditions.
To evaluate the presence of the isolates, under conditions selecting for highly active fast‐growing MOB in a mixed community, a sequence batch set‐up with a relatively low sludge retention time of 90 h was chosen. All sequence batch enrichments showed methane oxidation activity, although the moment that a significant methane oxidation (a drop below the initial average methane concentration subtracted by five times the standard deviation) was observed differed between samples: 72 h for the WWTP cultures, 96 h for the wetland cultures and 144 h for the cultures inoculated with samples from the slurry pit or the biofilter material (Fig. S2). A steep rise in the methane oxidation rate (MOR) over time was observed, with a maximum after 144 h of 169 ± 28 mg CH4 l−1 day−1 and 184 ± 24 mg CH4 l−1 day−1 for the WWTP and wetland cultures respectively. The maximal MOR was lower for the two other cultures, with 83 ± 3 mg CH4 l−1 day−1 after 192 h and 39 ± 45 mg CH4 l−1 day−1 after 216 h for the slurry pit cultures and the biofilter cultures respectively. When the oxygen concentration in the reactors became limiting, a decrease in the MOR was observed and a second cycle was started. The observed daily MOR for the second cycle was of the same order for all four inocula with a minimum of 108 ± 16 (biofilter) and a maximum of 266 ± 7 (wetland) mg CH4 l−1 day−1 (Table S1).
PmoA DGGE analyses (Fig. 2) were performed on the 11 representative strains selected following rep‐PCR fingerprinting (Fig. 1) and SBR enrichments after the first cycle of each environmental sample. DGGE profiles of WWTP and wetland were more diverse than those of the biofilter and slurry pit. From the seven Methylomonas strains isolated from the WTTP only one (represented by isolate K1) could not be traced back to the complete profile of the SBR enrichment, while this was the case for the other six strains, suggesting that these were dominantly present. It is however clear that different strains (isolates C1, D1, G1, E1 and H1), as proven by rep analysis, did show a pmoA band at a similar height, indicating that one band of the complete profile from the SBR enrichment covered a diversity of different methanotrophic strains. From the wetland, the two retrieved Methylomonas strains (represented by isolate A6 and J1) could be traced back to the SBR enrichment, while this was not the case for the single Methylosinus strain (isolate F1). The two isolates retrieved from the biofilter formed a single stable rep cluster with isolates from wetland, represented by isolate A6. The pmoA band of this strain was also observed in the DGGE profile of the biofilter, again suggesting the dominant presence in the SBR enrichment of this sample. From the slurry pit, only one strain was isolated for which the matching pmoA band could not be observed in the DGGE profile of the SBR enrichment. However, the pmoA band of this strain had a similar GC content as isolate B1 from WWTP, of which rep analysis already showed that they represented different strains.
Figure 2.
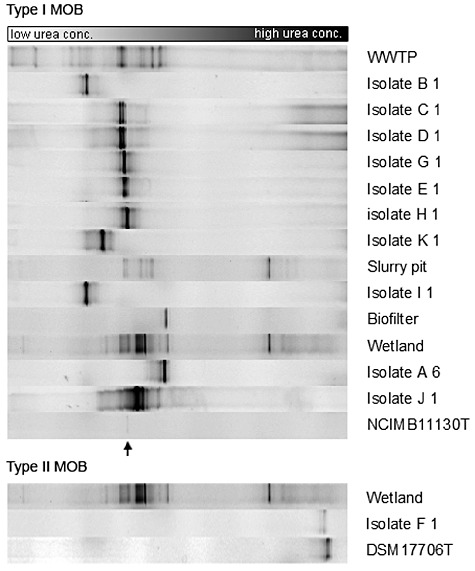
DGGE analysis, based on the pmoA primer set A189fGC/mb661r for type I and type II MOB respectively. The community in the active enrichment from one sequence batch reactor is shown after the first cycle for each of the four samples, i.e. a sample from the wastewater treatment plant (WWTP), a slurry pit, biofilter material and a wetland. The 11 representative strains selected based on rep‐PCR fingerprinting (Fig. 1) are shown in relation to their corresponding active enrichment. Strains NCIMB11130T (Methylomonas methanica, band indicated by arrow) and DSM17706T (Methylosinus sporium) are shown as reference. Based on the band position of the strains, eight out of 11 representative strains could be linked to their active community, demonstrating the abundance of these strains in a mixed community selecting for fast‐growing methane‐oxidizing bacteria.
