Abstract
A cocktail of six lytic bacteriophages, SalmoFresh™, significantly (p < 0.05) reduced the number of surface-applied Salmonella Kentucky and Brandenburg from stainless steel and glass surfaces by > 99% (2.1–4.3 log). Both strains were susceptible to SalmoFresh™ in the spot-test assay. Conversely, SalmoFresh™ was unable to reduce surface contamination with a Salmonella Paratyphi B strain that was not susceptible to the phage cocktail in the spot-test assay. However, by replacing two SalmoFresh™ component phages with two new phages capable of lysing the Paratyphi B strain in the spot-test assay, the target range of the cocktail was shifted to include the Salmonella Paratyphi B strain. The modified cocktail, SalmoLyse™, was able to significantly (p < 0.05) reduce surface contamination of the Paratyphi B strain by > 99% (2.1–4.1 log). The data show that both phage cocktails were effective in significantly reducing the levels of Salmonella on hard surfaces, provided the contaminating strains were susceptible in the spot-test (i.e., spot-test susceptibility was indicative of efficacy in subsequent surface decontamination studies). The data also support the concept that phage preparations can be customized to meet the desired antibacterial application.
Keywords: Salmonella, SalmoFresh™, SalmoLyse™, bacteriophage, food safety, phage, surface decontamination
Introduction
Illnesses caused by Salmonella constitute a substantial health burden in the United States and around the world. The Centers for Disease Control (CDC) estimates that each year, in the US, infection by foodborne Salmonella causes over one million people to become sick, approximately 19,000 of whom are hospitalized and an estimated 400 individuals die.1 In the US alone, these illnesses result in nearly $2.4 billion in medical costs annually, which constitutes a substantial economic hardship on national and local economies.2 Salmonella are also an international health risk, causing an estimated 93.8 million illnesses globally, and approximately 155,000 deaths, each year.3
According to the Food Safety and Inspection Service (FSIS) of the US Department of Agriculture (USDA), about 75% of the annual cases of human salmonellosis are due to the consumption of contaminated poultry, beef and egg products.4 Although this bacterium is inactivated when these foods are properly cooked, Salmonella may survive for several days on the surfaces on which the foods are processed and handled before cooking.5 If other foods, especially ready-to-eat foods (e.g., fruits and vegetables), come into contact with those contaminated surfaces, Salmonella may contaminate them and cause foodborne salmonellosis.5 Strategies currently employed to manage Salmonella and other foodborne pathogenic bacteria focus on using chemical disinfectants as well as physical disruption, including heat, steam and UV-light irradiation to non-selectively reduce the microbial burden on food contact surfaces.6,7 These strategies, while effective, have significant drawbacks, such as corrosion of equipment, toxic chemical residues, and damage to the quality of foods.
A novel approach to reduce the need for harsh chemical sanitizers is the use of lytic bacteriophages (or phages) as biocontrol agents.8-10 This method relies upon the activity of environmentally-isolated lytic bacteriophages that are capable of exclusively infecting and killing specific bacterial strains or a subgroup of strains, usually within the same genus. The concept of using lytic bacteriophage as a pathogen control strategy has been the subject of several recent reviews in the scientific literature.11-13 This communication reports that (1) two lytic bacteriophage cocktails, each containing six Salmonella-specific phages, significantly reduced the levels of Salmonella on selected hard surfaces, (2) the contaminating strain must be susceptible to the lytic bacteriophage cocktails, as determined by the spot-test assay,14 for the cocktail to be able to reduce Salmonella contamination on hard surfaces, and (3) a lytic bacteriophage cocktail specific for Salmonella could be rapidly customized and adapted to achieve lytic activity against Salmonella strains/serotypes not previously lysed. This is an important consideration for enabling continued effectiveness of the approach, as it allows updating phage preparations to keep up with the emergence of bacterial resistance against a given phage cocktail as the result of selective pressure caused by the continued use of such phage preparations and/or natural shifts in bacterial populations.
