Summary
In the gut ecosystem, nitric oxide (NO) has been described to have damaging effects on the energy metabolism of colonocytes. Described mechanisms of NO production are microbial reduction of nitrate via nitrite to NO and conversion of l‐arginine by NO synthase. The aim of this study was to investigate whether dietary compounds can stimulate the production of NO by representative cultures of the human intestinal microbiota and whether this correlates to other processes in the intestinal tract. We have found that the addition of a reduced sulfur compound, i.e. cysteine, contributed to NO formation. This increase was ascribed to higher sulfide concentrations generated from cysteine that in turn promoted the chemical conversion of nitrite to NO. The NO release from nitrite was of the order of 4‰ at most. Overall, it was shown that two independent biological processes contribute to the chemical formation of NO in the intestinal tract: (i) the production of sulfide by fermentation of sulfur containing amino acids or reduction of sulfate by sulfate reducing bacteria, and (ii) the reduction of nitrate to nitrite. Our results indicate that dietary thiol compounds in combination with nitrate may contribute to colonocytes damaging processes by promoting NO formation.
Introduction
Nitric oxide (NO) is one of the smallest and simplest biologically active molecules in the human body. As one of the most widespread signalling molecules in mammals, NO is a major player in controlling nearly every cellular and organ function in the body (Ignarro, 2000). Particularly in the colon, NO plays an important role in the first defence of the host by exerting antimicrobial effects on the invading pathogens. NO is produced by the conversion of l‐arginine by an inducible isoform of NO synthase (iNOS) which is expressed by inflammatory cells, such as activated macrophages. iNOS has been described to be upregulated in the colon of patients with inflammatory bowel diseases (IBD) (Boughton‐Smith et al., 1993; Middleton et al., 1993; Rachmilewitz et al., 1995). Moreover, dysbiosis in the NO metabolism has been linked to ulcerative colitis (UC), one of the main forms of IBD (Roediger, 2008). Co‐action of NO and sulfide has even been suggested to be involved in the initiation of UC, although this has not been confirmed (Roediger and Babidge, 2000).
Besides the production of NO by epithelial cells, microbial and chemical processes have been described with NO as intermediate or end‐product. First, some Gram‐positive bacteria, i.e. Staphylococcus aureus, Bacillus anthracis and Bacillus subtilis, producing NO from arginine by a bacterial NOS, have been shown to produce NO as a cytoprotective against oxidative stress and antibiotics (Gusarov and Nudler, 2005; Shatalin et al., 2008; Gusarov et al., 2009). Second, NO is produced as an intermediate in the denitrification, a four‐step reduction process of nitrate over nitrite, NO and N2O to N2 gas. A broad range of denitrifying bacteria, mainly Gram‐negative, have been described and the genes and enzymes involved in the process have been thoroughly investigated (Zumft, 1997). Denitrification has been suggested to have a minor role in the gastrointestinal tract (Allison and Macfarlane, 1988). Thirdly, conditions in the intestinal tract have been described to stimulate dissimilatory nitrate reduction to ammonium (DNRA), the reduction of nitrate/nitrite to ammonium. The functional gene of DNRA, nrfA, has been identified in Bacteroides species (Mohan et al., 2004) and gamma‐, delta‐ and epsilon‐subclasses of the Proteobacteria (Smith et al., 2007) and even sulfate reducing bacteria (SRB) (Mitchell et al., 1986; Seitz and Cypionka, 1986; Dannenberg et al., 1992; Pereira et al., 1996). Escherichia coli, capable of DNRA but not denitrification has been shown to generate substantial amounts of NO (Ji and Hollocher, 1989). Moreover, DNRA has been shown to be the most prevalent nitrate reduction process in the colon and NO was suggested as intermediate or side‐product during this process (Vermeiren et al., 2009). Finally, non‐enzymatic NO production from nitrite at a pH below 5.5 has been described and is considered to occur in the stomach, on the surface of the skin, in the ischaemic heart, and in infected nitrite‐containing urine (Weitzberg and Lundberg, 1998).
Nitrate has previously been shown to be positively correlated to concentrations of NO produced by gut microbial species (Sobko et al., 2005; Vermeiren et al., 2009). In this paper, in vitro experiments inoculated with intestinal microbial communities were performed in order to identify the influence of dietary compounds on the formation of NO. Finally, it was investigated which microbial or chemical processes occurring in the gut environment, can be correlated to the formation of NO.
Results and discussion
Nitric oxide production by the human intestinal microbial community
NO concentrations produced by a representative human intestinal microbial community in feed medium were measured after 24 h of batch incubation. The community produced 211.9 ± 5.6 ng NO‐N l−1 when incubated anaerobically in the feed medium (Fig. 1). The initial concentration of nitrate present in the feed medium was 0.12 mg NO3‐N l−1. In healthy persons, it is calculated that levels in the order of a few mg NO3‐N l−1 reach the large intestine (Xu, 2001) and nitrate concentrations in faeces were shown to be around 0.15 mg NO3‐N l−1 (Reinders et al., 2007). Therefore, the nitrate concentrations of the feed medium are representative for the intestinal environment.
Figure 1.