Discussion
Methane‐oxidizing bacteria can serve as important sinks for the greenhouse gas methane or as key players in different biotechnological industries (Semrau et al., 2010). Currently, their applicability is limited by the number of suitable strains readily available, which were not specifically isolated for this purpose, and the lack of the necessary properties for efficient use in large‐scale industrial applications (Jiang et al., 2010). Therefore, novel methanotrophic strains need to be easily obtained in culture and characterized. However, the isolation of MOB with the conventional plating approach is laborious and time‐consuming, requiring one or more liquid enrichment steps followed by serial dilution plating and extensive purification (Whittenbury et al., 1970; Bowman et al., 1993; Bodrossy et al., 1995; Dedysh et al., 1998b; Iguchi et al., 2010). Therefore, in this study, a simple and miniaturized isolation protocol was applied to efficiently isolate MOB by a combination of several recently developed cultivation procedures: a prolonged initial enrichment at low temperatures (Song et al., 2009) and adaptation of incubation conditions for desired MOB (Dedysh et al., 1998a), followed by high‐throughput extinction culturing (Button et al., 1993; Wise et al., 1999; Connon and Giovannoni, 2002; Bodelier et al., 2005) and subsequent purification using gellan gum plates (Janssen et al., 2002; Dedysh et al., 2007). Other high‐throughput culturing approaches were designed to favour the isolation of abundant bacteria in situ (Connon and Giovannoni, 2002; Rappe et al., 2002; Stingl et al., 2007; Song et al., 2009), while our protocol specifically targeted a certain sub‐population of a specific functional group with a custom‐made prolonged enrichment. This made the organisms of interest, in this case fast‐growing MOB, abundant ex situ before extinction culturing. This approach resulted in immediate pure cultures, avoiding elaborate purification, which is known to be problematic for the isolation of MOB (Bowman, 2006). Without this approach, no immediate MOB pure culture would be obtained since heterotrophic bacteria are more abundantly present in environmental samples (Wise et al., 1999). In perspective of biotechnological applications, varying the cultivation parameters of the initial enrichment in combination with miniaturization in 96‐well plates has the potential to select for MOB with specific desired characteristics. As such, these wanted MOB can become abundant, even if non‐abundant in situ, and can then be rapidly isolated via miniaturized extinction culturing. For example, the cultivation conditions chosen in this study, a diluted NMS medium under a high concentration of CH4 and a relatively low hydraulic retention time, are known to select for highly active fast‐growing MOB, a characteristic which is important for industrial use of bacteria (Begonja and Hrsak, 2001; Schrader et al., 2009; Wendlandt et al., 2010). Indeed, the mixed SBR communities showed a high methane removal rate from 108 ± 16 to 266 ± 7 mg CH4 l−1 day−1 respectively, which is in the range of reported highly active methane‐oxidizing communities (Melse and Van der Werf, 2005; Nikiema et al., 2005; Gebert and Grongroft, 2006; Scheutz et al., 2009). By band position analysis of pmoA‐targeted DGGE, most isolated MOB could be traced back to this active community, demonstrating that the isolated strains were able to rapidly oxidize methane and grow to higher densities in a competitive setting. Most isolated MOB were indeed closely related to Methylomonas (Type Ia), known to harbour fast‐growing MOB with a short generation time of 3.5 h (Whittenbury et al., 1970). Fast‐growing Type Ib MOB (i.e. Methylococcus) with similar doubling times (Whittenbury et al., 1970) were not expected to be retrieved because they require higher isolation temperatures, while type II MOB are generally known to grow slower, with generation times ranging from 5 h up to several days (Whittenbury et al., 1970; Dedysh et al., 2000; 2002; Vorobev et al., 2010). Since DGGE patterns mainly represent the major constituents of a community (Muyzer and Smalla, 1998), the few strains that could not be traced back by band position analysis probably were expected to have longer generation times and as such were not abundantly present in the highly active methane‐oxidizing communities. Not all pmoA bands in the mixed communities had isolated representatives, therefore an up‐scaling of the isolation campaign, with more extinction cultures per enrichment, would detect more novel MOB cultivable under the set conditions. This was confirmed as extinction series from initial enrichments did lead to the isolation of two different MOB strains (strains E1 & H1 and strains B1 & D1).
In total, 22 MOB isolates belonging to 11 distinct strains were obtained, which is a relatively large number compared with other studies (Dianou and Adachi, 1999; Auman et al., 2000; Bussmann et al., 2004; Miller et al., 2004). Members of Methylomonas (type I MOB) were isolated from all four samples, while Methylosinus representatives (type II MOB) were only obtained from the wetland sample. Despite that isolates were assigned to known genera, they represented at least 10 different Methylomonas strains, several of which belonging to novel species within the genus based on 16S rRNA gene sequence dissimilarity with Methylomonas type strains. Currently, four species have been validly described, although only the type strain of Methylomonas methanica (NCIMB 11130T) is still accessible for the scientific community. This issue on availability of fastidious microorganisms such as MOB is a widely recognized problem and greatly hampers the in‐depth investigation of their biotechnological potential, especially because properties such as substrate affinity, growth rate, substrate range or degradation of xenobiotics are strain‐dependent features. For example, six different Methylomonas strains isolated in this study possessed sMMO, in addition to pMMO, while the type strain of the genus does not possess sMMO (Koh et al., 1993). Both sMMO and pMMO are known to degrade pollutants, such as chlorinated hydrocarbons (Bowman et al., 1993; Jiang et al., 2010): sMMO has a broader substrate range and is known to rapidly degrade pollutants, pMMO degrades compounds at slower rates but over an extended time frame (Semrau et al., 2010). While common in type II & Ib MOB (Hanson and Hanson, 1996), few type Ia MOB such as Methylomonas contain sMMO although it has been reported (Koh et al., 1993; Shen et al., 1997; Auman et al., 2000; Bussmann et al., 2006). Since type I MOB have a higher efficiency in carbon conversion (Scheutz et al., 2009), these Methylomonas isolates, which possess both sMMO and pMMO, could particularly be of interest to screen for degradation of recalcitrant compounds.