Results and Discussion
Selection of bacteriophages for biocontrol cocktails
Standard enrichment techniques15 were used to isolate a total of 21 Salmonella-specific bacteriophages from various surface-water sources in Maryland (including Baltimore Inner Harbor and Chesapeake Bay). The ability of each bacteriophage to lyse Salmonella was determined by screening the bacteriophages against a collection of 916 Salmonella strains (representing a total of 43 known serotypes and 275 strains of undetermined serotype), using the classical spot-test assay14 (data not shown). After determining the lysis range for each bacteriophage, SalmoFresh™, a bacteriophage cocktail possessing lytic activity against a broad range of Salmonella strains, was created. The optimal lytic bacteriophages were chosen for inclusion after determining their (1) host ranges (i.e., the phages that killed the greatest percentage of our Salmonella isolates), and (2) abilities to kill Salmonella serotypes of high public health importance, including S. Typhimurium, S. Enteritidis, S. Heidelberg, S. Newport and S. Hadar. The resulting cocktail contained 6 bacteriophages (designated SPT-1, SBA-1781, SSE-121, STML-13-1, STML-198 and SKML-39) and, at a concentration of ca. 2 × 104 PFU/mL, it killed 780 (85%) of the 916 Salmonella isolates examined in this study. A summary of serotypes (those represented by 5 or more strains in our collection) examined, as well as the effectiveness of SalmoFresh™ against them is shown in Table S1. An alternative cocktail (designated SalmoLyse™) was also prepared as explained later in the text. This cocktail contained 4 of the 6 bacteriophages included in SalmoFresh™ (SPT-1, SBA-1781, SSE-121 and STML-198), but substituted two other phages (SEML-239-1 and SNN-387 in place of STML-13-1 and SKML-39). The reformulated cocktail lysed 756 (83%) of the 916 Salmonella test strains (at a concentration of ca. 2 × 104 PFU/mL). The number of serotypes (listed in Table S1) lysed was not affected.
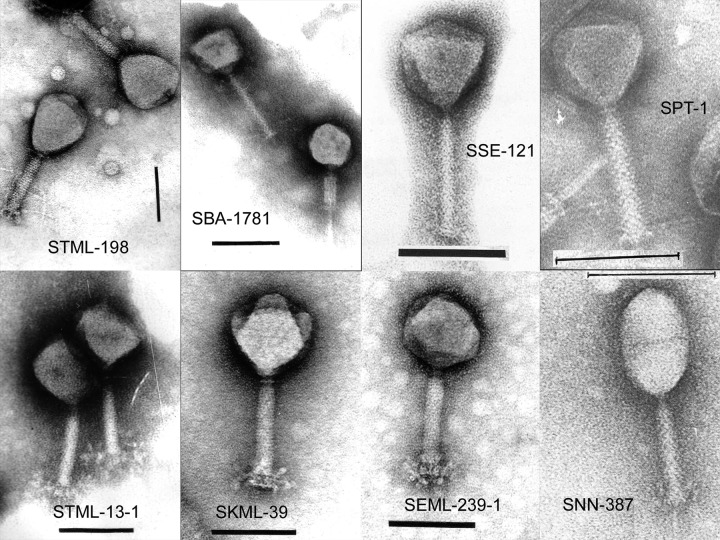
The specificity of both cocktails was examined by determining their lytic activities against 35 strains of seven bacterial species other than Salmonella (see Materials and Methods). Neither cocktail lysed any of the Gram-positive strains (Listeria spp, Staphylococcus aureus, or Enterococcus spp), nor did they lyse the Gram-negative Pseudomonas aeruginosa or Acinetobacter baumannii strains. However, SalmoFresh™ and SalmoLyse™ lysed two E. coli O157:H7 strains and SalmoLyse™ also lysed a single Shigella strain. The results support the idea that both preparations are relatively specific for Salmonella with slight cross-reactivity against a small number of strains of very closely related species. Electron microscopic images of the eight bacteriophages used to prepare SalmoFresh™ and SalmoLyse™ are contained in Figure 1. All of these bacteriophages belong to the Myoviridae family of double-stranded DNA bacteriophages, according to the classification scheme of Ackermann and Berthiaume.16
Figure 1. Electron micrographs of phages present in SalmoFresh and SalmoLyse. Black bars represent 0.1 μM.