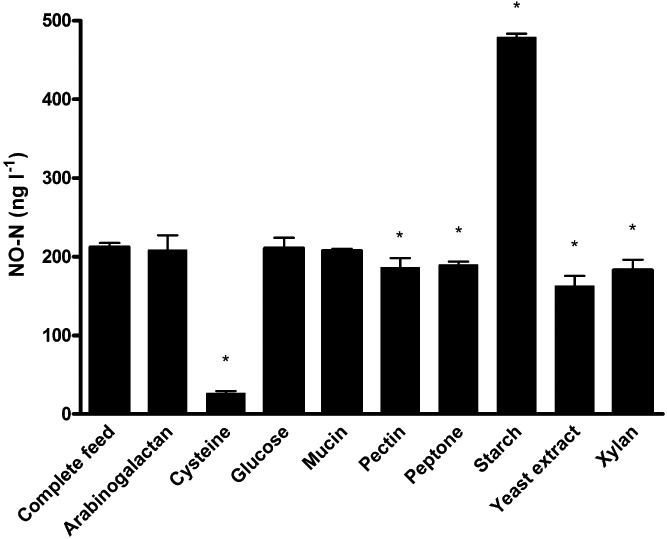
NO‐N (ng l−1) production by the human intestinal microbiota incubated in feed medium for 24 h of which all compounds were excluded separately, and the complete feed medium. The mean ± SD is shown. The asterisk indicates significant differences compared to the control feed medium (P < 0.05).
To further investigate which compounds, besides the trace amounts of nitrate, were necessary to induce NO production, all compounds were excluded one by one from the feed medium and the NO production was measured after 24 h of incubation. Most remarkably, when cysteine or starch were excluded, NO concentrations reached significantly lower levels, i.e. 26.7 ± 2.7 ng NO‐N l−1 (factor 8 decrease) or significantly higher levels, i.e. 479.1 ± 4.4 ng NO‐N l−1 (factor 2 increase) respectively (Fig. 1). Still significant decreases were obtained when pectin (factor 1.1), peptone (factor 1.1), yeast extract (factor 1.3) or xylan (factor 1.2) were excluded from the feed medium (Fig. 1). The concentrations of NO produced by the abiotic controls of the different media were below 0.5 ng NO‐N l−1. To our knowledge, no literature is available on the direct influence of cysteine, starch or any of these compounds on microbial NO production. As the results were most remarkable for cysteine, this compound was chosen for further investigation elucidating the NO formation mechanism.
Positive correlation between cysteine and NO production
Cysteine was added to the feed medium at two different concentrations to study the concentration‐dependent effect of cysteine on the NO production (Fig. 2). Feed medium without cysteine was considered as control and reached a NO concentration of 23.5 ± 1.4 ng NO‐N l−1. Cysteine added at 4 mmol l−1 and 20 mmol l−1 increased the production of NO significantly to concentrations of 411.8 ± 26.5 ng NO‐N l−1 (factor 18 increase) and 1303.8 ± 85.4 ng NO‐N l−1 (factor 56 increase) respectively. In the abiotic controls, NO concentrations of 2.0 and 8.0 ng NO‐N l−1 were measured for 4 mmol l−1 and 20 mmol l−1 cysteine respectively. A positive linear correlation between cysteine and NO was found (r = 0.99). Three possible explanations for this correlation were suggested, i.e. redox potential, sulfide production or DNRA.
Figure 2.
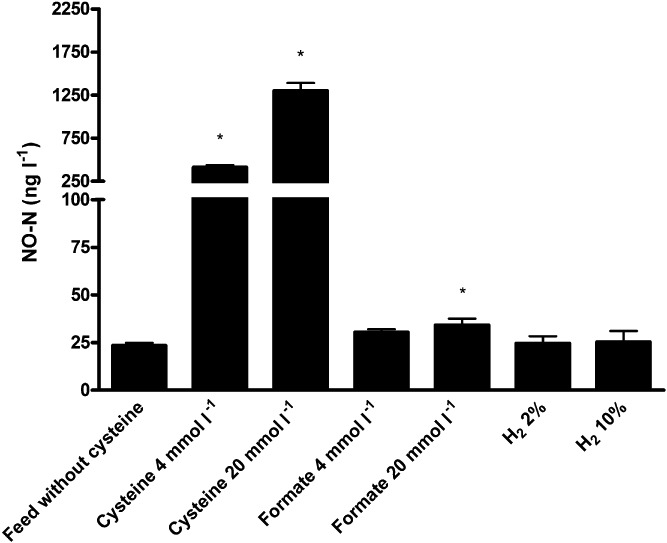
NO‐N (ng l−1) production by the human intestinal microbiota incubated for 24 h in feed medium without cysteine (control), supplemented with cysteine (4 or 20 mmol l−1), formate (4 or 20 mmol l−1) or H2 gas (2 % or 10 % in headspace). The mean ± SD is shown. The asterisk indicates significant differences compared to the feed without cysteine (P < 0.05).
The redox reducing potential of cysteine
As cysteine is frequently used during anaerobic incubations for its redox reducing potential, this characteristic might have changed the growth conditions for the microbiota and concomitant NO production. Cysteine has been shown to influence the growth and hydrogen production of Enterobacterium bacterium. In the latter study, l‐cysteine concentrations above 4 mmol l−1 showed a marked decrease in the redox status, hydrogen production and biomass growth (Chen et al., 2010). To investigate whether the reducing power of cysteine was responsible for the increased NO concentrations in our study, the redox potentials were measured after 24 h of incubation. The redox potential of the inoculated feed medium was around −174 ± 3 mV, while addition of 4 mmol l−1 or 20 mmol l−1 cysteine decreased this value to −205 ± 9 mV (P = 0.17) and −223 ± 4 mV (P < 0.05) respectively. Yet, the redox potential of both cysteine concentrations were not significantly different from each other (P = 0.41). To conclude, a five times increase in the cysteine concentration resulted in a 20 times increase in the NO concentration. Thus, the redox reducing potential of cysteine was not considered to cause the significant increases in NO production.