In this study, the isolation efficiency for the retrieval of fast‐growing MOB between miniaturized extinction culturing and conventional dilution plating was compared. To our knowledge, such a comparison has not been previously reported. The plating approach, which is the most applied method for isolation of MOB (Dunfield et al., 2003; Dedysh et al., 2004; Heyer et al., 2005; Tsubota et al., 2005; Wartiainen et al., 2006), did not render any MOB in this study when performed in parallel with extinction culturing from the same initial enrichments. However, the applied methodology should allow their isolation since MOB isolates obtained from extinction culturing and reference strains from public bacteria collections were cultivated successfully on solid medium in the same manner. Therefore, it is likely that additional plating trials investigating more colonies would allow the isolation of MOB. However, even then miniaturized extinction culturing from initial enrichments will be more time and labour efficient than conventional plating in retrieving numerous methane‐oxidizing bacteria with specific desired characteristics.
Experimental procedures
Isolation of methane‐oxidizing bacteria
Samples were taken from (i) the top layer of a denitrification tank of a wastewater treatment plant (WWTP, Ossemeersen, Gent, Belgium), (ii) a covered but aerobic slurry pit of a cow stable (Melle, Belgium), (iii) the top litter layer of a wetland (Bourgoyen, Gent, Belgium), and (iv) the biofilter of an anaerobic digester (DRANCO‐process, Brecht, Belgium).
On the day of sampling, 3 g of each sample was homogenized in 27 ml of a 5 times dNMS medium (Dunfield et al., 2003), with a modified copper concentration (0.8 µM Cu2+) and a 2 mM Na2HPO4/KH2PO4 buffer. The pH of the medium was adjusted to the pH of each sample (biofilter material: pH 7.8; other samples: pH 6.8). Dilution series of samples (10−2 up to 10−11) were prepared in dNMS medium in triplicate. The resulting 120 vials were sealed and 20% (v/v) CH4 was added to the headspace. The cultures were incubated for 5 weeks at 20°C while shaken (90 r.p.m.). Weekly, concentrations of CH4, O2, CO2 and N2O in the headspace were analysed with a Compact GC (Global Analyser Solutions, Belgium). Dilutions were considered positive for methanotrophic growth when CH4 and O2 were consumed (drop below initial average subtracted by five times the standard deviation), with subsequent rise in CO2 levels, and observation of turbidity. Available in triplicate for each dilution, this information was used to estimate the abundance of cultivable MOB using Most Probable Number (MPN) tables (Anonymous, 2008).
To retrieve MOB from the enrichment cultures showing methanotrophic activity, dilution plating and extinction culturing were carried out in parallel. A schematic overview of the followed methodology is shown in Fig. 3.
Figure 3.
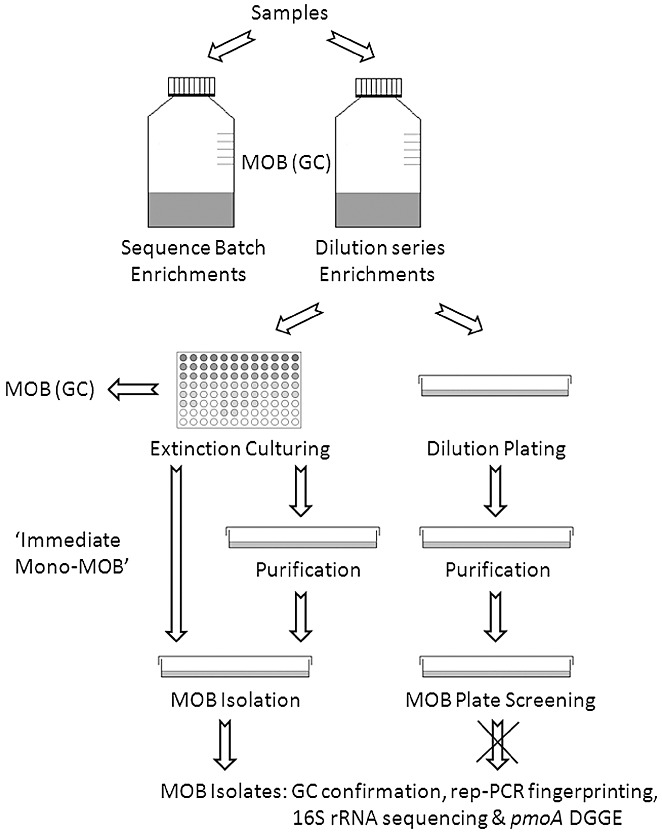
Flow chart of the followed methodology.