Surface decontamination studies
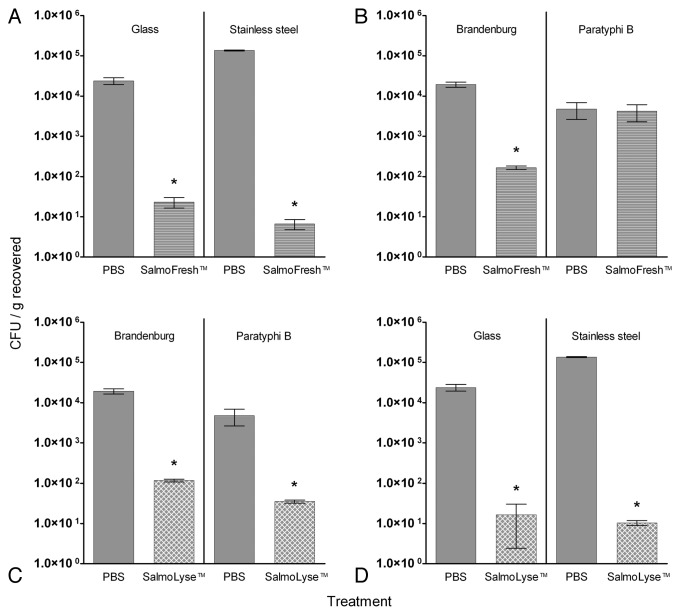
The ability of the bacteriophage cocktails to reduce Salmonella on artificially contaminated hard surfaces was evaluated as described previously.8,9 Briefly, stainless steel coupons or glass coverslips were contaminated with S. enterica serovar Kentucky strain S800 at a concentration of ca. 1 × 106 CFU/surface and then treated for 5 min with either phosphate buffered saline (PBS) or SalmoFresh™ at a concentration of ca. 1 × 107 PFU/surface. Strain S800 was randomly selected from the Salmonella strains that were susceptible to SalmoFresh™ in the spot-test assay. A contact time of 5 min was selected to mimic the contact time of another phage preparation (ListShield™) previously approved by the Environmental Protection Agency (EPA) for similar surface decontamination applications (EPA registration number 74234-1). The “free phages” (i.e., phages unattached to Salmonella adhering to the coverslips) were removed by washing with peptone water, and the concentrations of viable Salmonella remaining on the coverslips were determined by a standard colony counting assay (see Materials and Methods). Recoveries of Salmonella from PBS-treated controls ranged from ca. 5 × 103 to 1 × 105 CFU (Fig. 2). SalmoFresh™ treatment significantly (p < 0.05, unpaired t-test) reduced Salmonella contamination on the stainless steel and glass surfaces by 4.3 log CFU/surface (99.995%) and 3.0 log CFU/surface (99.90%), respectively (Fig. 2A), vs. the PBS-treated controls. This reduction is similar to that recently reported for Yersinia pestis-specific bacteriophages (> 99%) and Escherichia coli O157:H7-specific bacteriophages (94%).8,9
Figure 2. Efficacy of SalmoFresh™ or SalmoLyse™ on Salmonella-contaminated surfaces. Error bars represent standard error of the mean. Asterisks denote values that are significantly different from PBS controls (p < 0.05, t-test). (A) SalmoFresh™ treated stainless steel and glass surfaces contaminated with serotype Kentucky S800; (B) SalmoFresh™ treated glass surfaces contaminated with serotypes Brandenburg S806 or Paratyphi B S661; (C) SalmoLyse™ treated glass surfaces contaminated with serotypes Brandenburg S806 or Paratyphi B S661; (D) SalmoLyse™ treated stainless steel and glass surfaces contaminated with serotype Kentucky S800.