Increased NO production by reduced sulfur compounds
Second, as bacterial degradation of cysteine to H2S has been described (Forsberg, 1980) and H2S has been shown to be an electrondonor for DNRA (Eisenmann et al., 1995), a comparison was made between the NO and sulfide produced when equimolar concentrations of cysteine or Na2S were added to the feed medium. The feed medium without cysteine produced 72.7 ± 6.4 ng NO‐N l−1 and these concentrations increased significantly to 730.2 ± 30.3 ng NO‐N l−1 (factor 10 increase) and 2832.5 ± 181.7 ng NO‐N l−1 (factor 39 increase) for 4 and 20 mmol l−1 cysteine respectively. It was shown that Na2S stimulated the NO production in a similar way as cysteine. Na2S addition resulted in significantly higher NO concentrations of 684.0 ± 52.8 ng NO‐N l−1 (factor 10 increase) and 2881.4 ± 99.2 ng NO‐N l−1 (factor 40 increase) for 4 and 20 mmol l−1 respectively (Fig. 3A). The NO concentrations measured in feed with cysteine or Na2S added at 4 mmol l−1 or 20 mmol l−1 respectively, were not significantly different (P = 1.00 for both concentrations). Sulfide concentrations after 24 h in feed without cysteine reached concentrations around 9.3 ± 0.5 mg l−1. Addition of 4 mmol l−1 cysteine or Na2S increased this sulfide concentration significantly to 72.8 ± 6.2 mg l−1 and 55.9 ± 1.9 mg l−1 respectively. Higher concentrations resulted in sulfide concentrations above the upper detection limit (Fig. 3B). In the abiotic controls, NO concentrations of 0.08 ± 0.04 and 7.4 ± 1.5 ng NO‐N l−1 were measured for 4 mmol l−1 and 20 mmol l−1 cysteine respectively. Remarkably, in the abiotic controls of Na2S, NO concentrations of 288.8 ± 13.5 ng NO‐N l−1 (factor 4 increase, P < 0.05) and 2070.2 ± 48.5 ng NO‐N l−1 (factor 29 increase, P < 0.05) were measured for 4 mmol l−1 and 20 mmol l−1 Na2S (Fig. 3A). These abiotic controls were however still significantly lower than their biotic counterpart (P < 0.05) indicating that NO formation resulted from a combination of chemical and biological processes.
Figure 3.
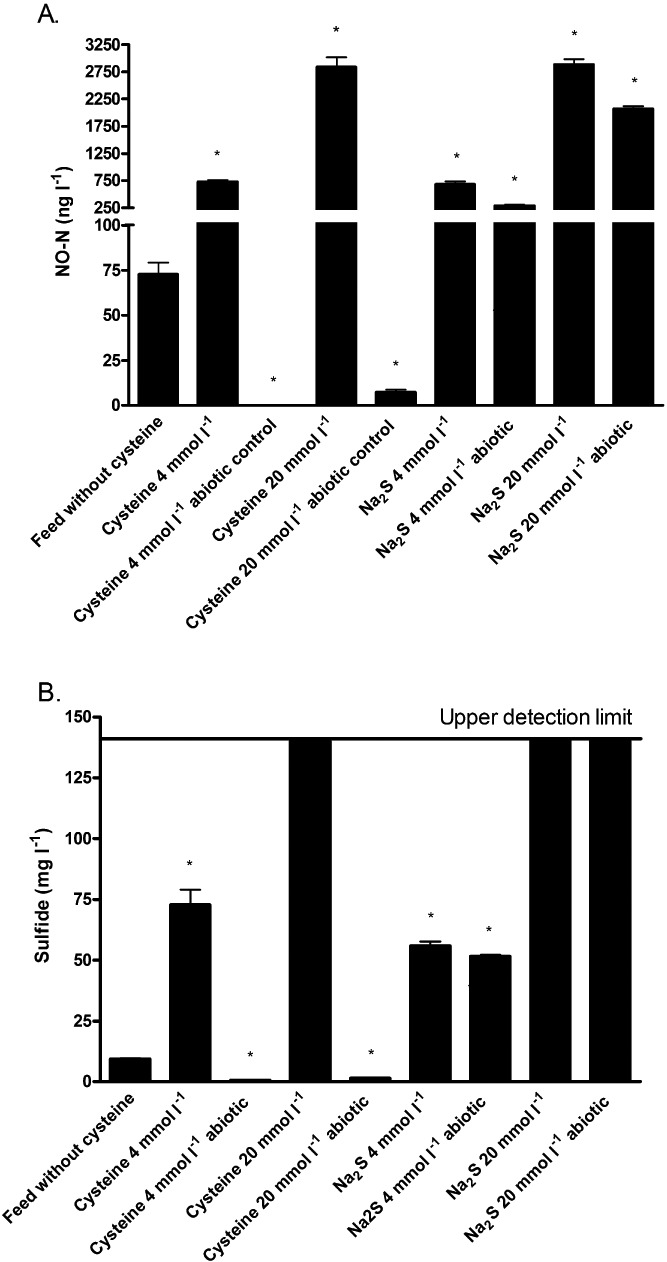
(A) NO‐N (ng l−1) and (B) sulfide (mg l−1) production in feed medium with or without cysteine or Na2S supplemented (4 or 20 mmol l−1) inoculated or abiotic after 24 h of incubation. The mean ± SD is shown. The asterisk indicates significant differences compared to the feed without cysteine (P < 0.05).