Dilution plating was performed by inoculation of the enrichment cultures (10−2, 10−4 and 10−6; 100 µl per plate) on dNMS medium with 0.9% gellan gum and 1% MgSO4·7H2O. After incubation for 2 weeks at 20°C in gas‐tight jars under a CH4 : air (1:1) atmosphere, 200 colonies were randomly selected and subcultured to purity. The isolates were checked for methanotrophy by (i) growth on solid dNMS with methane in the headspace, (ii) absence of growth on solid dNMS under air, and (iii) absence of growth on 1/10 Trypticase Soy Agar (TSA) under air.
Extinction culturing was performed in duplicate by serially diluting the enrichment cultures (10−2 to 10−9) with liquid dNMS medium (dilution‐to‐extinction) in sterile 96‐well microtitre plates. After incubation for 2 weeks at 20°C under a CH4 : air (1:1) atmosphere, turbidity indicative of growth was checked visually and by measuring the optical density at 600 nm. For each dilution series, the highest dilution showing growth was (i) confirmed for methane oxidation by GC analysis (see above), and (ii) plated on solid dNMS medium (with gellan gum) and subcultured to purity if necessary. MOB purity was evaluated by (i) colony morphology, (ii) phase‐contrast microscopy, and (iii) absence of growth on 1/10 TSA and dNMS plates supplemented with 0.1% glucose, 0.1% fructose and 0.1% yeast extract under air. The isolates were confirmed for methane oxidation by GC analysis.
Methane oxidation rate of sequence batch enrichments
In parallel with the dilution series enrichments used for MOB isolation, sequence batch enrichments using the same cultivation conditions were set up in triplicate on the day of sampling for each of the four original samples, allowing the estimation of MORs of the methanotrophic communities cultivable under set conditions. Gastight Schott bottles with a total volume of 1150 ml were filled with 200 ml dNMS medium. Inoculation was performed with 2 ml of the original sample. For the sample of biofilter material, 0.5 g of inoculum was added. After sealing the reactors, 20% (v/v) CH4 was added to the headspace (950 ml). The cultures were placed on a shaker (100 r.p.m.) at 20°C and GC analysis of the headspace was performed daily. When the activity dropped to almost zero, reactors were opened under non‐sterile conditions after which 160 ml of liquid phase was removed for physicochemical analysis or stored at −20°C for pmoA gene DGGE analysis (see further). The remainder of the liquid phase (40 ml) of the triplicate sequence batch enrichments was merged together and subsequently distributed equally over the three reactors. Freshly made dNMS medium was added to a total volume of 200 ml after which 20% (v/v) CH4 was again added to the headspace. In total, this cycle was repeated two times. The hydraulic retention time and sludge retention time for each cycle were 90 h.
Identification and characterization of MOB isolates
Dereplication of all isolates was performed to assess genetic heterogeneity and group isolates with identical genomic fingerprints, further referred to as strains. Rep‐PCR was performed as described by Ghyselinck and colleagues (2011) with a (GTG)5‐primer (Versalovic et al., 1994). The clustering method was supported by visual inspection: isolates were considered as genomically identical when they demonstrated identical fingerprints, which led to a cut‐off value of 93%. For each group, a representative strain was selected randomly and deposited in the BCCM/LMG Culture Collection (LMG 26258–26263 & LMG 26612–26616).
Each strain was identified to the genus level through 16S rRNA gene sequence analysis. PCR amplification and sequencing of the 16S rRNA gene was performed as described by Heyrman and Swings (2001). The sequences were analysed using a 3130 XL Genetic Analyzer (Applied Biosystems, USA) and assembled with BioNumerics 5.1 software (Applied Maths, Belgium). A reliable genus identification was obtained in two steps: (i) query in the ‘Classifier’ program of the Ribosomal Database Project II (Cole et al., 2005) of the 16S rRNA gene sequence of each new strain; and (ii) all type strains of all species of all genera mentioned in the Classifier report were compared in an exhaustive pairwise manner with the query sequence of each new strain in BioNumerics 5.1. Strains were provisionally assigned to the genus of their closest type strain based on the obtained 16S rRNA gene sequence.
A slightly modified version of the naphthalene oxidation assay of Brusseau and colleagues (1990) was used to measure sMMO activity of the MOB strains. A crystal of naphthalene was added to 5 ml freshly grown culture, in dNMS without copper addition, and incubated at 28°C on a shaker (150 r.p.m.) for 2 h. After incubation, 20 µl of freshly prepared tetrazotized‐o‐dianisidine solution (2.68 g l−1) was added to 180 µl of each cell suspension in duplicate in microtitre plates, and the formation of a coloured diazo‐dye was immediately monitored by recording the absorbance at a wavelength of 525 nm via spectrophotometry. The assay was validated using four MOB reference type strains (Table S2) that possess sMMO (DSM 17706T, DSM 15673T, DSM 18500T, NCIMB 11131T) and four that only possess pMMO (DSM 13736T, DSM 17261T, NCIMB 11914T, NCIMB 11130T). Primers described in literature for amplification of the mmoX gene were also tested using these type strains (Table S2). Primers described by Hutchens and colleagues (2004) were selected for mmoX gene amplification of the isolates positive for the sMMO activity assay. Amplification was confirmed by subsequent sequencing of the mmoX gene.