The challenge strain used during these studies, Salmonella Kentucky S800, was selected because it was susceptible to SalmoFresh™, as determined by a spot-test assay.14 The susceptibility of a bacterial strain to a lytic bacteriophage is typically assumed to be required for bacteriophage efficacy as an antibacterial for various practical applications. However, rigorous experimental evidence establishing that idea for food safety or surface decontamination applications have not yet been reported. Therefore, to test this assumption, we repeated the study using a susceptible strain, Salmonella Brandenburg S806, and a non-susceptible strain, Salmonella Paratyphi B S661, as the challenge organisms. S. Brandenburg S806 was used instead of the previously used susceptible S. Kentucky S800 to examine the robustness of the approach and to confirm that the observed efficacy of SalmoFresh™ is not limited to one susceptible test strain. S. Paratyphi B S661 was selected because it is resistant to SalmoFresh™ at both 2 × 104 and 1 × 109 PFU/mL when examined in the spot-test assay. Using the glass surface in the surface decontamination study, SalmoFresh™ treatment significantly reduced (p < 0.05) the Salmonella load on coverslips experimentally contaminated with the susceptible strain, S. Brandenburg S806, by 2.1 log CFU/surface (99.14%), but it did not significantly reduce the levels of the non-susceptible strain, S. Paratyphi B S661 (Fig. 2B). These data support the idea that the susceptibility of the presumptive bacterial pathogen in a spot-test is indicative of bacteriophage efficacy in decontaminating hard surfaces.
To further test the hypothesis that susceptibility in the spot-test assay is a requirement for the ability of phage preparation to reduce Salmonella levels on hard surfaces, we formulated a new six-phage cocktail (designated SalmoLyse™) lytic for the non-susceptible Paratyphi B strain but still possessing broad activity against other Salmonella isolates, including the S. Kentucky S800 and S. Brandenburg S806 strains used during the initial experiments. After formulating SalmoLyse™, the surface decontamination studies were repeated using that cocktail. Glass coverslips were contaminated with S. Paratyphi B S661 and S. Brandenburg S806 strains (both susceptible to SalmoLyse™ in the spot-test assay) and were treated with either PBS or SalmoLyse™ at a concentration of ca. 1x107 PFU/surface. SalmoLyse significantly reduced (p < 0.05, unpaired t-test) contamination of the glass surfaces by both strains by 2.1 logs (99.27%) and 2.2 logs (99.40%), respectively (Fig. 2C). The cocktail also significantly reduced S. Kentucky strain S800 contamination of glass and stainless steel surfaces by 3.2 logs (99.93%) and 4.1 logs (99.99%), respectively (Fig. 2D). These results were similar to the reductions observed when SalmoFresh™ was used against the S. Kentucky strain on glass and stainless steel surfaces (Fig. 2A).
In all our surface decontamination studies, the Salmonella were exposed to the phage preparations on the hard surfaces for 5 min, after which the phages were washed away and the levels of residual Salmonella on those surfaces were enumerated. The normal lytic cycle of a bacteriophage requires about 20–40 min,17 thus, the significant reductions in Salmonella counts we observed during our studies (with the Salmonella strains susceptible to SalmoFresh and SalmoLyse in the spot test assay) were not likely to be the endpoints of the full lytic process on the hard surfaces but were rather the result of initial adsorption of the phage particles to the bacterial membrane and subsequent lysis of the bacteria. In this context, most lytic phage-infected bacteria are destined for lysis by ≤ 1 min post-injection of phage DNA.8,17
Concluding Remarks
The use of lytic bacteriophages to remove specific bacteria from hard surfaces has been gaining increased attention, as indicated by studies focusing on bacteriophages targeting some major foodborne bacterial pathogens, such as L. monocytogenes18,19 and E. coli O157:H7,8,10 as well as some select bacterial agents of bioterrorism concern.9 Additionally, at least one bacteriophage preparation has been registered by the US Environmental Protection Agency (EPA registration number 74234-1) as a “microbial pesticide” suitable for reducing L. monocytogenes contamination of food processing plants and food handling establishments. Our current data support the idea that a bacteriophage cocktail-based approach also has merit for significantly reducing Salmonella contamination of glass and stainless steel surfaces.