Small increase in NO production by stimulation of DNRA
Thirdly, DNRA is believed to be the most important microbial process for the removal of nitrate in the intestinal tract (Allison and Macfarlane, 1988; Vermeiren et al., 2009). H2S has been shown to be an electrondonor for DNRA (Eisenmann et al., 1995), and we have found that Na2S stimulated the formation of NO. Therefore, the influence of two alternative DNRA stimulating electron donors, i.e. formate and H2 (Simon, 2002) on NO formation was further investigated. The addition of 4 and 20 mmol l−1 formate slightly increased the NO concentrations to 30.4 ± 1.5 ng NO‐N l−1 (factor 1.3 increase) and 34.2 ± 3.4 ng NO‐N l−1 (factor 1.5 increase, P < 0.05) respectively (Fig. 2). H2 gas added to the headspace to final concentrations of 2% and 10% did not show an increase in the production of NO (Fig. 2). The NO concentrations of all abiotic controls were below 0.6 ng NO‐N l−1. Stimulation of DNRA by formate increased the NO concentrations significantly; however, the impact of formate was very small compared to the impact cysteine had on the NO production. Thus, stimulation of DNRA was not considered as one of the main driving forces for NO formation under intestinal conditions.
Abiotic NO production from nitrite, but not nitrate or hydroxylamine
Considering our previous results, the abiotic controls of Na2S showed remarkably high NO concentrations. To identify the specific N‐compound from which NO is formed abiotically, solutions of nitrate, nitrite and hydroxylamine (5 mg N l−1) were incubated abiotically for 24 h at 37°C with or without 20 mmol Na2S l−1. Nitrate and hydroxylamine did not produce NO at either condition, while concentrations of 13 ng NO‐N l−1 were measured for nitrite after 24 h. Remarkably, these concentrations increased significantly with a factor 1000 when Na2S was added (data not shown).
Release of NO from nitrite is pH‐dependent
As non‐enzymatic NO release from nitrite at a pH below 5.5 has been described (Weitzberg and Lundberg, 1998), the influence of pH on the chemical production of NO from nitrite with or without Na2S was studied. As expected, NO concentrations around 5100 ng NO‐N l−1, 15 ng NO‐N l−1 and 8 ng NO‐N l−1 were measured for pH 5, 7 and 9 respectively, confirming previous reports on chemical decomposition of nitrite at a pH < 5.5. The NO released was only a minor fraction of the nitrite added, i.e. 1‰ at pH 5. When Na2S was added, concentrations around 19000 ng NO‐N l−1, 12700 ng NO‐N l−1 and 50 ng NO‐N l−1 were measured for pH 5, 7 and 9 respectively (Fig. 4A). Under these conditions, 3.8‰, 2.5‰ and 0.01‰ of nitrite was converted to NO at pH 5, 7 and 9 respectively. Although literature is limited concerning this process, Roediger and Babidge (2000) have suggested a similar process. They described an increased release of NO, with a factor 3 to 4, from the NO donor nitrosoglutathione (GSNO) when adding Na2S. Moreover, our results corroborate a recent publication describing the production of NO from nitrite induced by H2S (Grossi, 2009). The latter author describes a NO release from the NO donor sodium nitroprusside (SNP) or nitrite mediated by HS‐/H2S, particularly at a pH lower than 7. An increased NO formation with a decreasing pH can be explained by the fact that only the HNO2 form of nitrite can be reduced to NO.
Figure 4.
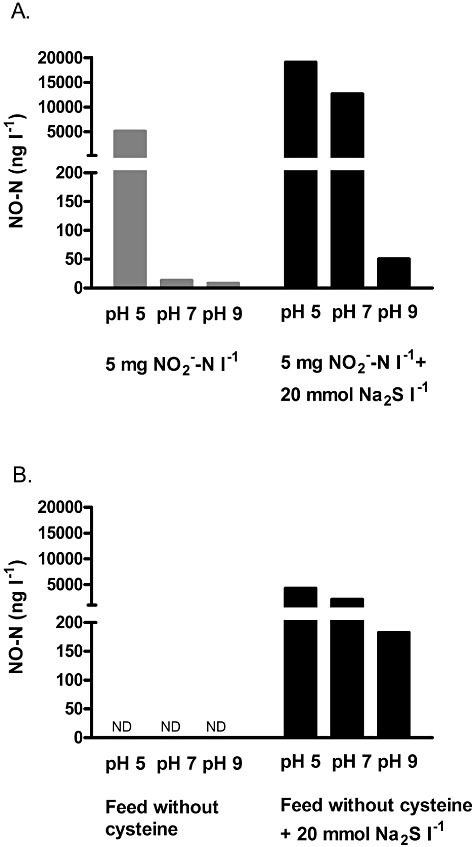
Abiotic NO‐N (ng l−1) production from (A) 5 mg NO2‐N l−1 solution and (B) feed without cysteine incubated with or without 20 mmol Na2S l−1 at pH 5, 7 or 9 for 24 h at 37°C. ND indicates that the values were below the detection limit.