pmoA gene DGGE analysis
Primers described in literature were tested for suitability for DGGE analysis targeting either 16S rRNA or pmoA, since most known MOB, except for members of Methylocella and Methyloferula (Dedysh et al., 2005; Vorobev et al., 2010), as well as all novel strains from this study contained pmoA. Thirteen type strains (six type I and seven type II MOB) were used as positive controls for the evaluation. These strains, their main properties, the primers tested and results obtained are listed in Table S2. The PCR mix and temperature‐time profiles from the original description were tested, as well as the PCR mix used for 16S rRNA gene amplification (Heyrman and Swings, 2001). Only one pmoA primer set (A189f/mb661r) could correctly detect the pMMO gene in all tested strains. The 16S rRNA gene could be amplified in all strains with both the Type IF/Type IR and Type IIF/Type IIR sets of Chen and colleagues (2007); however, amplicons were too long for DGGE analysis. The pmoA set A189f/mb661r (Costello and Lidstrom, 1999) with GC clamp 5′‐CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG‐3′ was selected for further DGGE analysis.
The DNA extraction procedure was adapted from Gevers and colleagues (2001) and El Fantroussi and colleagues (1999). DGGE analysis of the PCR amplicons was performed with an INGENY phorU2X2 DGGE‐system (Goes, The Netherlands). A 6.5% (w/v) polyacrylamide gel with a 30–80% denaturing gradient [a 100% denaturant solution contains 7 M urea and 40% (w/v) formamide] was applied. Gels were run in 1× TAE buffer for 16 h at 150 V and stained afterwards with SYBR Green I nucleic acid gel stain. The resulting DGGE patterns were processed using BioNumerics 5.1. Band position analysis was used to track the obtained isolates back to the sequence batch cultures and the dilution series enrichments used for MOB isolation and was performed by visual comparison of band location in the gel.
Nucleotide sequence accession numbers
The 16S rRNA gene sequence data generated in this study has been deposited in GenBank/EMBL/DDBJ with accession numbers FR798952 to FR798973.
Acknowledgments
This work was funded by the Geconcerteerde Onderzoeksactie (GOA) of Ghent University (BOF09/GOA/005) and a PhD grant for David van der Ha from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT‐Vlaanderen, SB‐83259). The authors gratefully thank Willy Verstraete, Suzanne Read and Jonas Ghyselinck for proof reading the manuscript. Tim Lacoere and Joke Buyse are greatly acknowledged for their help with the molecular analyses.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Average methane (A, D, G, J), oxygen (B, E, H, K) and carbon dioxide (C, F, I, L) levels (%) during 5 weeks incubation in dNMS at 20°C (90 r.p.m.) under a CH4 : air (1:4) atmosphere for the WWTP (A, B, C) slurry pit (D, E, F), wetland (G, H, I) and biofilter sample (J, K, L) for the dilution series 10−2 (white square), 10−3 (white triangle), 10−4 (black triangle) 10−5 (black square) and 10−6 (black diamond) in triplicate (error bars not shown for clarity).
The average methane oxidation rate (n = 3, mg CH4 l−1liquidphase day−1) of sequence batch enrichments of the original samples during the first cycle, for the sample of the wastewater treatment plant (black circle), slurry pit (white circle), wetland (black triangle) and biofilter material (white triangle). One‐sided error bars are shown for clarity, with the same line type.
Table S1. Overview of activity parameters of enriched cultures during the second cycle in sequence batch reactors: the methane oxidation rate (mg CH4 l−1 liquid day−1) during the first and second day of the second cycle for the four samples, the ratio of produced CO2 over consumed CH4 (mg CO2‐C mg−1 CH4‐C), the ratio of produced volatile suspended solids over consumed CH4 (mg VSS mg−1 CH4‐C) and the ratio of consumed CH4 over consumed NO3‐ (mg CH4‐C mg−1 NO3‐N).