Our data also indicate that updating of bacteriophage cocktail formulations to address the issues of bacterial population dynamics and potential emergence of bacteriophage insensitive mutants (BIMs) is technically feasible. In real life settings, this may be achieved by continuous testing of all recovered/surviving Salmonella isolates from a processing plant setting for their sensitivity to the phage preparation used in that facility, and to updating the phage preparation when the predominant Salmonella isolates in that facility are no longer susceptible to the original phage cocktail. This approach of customization of bacteriophage cocktails may provide flexibility for maintaining an effective long-term pathogen control strategy in various settings. However, several important factors should be considered concerning the implementation of an effective phage-based biocontrol approach in real-life situations. For example: (1) Lytic phages, because of their specificity, will only kill their specific bacterial hosts (in the case of this study, Salmonella). Thus, bacteriophage strategies should be considered a supplement, rather than a substitute, for chemical sanitizers and disinfectants that typically have a very broad spectrum of activity. (2) With some rare exceptions,19 the typical concentrations of commonly used chemical sanitizers and disinfectants will rapidly inactivate phages. Therefore, application of bacteriophage should be coordinated with the use of common chemical sanitizers, to ensure optimization of both strategies. For example, after treating surfaces with a phage cocktail, allowing a sufficient time to elapse before applying a chemical sanitizer or disinfectant to the surfaces. (3) To facilitate rapid updating of phage-based biocontrol agents, several regulatory issues need to be addressed before such updates can be implemented in real-life situations. The approach is a clear departure from the traditional approval process for other antimicrobials. However, a positive development in that regard is the FDA’s flexibility regarding its recent approval of a bacteriophage cocktail specific for L. monocytogenes, whereby the agency allowed for future updates to the cocktail with new replacement phages, if and when necessary to maintain efficacy (21 CFR §172.785). All new phages will need to meet the same stringent safety and efficacy criteria as the original phages in the six-phage cocktail, and the manufacturing process and all quality control protocols must be strictly adhered to for all new phages—but these are logical and technically-feasible requirements. It is possible that similar strategy could be used by the EPA when regulating phage preparations for surface applications.
The data obtained during our studies demonstrate that treatment with bacteriophage cocktails lytic for Salmonella significantly reduced the levels of Salmonella contaminating the two hard surfaces examined. If the observed efficacy of SalmoFresh™ and SalmoLyse™ is reproducible in food processing facilities, a bacteriophage cocktail-based approach may help to reduce the immediate Salmonella levels on surfaces, potentially enabling these facilities to reduce the use of chemical sanitizers for that purpose. Additionally, using SalmoFresh™ or a similar bacteriophage-based preparation may also have more long-term implications for reducing Salmonella contamination. In this context, various sanitizing or disinfecting chemicals are routinely used while attempting to produce a bacteria-free environment in food processing facilities. However, such an environment is unsustainable, due to the rapid reintroduction of various microorganisms during food processing procedures (potentially including the pathogenic bacteria that were the targets of the original, chemical-based sanitation protocol). Thus, routine use of SalmoFresh™ or SalmoLyse™ (or any other technically equivalent phage preparation) may provide a subtle selective pressure that may make it increasingly difficult for Salmonella to re-establish themselves in that environment. Additional long-term studies are needed to investigate the validity of this phage-mediated “eco-management” approach.
Materials and Methods
Bacterial strains and growth media
A total of 916 Salmonella isolates from various research and public health laboratories in the United States were used in the studies described here, representing a total of 43 known serotypes and 275 strains of undetermined serotype. Also, a total of 35 strains of seven bacterial species other than Salmonella were used during the studies. They included three strains of L. monocytogenes, two strains of L. innocua, and five strains each of Staphylococcus aureus, Pseudomonas aeruginosa, Acinetobacter baumannii, Enterococcus spp, Shigella spp and E. coli. All strains were stored at −80°C in 70% LB broth-30% glycerol. The three Salmonella enterica serotypes used in our surface decontamination studies were S. Kentucky S800, S. Brandenburg S806 and S. Paratyphi B S661.