To confirm chemical NO formation in the feed medium, an additional experiment was performed incubating the feed medium abiotically without cysteine at pH 5, 7 and 9 for 24 h with or without 20 mmol l−1 Na2S. Without Na2S added to the feed medium, no NO was formed (Fig. 4B). With Na2S, NO was formed but the concentrations were around five times lower compared to NO produced in the nitrite solution described above. This can be explained by the very low concentrations of nitrite in the feed which are below 0.05 mg l−1. A negative correlation between NO and the pH was seen, similarly as for the nitrite solution (Fig. 4B).
NO production is regulated by dietary compounds and microbial processes
Taking these findings together, a combination of biological and chemical reactions lead to the production of NO by the human intestinal microbiota (Fig. 5). As such, dietary compounds may influence the NO production significantly. Nitrate in the lower parts of the gastrointestinal tract originates from dietary products, like leaf vegetables [accounting for 60 to 80% of the total dietary nitrate intake (Ysart et al., 1999)] and from endogenously synthesized nitrate, which mainly comes from the l‐arginine–NO pathway (Leaf et al., 1989). Nitrate is reduced to nitrite by the bacteria in the oral cavity or is mainly absorbed in the small intestine but levels of a few mg can reach the large intestine (Xu, 2001). Nitrite is used as a food additive for example in meat to prevent botulism and to stabilize the colour (Weiss et al., 2010), but concentrations in faecal samples are very low, i.e. around 70 µg N l−1. Considering the low levels of NO (in the range of ng l−1 to µg l−1), direct chemical reduction from nitrite cannot be excluded. Yet, many species residing in the intestinal tract have the ability to reduce nitrate to nitrite, thereby creating a major source of nitrite (Zumft, 1997).
Figure 5.
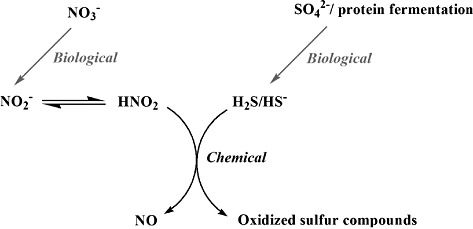
Hypothesis on the production of NO in the intestinal tract. The end‐products of two biological processes, i.e. the reduction of nitrate to nitrite and the reduction of sulfur compounds to H2S, are necessary for the chemical production of NO.
The second microbial process necessary to reduce nitrite to NO is the production of H2S which can be partly related to the reduction of sulfate by SRB. Sources of inorganic sulfate include bread, dried fruits, nuts, fermented beverages, garlic and brassica vegetable (Florin et al., 1991). Second, dietary protein, especially meat, has been shown to be an important substrate of H2S. Sulfur amino acids, such as cysteine and methionine, rather than sulfate have been suggested to be a more readily utilizable sulfur source (Florin et al., 1991; Levine et al., 1998; Magee et al., 2000). H2S has been shown to reach concentrations of up to 3 to 4 mmol l−1 in healthy individuals (Florin et al., 1991; Magee et al., 2000).
To come back to the results of the first experiment, exclusion of starch from the feed medium increased the concentrations of NO significantly. We suggest that due to a lack of starch, the fermentation of sulfur‐containing amino acids is increased leading the higher H2S and concomitant NO concentrations. Indeed, when measuring the sulfide concentrations after 24 h, feed medium without starch showed significantly higher sulfide concentrations (factor 1.2) than the feed with cysteine (data not shown).
Due to the fact that inocula were used from different SHIME runs, we found variability in the concentrations of NO produced during the batch experiments. This interindividual variation can be ascribed to variability in the microbial production of nitrite and sulfide. Also Sobko and colleagues (2005) have described large individual variation in the NO production between the faecal samples.
Physiological consequences for colonocytes
Co‐action of NO and sulfide has been described to be more damaging for colonocytes than either of the compounds (Roediger and Babidge, 2000). NO impairs the energy metabolism of colonocytes by reducing the acyl‐CoA production, necessary for the generation of energy from butyrate. Co‐incubation with sulfide showed an increased generation of NO from the NO donor GSNO which further reduced CoA‐metabolism in the colonocytes (Roediger and Babidge, 2000). In our study, we have shown that the production of NO can be stimulated directly by H2S and indirectly by cysteine. To what extent H2S or thiol compounds indirectly contribute to the NO induced damage to colonocytes, should be further investigated. This may relate to the genotoxicity of sulfide, which is mediated by free radicals (Attene‐Ramos et al., 2007). In respect to our findings, further studies may investigate whether sulfide genotoxicity may indeed be related to the formation of NO radicals from nitrite in the medium.
Conclusions
To conclude, this study has shown that the combination of two biological and one chemical process contributes to the reduction of nitrate to NO in the intestinal tract. Dietary compounds and the microbial community composition determine the conditions in the colon and hence the chemical production of NO. NO is formed by the reduction of nitrite with H2S, both products of the microbial metabolism in the intestinal tract. Only small fractions of NO are released from nitrite, yet these amounts may impact the metabolism of colonocytes (Roediger and Babidge, 2000). The significance of the chemical release of NO from nitrite, mediated by H2S and negatively correlated to the pH, might be underestimated in several biological processes.