Table S2. 16S rRNA, pmoA and mmoX gene primer set evaluation using 13 MOB reference type strains.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Anonymous. USDA‐FSIS; 2008. [Google Scholar]
- Auman A.J., Stolyar S., Costello A.M., Lidstrom M.E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol. 2000;66:5259–5266. doi: 10.1128/aem.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begonja A., Hrsak D. Effect of growth conditions on the expression of soluble methane monooxygenase. Food Technol Biotechnol. 2001;39:29–35. [Google Scholar]
- Bodelier P.L.E., Meima‐Franke M., Zwart G., Laanbroek H.J. New DGGE strategies for the analyses of methanotrophic microbial communities using different combinations of existing 16S rRNA‐based primers. FEMS Microbiol Ecol. 2005;52:163–174. doi: 10.1016/j.femsec.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Bodrossy L., Murrell J.C., Dalton H., Kalman M., Puskas L.G., Kovacs K.L. Heat‐tolerant methanotrophic bacteria from the hot‐water effluent of a natural‐gas field. Appl Environ Microbiol. 1995;61:3549–3555. doi: 10.1128/aem.61.10.3549-3555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. The methanotrophs – the families Methylococcaceae and Methylocystaceae. Chapter 3.1.14. Prokaryotes. 2006;5:266–289. [Google Scholar]
- Bowman J.P., Jimenez L., Rosario I., Hazen T.C., Sayler G.S. Characterization of the methanotrophic bacterial community present in a trichloroethylene‐contaminated subsurface groundwater site. Appl Environ Microbiol. 1993;59:2380–2387. doi: 10.1128/aem.59.8.2380-2387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusseau G.A., Tsien H.C., Hanson R.S., Wackett L.P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1:19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- Bussmann I., Pester M., Brune A., Schink B. Preferential cultivation of type II methanotrophic bacteria from littoral sediments (Lake Constance) FEMS Microbiol Ecol. 2004;47:179–189. doi: 10.1016/S0168-6496(03)00260-5. [DOI] [PubMed] [Google Scholar]
- Bussmann I., Rahalkar M., Schink B. Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol Ecol. 2006;56:331–344. doi: 10.1111/j.1574-6941.2006.00076.x. [DOI] [PubMed] [Google Scholar]
- Button D.K., Schut F., Quang P., Martin R., Robertson B.R. Viability and isolation of marine‐bacteria by dilution culture – theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Dumont M.G., Cebron A., Murrell J.C. Identification of active methanotrophs in a landfill cover soil through detection of expression of 16S rRNA and functional genes. Environ Microbiol. 2007;9:2855–2869. doi: 10.1111/j.1462-2920.2007.01401.x. [DOI] [PubMed] [Google Scholar]
- Cole J.R., Chai B., Farris R.J., Wang Q., Kulam S.A., McGarrell D.M. The Ribosomal Database Project (RDP‐II): sequences and tools for high‐throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connon S.A., Giovannoni S.J. High‐throughput methods for culturing microorganisms in very‐low‐nutrient media yield diverse new marine isolates. Appl Environ Microbiol. 2002;68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello A.M., Lidstrom M.E. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol. 1999;65:5066–5074. doi: 10.1128/aem.65.11.5066-5074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal R.C., Allen D.E. Greenhouse gas fluxes from natural ecosystems. Aust J Bot. 2008;56:369–407. [Google Scholar]
- Dedysh S.N. Exploring methanotroph diversity in acidic northern wetlands: Molecular and cultivation‐based studies. Microbiology. 2009;78:655–669. [Google Scholar]
- Dedysh S.N., Panikov N.S., Tiedje J.M. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl Environ Microbiol. 1998a;64:922–929. doi: 10.1128/aem.64.3.922-929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh S.N., Panikov N.S., Liesack W., Grosskopf R., Zhou J.Z., Tiedje J.M. Isolation of acidophilic methane‐oxidizing bacteria from northern peat wetlands. Science. 1998b;282:281–284. doi: 10.1126/science.282.5387.281. [DOI] [PubMed] [Google Scholar]
- Dedysh S.N., Liesack W., Khmelenina V.N., Suzina N.E., Trotsenko Y.A., Semrau J.D. Methylocella palustris gen. nov., sp. nov., a new methane‐oxidizing acidophilic bacterium from peat bags, representing a novel subtype of serine‐pathway methanotrophs. Int J Syst Evol Microbiol. 2000;50:955–969. doi: 10.1099/00207713-50-3-955. et al. [DOI] [PubMed] [Google Scholar]
- Dedysh S.N., Khmelenina V.N., Suzina N.E., Trotsenko Y.A., Semrau J.D., Liesack W., Tiedje J.M. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane‐oxidizing and dinitrogen‐fixing acidophilic bacterium from Sphagnum bog. Int J Syst Evol Microbiol. 2002;52:251–261. doi: 10.1099/00207713-52-1-251. [DOI] [PubMed] [Google Scholar]
- Dedysh S.N., Berestovskaya Y.Y., Vasylieva L.V., Belova S.E., Khmelenina V.N., Suzina N.E. Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int J Syst Evol Microbiol. 2004;54:151–156. doi: 10.1099/ijs.0.02805-0. et al. [DOI] [PubMed] [Google Scholar]