Bacteriophage preparations
Both of the bacteriophage cocktails formulated each contained a total of 6 phages. Four of the phages were common to SalmoFresh™ and SalmoLyse™: SPT-1, STML-198, SSE-121 and SBA-1781. The remaining two phages were unique for each cocktail: SalmoFresh™ contained SKML-39 and STML-13-1, while SalmoLyse™ contained SEML-239-1 and SNN-387. Electron micrographs of the eight monophages used to prepare the phage cocktails are shown in Figure 1.
Spot-test assay
The classical spot-test assay14 was used to determine susceptibility of the test strains to the individual phages and the two bacteriophage cocktails. Briefly, the test strain was grown to early log phase, a 0.100 mL aliquot was combined with top agar (LB + 0.7% agar), and plated onto LB agar plates. Once set, 10 μL of the test phage or cocktail was spotted onto the lawn and allowed to dry before incubating at the appropriate temperature for the strain (e.g., 35 ± 2°C for Salmonella) for 24 ± 4 h. A strain was deemed susceptible if zone of lysis developed at the spot where phage was applied.
General design of studies utilizing Salmonella-contaminated hard surfaces
The studies were performed as described previously.8,9 All tests were performed in triplicate. Square glass coverslips and stainless steel coupons (both ca. 25 × 25 mm) were used as examples of the hard surfaces commonly found in food preparation settings. After cleaning with 70% ethanol and rinsing with deionized water, the substrates were sequentially (1) placed in glass petri dishes (one item per dish), (2) sterilized in an autoclave, (3) treated with 10 μL of a dried skim milk solution (5% w/v) to simulate a dirty surface since food processing surfaces are often covered with organic matter, and (4) stored at room temperature, in a laminar flow biosafety hood, until completely dry (typically 20–30 min). After drying, the surfaces of the matrices were contaminated by applying the appropriate Salmonella serotype (10 μL of a suspension of ca. 1 × 108 CFU/mL), and allowed to dry 30 min in a laminar flow biosafety hood to enhance bacterial attachment. After drying and attachment, 0.1 mL of the appropriate phage cocktail (ca. 1 × 108 PFU/mL; MOI = 10) or PBS were applied on top of the Salmonella-contaminated surfaces. The test and control matrices were stored 5 min at room temperature, and the excess bacteriophage preparations and PBS were removed by holding the surfaces vertically and allowing the excess liquid to drain onto paper towels. The test and control matrices were then mixed gently for 30 s in separate conical tubes (50-mL capacity) containing 20 mL peptone water. Serial 10-fold dilutions (10−1, 10−2 and 10−3) of the mixtures were prepared in peptone water, and the undiluted and diluted mixtures were immediately passed through separate membrane filters (0.45-μm pore size; Nalgene). The filters were then washed with 20 mL peptone water to remove unattached phages. The washed filters were placed upside down in separate petri dishes containing Salmonella Shigella Agar (SSA; Becton-Dickinson) and the number of recovered Salmonella was enumerated by counting the Salmonella colonies that grew on the filters during incubation (30°C, 24–48 h). Since the entire 20 mL of peptone water was filtered through the Nalgene membrane filters, the resulting counts represented the total colony-forming units (CFU) recovered from each of the tested surfaces. The efficacy of phage treatment was evaluated by comparing the number of Salmonella recovered from PBS-treated control samples vs. phage-treated samples. Unpaired t-test was used to determine whether the observed differences were statistically significant, with p < 0.05 considered significant.
Supplementary Material
Acknowledgments
We thank Hans W. Ackermann for his electron microscopic examination of the Salmonella bacteriophages described in this report. The studies reported in this communication were supported, in part, by the Procter and Gamble Company and by SBIR award 2012-33610-19947 from the National Institute of Food and Agriculture, United States Department of Agriculture (to AS).