Experimental procedures
Growth media
Unless stated otherwise, all products were ordered from Sigma Aldrich (Bornem, Belgium). Feed medium was composed of arabinogalactan (1 g l−1), pectin (2 g l−1), xylan (1 g l−1), potato starch (3 g l−1; Anco, Roeselare, Belgium), glucose (0.4 g l−1), yeast extract (3 g l−1; Oxoid, Aalst, Belgium), special peptone (1 g l−1; Oxoid, Aalst, Belgium) and mucin from porcine stomach type II (4 g l−1). l‐cysteine (0.5 g l−1) was added to scavenge‐dissolved oxygen and to lower the initial redox potential. Feed medium is based on a diet frequently used during in vitro incubation experiments of the gastrointestinal microbiota (Molly et al., 1993; Possemiers et al., 2004; Van de Wiele et al., 2004; Grootaert et al., 2009). Analyses of the feed medium indicated that very low concentrations of nitrate (0.115 ± 0.009 mg NO3‐N l−1) and nitrite (0.053 ± 0.004 mg NO2‐N l−1) were present.
Inocula
The Simulator of the Human Intestinal Microbial Ecosystem (SHIME) is a dynamic in vitro model of the human gastrointestinal tract. It is composed of five double‐jacketed vessels, simulating the stomach, small intestine and the three colon regions (ascending, transverse and descending colon). The first two compartments are a fill‐and‐draw setup and represent the stomach and small intestine. Peristaltic pumps add a defined amount of feed medium (140 ml, three times per day) and pancreatic and bile liquid (60 ml, three times per day), respectively, to the stomach and duodenum compartments and empty the respective compartments after specified intervals. The last three compartments are continuously stirred reactors with constant volume and pH control, specific for each compartment. Isolation of the bacterial inoculum, retention time, pH, temperature settings and reactor feed composition were described previously (Molly et al., 1993; Possemiers et al., 2004). For the batch experiments, samples from the colon ascendens were chosen as inocula as in this vessel the highest levels of NO were measured previously (Vermeiren et al., 2009). Three different SHIME runs, inoculated with faecal samples from three healthy individuals (male; 24, 25 and 27 years) who had no history of antibiotic intake 6 months prior to the study, were used as inocula for the batch experiments. Samples were taken at least 2 weeks after the stabilization period. SHIME samples were preferred over faecal samples because of the stability of the microbial community composition in this continuous system.
Batch experiments
Anaerobic incubations were performed in penicillin flasks with a total volume of 120 ml. The bottles were filled with 42.5 ml feed medium and 7.5 ml of a 1 M phosphate buffer (88 g K2HPO4 l−1 and 68 g KH2PO4 l−1). The pH was set between 6.8 and 7.0 using a 1 mol l−1 NaOH solution at the beginning of the incubation and decreased maximum one unit during the 24 h of incubation (data not shown). To obtain anaerobic conditions the bottles were closed with butyl rubber stops and flushed with Argon during 15 cycles of 2 min each at 800 mbar overpressure and 900 mbar underpressure (De Weirdt et al., 2010). Before starting the incubations, bottles were put on atmospheric pressure. To perform the experiments with a representative microbial community of the intestinal tract, 1 ml of SHIME suspension from the colon ascendens was used as inoculum. The penicillin flasks were incubated at 37°C for 24 h while shaking at 100 r.p.m.
To investigate chemical NO production, a nitrite solution of 0.3 mM NO2‐N l−1 or feed medium without buffer was incubated sterile at pH 5, 7 or 9 with or without 20 mM Na2S for 24 h at 37°C while shaking at 100 r.p.m.
Electron donors
Formate, Na2S, cysteine and H2 were separately added to the feed medium from which cysteine was excluded in two different concentrations. Cysteine, Na2S and formate were added to final equimolar concentrations of 4 and 20 mmol l−1. This corresponds to final concentrations of 0.5 and 2.5 g l−1 for cysteine, 0.31 and 1.56 g l−1 for Na2S, and 0.27 and 1.36 g l−1 for sodium formate respectively. A defined volume of the headspace of the penicillin flasks was replaced with 100% (v/v) H2 gas to final concentrations of 2% and 10% H2.
Nitrogen compound analysis
NO3‐, NO2‐ and NO concentrations were measured as described previously (Vermeiren et al., 2009). In short, NO3‐ and NO2‐ concentrations were measured by first reducing NO3‐ to NO2‐ by a copper‐cadmium reductor coil at pH of 8.0. NO2‐ concentrations were determined colorimetrically by an imidazole buffered reaction with N‐1‐naphtylethylenediamine. The NO3‐ concentration was quantified by subtraction of the concentrations of NO2‐ from the concentration of (NO3‐ + NO2‐) (Hauck, 1982). The samples were centrifuged at 4000 g for 3 min, filtered using 0.45 µM membrane filters (Millipore, NYSE:MIL, USA) and diluted in milli‐Q water before analysis. In short, NO measurements were based on the principle of chemiluminescence using Eco Physics CLD 77 AM (Eco Physics AG, Duernten, Switzerland) with a detection limit of 1 ppbv. A standard curve prepared with NO standard gas (9.8 ± 0.05 ppmv) diluted in air.
Sulfide analysis
Sulfide concentrations of the samples were measured using standard sulfide cuvette tests (range 0.1–2.0 mg l−1) (Hach Lange, Mechelen, Belgium). This method accounted for all dissolved sulfide species (H2S, HS‐ and S2‐).