- Dedysh S.N., Knief C., Dunfield P.F. Methylocella species are facultatively methanotrophic. J Bacteriol. 2005;187:4665–4670. doi: 10.1128/JB.187.13.4665-4670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh S.N., Belova S.E., Bodelier P.L.E., Smirnova K.V., Khmelenina V.N., Chidthaisong A. Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing ‘signature’ fatty acids of type I methanotrophs. Int J Syst Evol Microbiol. 2007;57:472–479. doi: 10.1099/ijs.0.64623-0. et al. [DOI] [PubMed] [Google Scholar]
- Dianou D., Adachi K. Characterization of methanotrophic bacteria isolated from a subtropical paddy field. FEMS Microbiol Lett. 1999;173:163–173. [Google Scholar]
- Dunfield P.F., Khmelenina V.N., Suzina N.E., Trotsenko Y.A., Dedysh S.N. Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int J Syst Evol Microbiol. 2003;53:1231–1239. doi: 10.1099/ijs.0.02481-0. [DOI] [PubMed] [Google Scholar]
- El Fantroussi S., Verschuere L., Verstraete W., Top E.M. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community‐level physiological profiles. Appl Environ Microbiol. 1999;65:982–988. doi: 10.1128/aem.65.3.982-988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert J., Grongroft A. Performance of a passively vented field‐scale biofilter for the microbial oxidation of landfill methane. Waste Manag. 2006;26:399–407. doi: 10.1016/j.wasman.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Gevers D., Huys G., Swings J. Applicability of rep‐PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol Lett. 2001;205:31–36. doi: 10.1111/j.1574-6968.2001.tb10921.x. [DOI] [PubMed] [Google Scholar]
- Ghyselinck J., Van Hoorde K., Hoste B., Heylen K., De Vos P. Evaluation of MALDI‐TOF MS as a tool for high‐throughput dereplication. J Microbiol Methods. 2011;86:327–336. doi: 10.1016/j.mimet.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Hanson R.S., Hanson T.E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer J., Berger U., Hardt M., Dunfield P.F. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int J Syst Evol Microbiol. 2005;55:1817–1826. doi: 10.1099/ijs.0.63213-0. [DOI] [PubMed] [Google Scholar]
- Heyrman J., Swings J. 16S rDNA sequence analysis of bacterial isolates from biodeteriorated mural paintings in the Servilia tomb (necropolis of Carmona, Seville, Spain) Syst Appl Microbiol. 2001;24:417–422. doi: 10.1078/0723-2020-00048. [DOI] [PubMed] [Google Scholar]
- Hutchens E., Radajewski S., Dumont M.G., McDonald I.R., Murrell J.C. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol. 2004;6:111–120. doi: 10.1046/j.1462-2920.2003.00543.x. [DOI] [PubMed] [Google Scholar]
- Iguchi H., Yurimoto H., Sakai Y. Methylovulum miyakonense gen. nov., sp. nov., a novel type I methanotroph from a forest soil in Japan. Int J Syst Evol Microbiol. 2010;61:810–815. doi: 10.1099/ijs.0.019604-0. [DOI] [PubMed] [Google Scholar]
- Janssen P.H., Yates P.S., Grinton B.E., Taylor P.M., Sait M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol. 2002;68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Chen Y., Jiang P., Zhang C., Smith T.J., Murrell J.C., Xing X.‐H. Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering. Biochem Eng J. 2010;49:277–288. [Google Scholar]
- Koh S.C., Bowman J.P., Sayler G.S. Soluble methane monooxygenase production and trichloroethylene degradation by a Type‐I methanotroph, Methylomons methanica‐68‐1. Appl Environ Microbiol. 1993;59:960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melse R.W., Van der Werf A.W. Biofiltration for mitigation of methane emission from animal husbandry. Environ Sci Technol. 2005;39:5460–5468. doi: 10.1021/es048048q. [DOI] [PubMed] [Google Scholar]
- Miller D.N., Yavitt J.B., Madsen E.L., Ghiorse W.C. Methanotrophic activity, abundance, and diversity in forested swamp pools: spatiotemporal dynamics and influences on methane fluxes. Geomicrobiol J. 2004;21:257–271. [Google Scholar]
- Muyzer G., Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Anton Leeuw Int J G. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- Nikiema J., Bibeau L., Lavoie J., Brzezinski R., Vigneux J., Heitz M. Biofiltration of methane: an experimental study. Chem Eng J. 2005;113:111–117. [Google Scholar]
- Rappe M.S., Connon S.A., Vergin K.L., Giovannoni S.J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Scheutz C., Kjeldsen P., Bogner J.E., De Visscher A., Gebert J., Hilger H.A. Microbial methane oxidation processes and technologies for mitigation of landfill gas emissions. Waste Manag Res. 2009;27:409–455. doi: 10.1177/0734242X09339325. et al. [DOI] [PubMed] [Google Scholar]
- Schrader J., Schilling M., Holtmann D., Sell D., Villela M., Marx A., Vorholt J.A. Methanol‐based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 2009;27:107–115. doi: 10.1016/j.tibtech.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Schut F., Devries E.J., Gottschal J.C., Robertson B.R., Harder W., Prins R.A., Button D.K. Isolation of typical marine‐bacteria by dilution culture – growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau J.D., DiSpirito A.A., Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- Shen R.N., Yu C.L., Ma Q.Q., Li S.B. Direct evidence for a soluble methane monooxygenase from type I methanotrophic bacteria: Purification and properties of a soluble methane monooxygenase from Methylomonas sp. GYJ3. Arch Biochem Biophys. 1997;345:223–229. doi: 10.1006/abbi.1997.0239. [DOI] [PubMed] [Google Scholar]