Glossary
Abbreviations:
- ca.
approximately
Disclosure of Potential Conflicts of Interest
CC, TA, JW, BA, ML and AS hold an equity stake in Intralytix, Inc., a Maryland corporation developing bacteriophage preparations (including SalmoFresh™ and SalmoLyse™) for various applications.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/bacteriophage/article/25697
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/25697
References
- 1.CDC. Estimates of foodborne illness in the United States. 2011. Available at: http://www.cdc.gov/foodborneburden/PDFs/FACTSHEET_A_FINDINGS_updated4-13.pdf
- 2.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–9. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 4.CDC. Making food safer to eat. CDC Vital Signs 2011. Available at: http://www.cdc.gov/VitalSigns/FoodSafety/
- 5.Kusumaningrum HD, Riboldi G, Hazeleger WC, Beumer RR. Survival of foodborne pathogens on stainless steel surfaces and cross-contamination to foods. Int J Food Microbiol. 2003;85:227–36. doi: 10.1016/S0168-1605(02)00540-8. [DOI] [PubMed] [Google Scholar]
- 6.Delaquis P, Bach S. Resistance and sublethal damage. In: Gomez-Lopez Vm, ed. Decontamination of fresh and minimally processed produce. Ames, IA:Wiley-Blackwell, 2012:77-86 [Google Scholar]
- 7.Stanfield P. Cleaning and sanitizing a food plant. In: Hui Yh, et al., ed(s). Food plant sanitation. New York:Marcel Dekker, Inc., 2003:110-125 [Google Scholar]
- 8.Abuladze T, Li M, Menetrez MY, Dean T, Senecal A, Sulakvelidze A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by Escherichia coli O157:H7. Appl Environ Microbiol. 2008;74:6230–8. doi: 10.1128/AEM.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rashid MH, Revazishvili T, Dean T, Butani A, Verratti K, Bishop-Lilly KA, Sozhamannan S, Sulakvelidze A, Rajanna C. A Yersinia pestis-specific, lytic phage preparation significantly reduces viable Y. pestis on various hard surfaces experimentally contaminated with the bacterium. Bacteriophage. 2012;2:168–77. doi: 10.4161/bact.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma M, Ryu J-H, Beuchat LR. Inactivation of Escherichia coli O157:H7 in biofilm on stainless steel by treatment with an alkaline cleaner and a bacteriophage. J Appl Microbiol. 2005;99:449–59. doi: 10.1111/j.1365-2672.2005.02659.x. [DOI] [PubMed] [Google Scholar]
- 11.Goodridge LD, Bisha B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage. 2011;1:130–7. doi: 10.4161/bact.1.3.17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greer GG. Bacteriophage control of foodborne bacteriat. J Food Prot. 2005;68:1102–11. doi: 10.4315/0362-028x-68.5.1102. [DOI] [PubMed] [Google Scholar]
- 13.Sulakvelidze A, Barrow PA. Phage therapy in animals and agribusiness. In: Kutter E and Sulakvelidze A, ed(s). Bacteriophages: Biology and applications. Boca Raton, FL:CRC Press, 2005:335-380 [Google Scholar]
- 14.Adams MH. Host specificity. In: Benzer S, et al., ed(s). Bacteriophages. London:Interscience Publishers Ltd., 1959:121-135 [Google Scholar]
- 15.Adams MH. Appendix: Isolation of bacterial viruses. In: Benzer S, et al., ed(s). Bacteriophages. London:Interscience Publishers, 1959:447-449 [Google Scholar]
- 16.Ackermann H-W, Berthiaume L. A summary of virus classification. In: ed(s). Atlas of virus diagrams. Boca Raton, FL:CRC Press, 1995:3-6. [Google Scholar]
- 17.Mathews CK. An overview of the T4 developmental program. In: Karam JD, et al., eds. Molecular biology of bacteriophage T4. Washington, DC:ASM Press, 1994:1-10 [Google Scholar]
- 18.Hibma AM, Jassim SA, Griffiths MW. Infection and removal of L-forms of Listeria monocytogenes with bred bacteriophage. Int J Food Microbiol. 1997;34:197–207. doi: 10.1016/S0168-1605(96)01190-7. [DOI] [PubMed] [Google Scholar]
- 19.Roy B, Ackermann HW, Pandian S, Picard G, Goulet J. Biological inactivation of adhering Listeria monocytogenes by listeriaphages and a quaternary ammonium compound. Appl Environ Microbiol. 1993;59:2914–7. doi: 10.1128/aem.59.9.2914-2917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.