Calculations
All headspace NO measurements were subsequently converted to concentrations present in the feed medium. The NO concentrations measured in the gas phase (ppbv) were converted to molar concentrations using the ideal gas law pV = nRT with T = 294.65 K. Taking into account [NO]g/[NO]ag ≅ 20 at equilibrium, the NO concentrations in the gas phase were converted to the concentrations in the liquid phase and expressed as ng NO‐N l−1.
Statistical analysis
All experiments were performed in triplicate. spss version 16.0 (SPSS, Chicago, USA) was used to carry out the statistical analyses. Normality of the data and homogeneity of variances was assessed using the Kolmogorov–Smirnov and the Levene test respectively. Comparison of normally distributed data was performed with anova or Dunnett T3; when anova indicated significant differences, means were compared using the Bonferroni comparison test. Comparison of means of not normally distributed data was evaluated with the non‐parametric Kruskal–Wallis test; when Kruskal–Wallis indicated significant differences, means were compared with the Behrens–Fisher comparison test using the npmc package of R software (http://www.r‐project.org/) (Munzel and Hothorn, 2001). A P‐value of less than 0.05 was considered significant.
Acknowledgments
The authors would like to thank Katja Van Nieuland and Samuel Bodé for their assistance with the nitrate, nitrite and NO analyses; Frederiek‐Maarten Kerckhof for his help with the statistical analyses. This work was financially supported by a Concerted Research Action of the Flemish Community (GOA) (BOF07/GOA/002) and by a grant of the ‘Strategisch Basisonderzoek – SBO’ of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT‐Vlaanderen, project nr. 100016).
References
- Allison C., Macfarlane G.T. Effect of nitrate on methane production and fermentation by slurries of human faecal bacteria. J Gen Microbiol. 1988;134:1397–1405. doi: 10.1099/00221287-134-6-1397. [DOI] [PubMed] [Google Scholar]
- Attene‐Ramos M.S., Wagner E.D., Gaskins H.R., Plewa M.J. Hydrogen sulfide induces direct radical‐associated DNA damage. Mol Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- Boughton‐Smith N.K., Evans S.M., Hawkey C.J., Cole A.T., Balsitis M., Whittle B.J.R., Moncada S. Nitric‐oxide synthase activity in ulcerative colitis and Crohn's disease. Lancet. 1993;342:338–340. doi: 10.1016/0140-6736(93)91476-3. [DOI] [PubMed] [Google Scholar]
- Chen H.Q., Ma X.X., Fan D.D., Luo Y.E., Gao P.F., Yang C.Y. Influence of l‐cysteine concentration on oxidation‐reduction potential and biohydrogen production. Chin J Chem Eng. 2010;18:681–686. [Google Scholar]
- Dannenberg S., Kroder M., Dilling W., Cypionka H. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction to O2 or nitrate by sulfate reducing bacteria. Arch Microbiol. 1992;158:93–99. [Google Scholar]
- De Weirdt R., Possemiers S., Vermeulen G., Moerdijk‐Poortvliet T.C.W., Boschker H.T.S., Verstraete W., Van de Wiele T. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol Ecol. 2010;74:601–611. doi: 10.1111/j.1574-6941.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- Eisenmann E., Beuerle J., Sulger K., Kroneck P.M.H., Schumacher W. Lithotrophic growth of Sulfurospirillum deleyianum with sulfide as electron donor coupled to respiratory reduction of nitrate to ammonium. Arch Microbiol. 1995;164:180–185. [Google Scholar]
- Florin T., Neale G., Gibson G.R., Christl S.U., Cummings J.H. Metabolism of dietary sulfate – absorption and excretion in humans. Gut. 1991;32:766–773. doi: 10.1136/gut.32.7.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg C.W. Sulfide production from cysteine by Desulfovibrio desulfuricans. Appl Environ Microbiol. 1980;39:453–455. doi: 10.1128/aem.39.2.453-455.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootaert C., Van den Abbeele P., Marzorati M., Broekaert W.F., Courtin C.M., Delcour J.A. Comparison of prebiotic effects of arabinoxylan oligosaccharides and inulin in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 2009;69:231–242. doi: 10.1111/j.1574-6941.2009.00712.x. et al. [DOI] [PubMed] [Google Scholar]
- Grossi L. Hydrogen sulfide induces nitric oxide release from nitrite. Bioorg Med Chem Lett. 2009;19:6092–6094. doi: 10.1016/j.bmcl.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Gusarov I., Nudler E. NO‐mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci USA. 2005;102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I., Shatalin K., Starodubtseva M., Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck R.D. Nitrogen isotope ratio analysis. In: Page A.L., Miller R.A., Keeney D.R., editors. ASA and SSSA; 1982. pp. 735–779. [Google Scholar]
- Ignarro L.J., editor. Academic Press; 2000. [Google Scholar]
- Ji X.B., Hollocher T.C. Nitrate reductase of Escherichia coli as a NO producing nitrite reductase. Biochem Arch. 1989;5:61–66. [Google Scholar]
- Leaf C.D., Wishnok J.S., Tannenbaum S.R. l‐arginine is a precursor for nitrate biosynthesis in humans. Biochem Biophys Res Commun. 1989;163:1032–1037. doi: 10.1016/0006-291x(89)92325-5. [DOI] [PubMed] [Google Scholar]