- Söhngen N.L. Uber Bakterien, welche methan ab kohlenstoffnahrung und energiequelle gerbrauchen. Z Bakteriol Parasitenkd Infectionsk. 1906;15:513–517. [Google Scholar]
- Song J., Oh H.M., Cho J.C. Improved culturability of SAR11 strains in dilution‐to‐extinction culturing from the East Sea, West Pacific Ocean. FEMS Microbiol Lett. 2009;295:141–147. doi: 10.1111/j.1574-6968.2009.01623.x. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;8:6–9. [Google Scholar]
- Staley J.T., Konopka A. Measurement of in sity activities of non photysynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Stingl U., Tripp H.J., Giovannoni S.J. Improvements of high‐throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J. 2007;1:361–371. doi: 10.1038/ismej.2007.49. [DOI] [PubMed] [Google Scholar]
- Stott M.B., Crowe M.A., Mountain B.W., Smirnova A.V., Hou S.B., Alam M., Dunfield P.F. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ Microbiol. 2008;10:2030–2041. doi: 10.1111/j.1462-2920.2008.01621.x. [DOI] [PubMed] [Google Scholar]
- Svenning M.M., Wartiainen I., Hestnes A.G., Binnerup S.J. Isolation of methane oxidising bacteria from soil by use of a soil substrate membrane system. FEMS Microbiol Ecol. 2003;44:347–354. doi: 10.1016/S0168-6496(03)00073-4. [DOI] [PubMed] [Google Scholar]
- Tamaki H., Hanada S., Sekiguchi Y., Tanaka Y., Kamagata Y. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ Microbiol. 2009;11:1827–1834. doi: 10.1111/j.1462-2920.2009.01907.x. [DOI] [PubMed] [Google Scholar]
- Trotsenko Y.A., Murrell J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol. 2008;63:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- Tsubota J., Eshinimaev B.T., Khmelenina V.N., Trotsenko Y.A. Methylothermus thermalis gen. nov., sp. nov., a novel moderately thermophilic obligate methanotroph from a hot spring in Japan. Int J Syst Evol Microbiol. 2005;55:1877–1884. doi: 10.1099/ijs.0.63691-0. [DOI] [PubMed] [Google Scholar]
- Versalovic J., Schneider M., De Bruijn F.J., Lupski J.R. Genomic fingerprinting of bacteria using repetitive sequence‐based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- Vorobev A.V., Baani M., Doronina N.V., Brady A.L., Liesack W., Dunfield P.F., Dedysh S.N. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium possessing only a soluble methane monooxygenase. Int J Syst Evol Microbiol. 2010;61:2456–2463. doi: 10.1099/ijs.0.028118-0. [DOI] [PubMed] [Google Scholar]
- Wartiainen I., Hestnes A.G., McDonald I.R., Svenning M.M. Methylobacter tundripaludum sp. nov., a methane‐oxidizing bacterium from Arctic wetland soil on the Svalbard islands, Norway (78 degrees N) Int J Syst Evol Microbiol. 2006;56:109–113. doi: 10.1099/ijs.0.63728-0. [DOI] [PubMed] [Google Scholar]
- Wendlandt K.D., Stottmeister U., Helm J., Soltmann B., Jechorek M., Beck M. The potential of methane‐oxidizing bacteria for applications in environmental biotechnology. Eng Life Sci. 2010;10:87–102. [Google Scholar]
- Whittenbury R., Phillips K.C., Wilkinson J.F. Enrichment, isolation and some properties of methane‐utilizing bacteria. J Gen Microbiol. 1970;61:205–217. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- Wise M.G., McArthur J.V., Shimkets L.J. Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture‐independent 16S ribosomal DNA analysis. Appl Environ Microbiol. 1999;65:4887–4897. doi: 10.1128/aem.65.11.4887-4897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.X., Xin J.Y., Chen L.L., Song H., Xia C.U. Biosynthesis of poly‐3‐hydroxybutyrate with a high molecular weight by methanotroph from methane and methanol. J Nat Gas Chem. 2008;17:103–109. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average methane (A, D, G, J), oxygen (B, E, H, K) and carbon dioxide (C, F, I, L) levels (%) during 5 weeks incubation in dNMS at 20°C (90 r.p.m.) under a CH4 : air (1:4) atmosphere for the WWTP (A, B, C) slurry pit (D, E, F), wetland (G, H, I) and biofilter sample (J, K, L) for the dilution series 10−2 (white square), 10−3 (white triangle), 10−4 (black triangle) 10−5 (black square) and 10−6 (black diamond) in triplicate (error bars not shown for clarity).
The average methane oxidation rate (n = 3, mg CH4 l−1liquidphase day−1) of sequence batch enrichments of the original samples during the first cycle, for the sample of the wastewater treatment plant (black circle), slurry pit (white circle), wetland (black triangle) and biofilter material (white triangle). One‐sided error bars are shown for clarity, with the same line type.
Table S1. Overview of activity parameters of enriched cultures during the second cycle in sequence batch reactors: the methane oxidation rate (mg CH4 l−1 liquid day−1) during the first and second day of the second cycle for the four samples, the ratio of produced CO2 over consumed CH4 (mg CO2‐C mg−1 CH4‐C), the ratio of produced volatile suspended solids over consumed CH4 (mg VSS mg−1 CH4‐C) and the ratio of consumed CH4 over consumed NO3‐ (mg CH4‐C mg−1 NO3‐N).
Table S2. 16S rRNA, pmoA and mmoX gene primer set evaluation using 13 MOB reference type strains.