- Levine J., Ellis C.J., Furne J.K., Springfield J., Levitt M.D. Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol. 1998;93:83–87. doi: 10.1111/j.1572-0241.1998.083_c.x. [DOI] [PubMed] [Google Scholar]
- Magee E.A., Richardson C.J., Hughes R., Cummings J.H. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr. 2000;72:1488–1494. doi: 10.1093/ajcn/72.6.1488. [DOI] [PubMed] [Google Scholar]
- Middleton S.J., Shorthouse M., Hunter J.O. Increased nitric‐oxide synthesis in ulcerative colitis. Lancet. 1993;341:465–466. doi: 10.1016/0140-6736(93)90211-x. [DOI] [PubMed] [Google Scholar]
- Mitchell G.J., Jones J.G., Cole J.A. Distribution and regulation of nitrate and nitrite reduction by Desulfovibrio and Desulfotomaculum species. Arch Microbiol. 1986;144:35–40. [Google Scholar]
- Mohan S.B., Schmid M., Jetten M., Cole J. Detection and widespread distribution of the nrfA gene encoding nitrite reduction to ammonia, a short circuit in the biological nitrogen cycle that competes with denitrification. FEMS Microbiol Ecol. 2004;49:433–443. doi: 10.1016/j.femsec.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Molly K., Woestyne M.V., Verstraete W. Development of a 5‐step multichamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- Munzel U., Hothorn L.A. A unified approach to simultaneous rank test procedures in the unbalanced one‐way layout. Biom J. 2001;43:553–569. [Google Scholar]
- Pereira I.C., Abreu I.A., Xavier A.V., LeGall J., Teixeira M. Nitrite reductase from Desulfovibrio desulfuricans (ATCC 27774) – a heterooligomer heme protein with sulfite reductase activity. Biochem Biophys Res Commun. 1996;224:611–618. doi: 10.1006/bbrc.1996.1074. [DOI] [PubMed] [Google Scholar]
- Possemiers S., Verthe K., Uyttendaele S., Verstraete W. PCR‐DGGE‐based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 2004;49:495–507. doi: 10.1016/j.femsec.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Rachmilewitz D., Stamler J.S., Bachwich D., Karmeli F., Ackerman Z., Podolsky D.K. Enhanced colonic nitric‐oxide generation and nitric‐oxide synthase activity in ulcerative colitis and Crohn's disease. Gut. 1995;36:718–723. doi: 10.1136/gut.36.5.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders C.I., Jonkers D., Jansson E.A., Stockbrugger R.W., Stobberingh E.E., Hellstrom P.M., Lundberg J.O. Rectal nitric oxide and fecal calprotectin in inflammatory bowel disease. Scand J Gastroenterol. 2007;42:1151–1157. doi: 10.1080/00365520701320505. [DOI] [PubMed] [Google Scholar]
- Roediger W.E.W. Review article: nitric oxide from dysbiotic bacterial respiration of nitrate in the pathogenesis and as a target for therapy of ulcerative colitis. Aliment Pharmacol Ther. 2008;27:531–541. doi: 10.1111/j.1365-2036.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- Roediger W.E., Babidge W.J. Nitric oxide effect on colonocyte metabolism: co‐action of sulfides and peroxide. Mol Cell Biochem. 2000;206:159–167. doi: 10.1023/a:1007034417320. [DOI] [PubMed] [Google Scholar]
- Seitz H.J., Cypionka H. Chemolithtrophic growth of Desulfovibrio desulfuricans with hydrogen coupled to ammonification of nitrate or nitrate. Arch Microbiol. 1986;146:63–67. [Google Scholar]
- Shatalin K., Gusarov I., Avetissova E., Shatalina Y., McQuade L.E., Lippard S.J., Nudler E. Bacillus anthracis‐derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Acad Sci USA. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol Rev. 2002;26:285–309. doi: 10.1111/j.1574-6976.2002.tb00616.x. [DOI] [PubMed] [Google Scholar]
- Smith C.J., Nedwell D.B., Dong L.F., Osborn A.M. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol. 2007;73:3612–3622. doi: 10.1128/AEM.02894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobko T., Reinders C.I., Jansson E.A., Norin E., Midtvedt T., Lundberg J.O. Gastrointestinal bacteria generate nitric oxide from nitrate and nitrite. Nitric Oxide. 2005;13:272–278. doi: 10.1016/j.niox.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Van de Wiele T., Boon N., Possemiers S., Jacobs H., Verstraete W. Prebiotic effects of chicory inulin in the simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 2004;51:143–153. doi: 10.1016/j.femsec.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Vermeiren J., Van de Wiele T., Verstraete W., Boeckx P., Boon N. Nitric oxide production by the human intestinal microbiota by dissimilatory nitrate reduction to ammonium. J Biomed Biotechnol. 2009 doi: 10.1155/2009/284718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J., Gibis M., Schuh V., Salminen H. Advances in ingredient and processing systems for meat and meat products. Meat Sci. 2010;86:196–213. doi: 10.1016/j.meatsci.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Weitzberg E., Lundberg J.O.N. Nonenzymatic nitric oxide production in humans. Nitric Oxide. 1998;2:1–7. doi: 10.1006/niox.1997.0162. [DOI] [PubMed] [Google Scholar]
- Xu J. Faculty of Agricultural and Applied Biological Sciences, Ghent University; 2001. [Google Scholar]
- Ysart G., Miller P., Barrett G., Farrington D., Lawrance P., Harrison N. Dietary exposures to nitrate in the UK. Food Addit Contam. 1999;16:521–532. doi: 10.1080/026520399283669. [DOI] [PubMed] [Google Scholar]
- Zumft W.G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


