Key Points
Identification of a miRNA panel as a biomarker for the diagnosis, prognosis, and prediction of acute graft-versus-host disease.
Abstract
Acute graft-versus-host disease (aGVHD) is the leading cause of morbidity and mortality after allogeneic hematopoietic cell transplantation (HCT). Approximately 35% to 50% of HCT recipients develop aGVHD; however, there are no validated diagnostic and predictive blood biomarkers for aGVHD in clinical use. Here, we show that plasma samples from aGVHD patients have a distinct microRNA (miRNA) expression profile. We found that 6 miRNAs (miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a) were significantly upregulated in the plasma of aGVHD patients (n = 116) when compared with non-GVHD patients (n = 52) in training and validation phases. We have developed a model including 4 miRNAs (miR-423, miR-199a-3p, miR-93*, and miR-377) that can predict the probability of aGVHD with an area under the curve of 0.80. Moreover, these elevated miRNAs were detected before the onset of aGVHD (median = 16 days before diagnosis). In addition, the levels of these miRNAs were positively associated with aGVHD severity, and high expression of the miRNA panel was associated with poor overall survival. Furthermore, the miRNA signature for aGVHD was not detected in the plasma of lung transplant or nontransplant sepsis patients. Our results have identified a specific plasma miRNA signature that may serve as an independent biomarker for the prediction, diagnosis, and prognosis of aGVHD.
Introduction
Acute graft-versus-host disease (aGVHD) is a common complication after allogeneic hematopoietic cell transplantation (HCT) that affects the skin, liver, and gastrointestinal tract and contributes to transplant-related morbidity and mortality. Following HCT, approximately 50% of patients will develop aGVHD and require systemic treatment.1-3 Accurate and early biomarkers for aGVHD would improve diagnosis and prognosis and help guide therapeutic interventions. Currently, the diagnosis of aGVHD is based on clinical symptoms in one or more of the main target organs and biopsy results. So far, there is no validated diagnostic or predictive blood biomarker for aGVHD in clinical use.4,5 Moreover, once aGVHD occurs, the most important predictor of long-term survival is the primary response to therapy. In patients who are resistant to initial therapy, the risk of morbidity and mortality increases significantly.6,7 Thus, there is a pressing need to identify noninvasive biomarkers to predict not only aGVHD development but also the survival and treatment outcome of aGVHD.
MicroRNAs (miRNAs) are a class of small noncoding RNA that negatively regulate gene expression by translational repression or induction of messenger RNA degradation.8 Increasing evidence suggests that miRNAs are present in plasma, serum, saliva, urine, and other body fluids in a remarkably stable form that is protected from endogenous RNase activity.9-11 Circulating miRNAs have the potential to serve as novel, noninvasive biomarkers for various diseases such as cancer, cardiovascular disease, sepsis, organ transplant rejection, liver injury, ectopic pregnancy, diabetes, and infection.12-20
In this study, we have identified a group of plasma miRNAs as potential biomarkers for aGVHD. We found that 6 miRNAs (miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a) were significantly upregulated in the plasma of aGVHD patients when compared with non-GVHD patients after HCT. We have developed a model including 4 miRNAs (miR-423, miR-199a-3p, miR-93*, and miR-377) to predict the probability of aGVHD. The levels of these miRNA biomarkers are positively associated with aGVHD severity. Moreover, elevated levels of these miRNAs can be detected before aGVHD diagnosis, and high expression of these miRNAs is associated with poor survival. Importantly, the aGVHD miRNA signature was not detected in plasma samples from lung transplant or nontransplant sepsis patients. Our results suggest that a panel of 4 circulating miRNAs can serve as an independent biomarker for the prediction, diagnosis, and prognosis of aGVHD.
Methods
Patients and samples
The study population consisted of 196 human subjects who underwent allogeneic HCT between 2008 and 2012 at Duke University Medical Center. The discovery set consisted of 4 HCT patients who developed aGVHD and 3 HCT patients who never developed aGVHD (non-GVHD). The training set consisted of 59 aGVHD and 19 non-GVHD patients, and the validation and blinded testing set consisted of 70 aGVHD and 41 non-GVHD patients. EDTA-anticoagulated blood was drawn 2 or 6 weeks after transplantation. Additionally, 45 patients who underwent lung transplantation (including 17 with biopsy-confirmed acute rejection and 28 without rejection), 38 patients who had sepsis with confirmed pathogens, and 10 healthy donors were enrolled as controls. The detailed information on sepsis patients and healthy donors is included in the supplemental Methods on the Blood Web site. Cell-free plasma was isolated from all blood samples using a 2-step centrifugation protocol (2000 rpm for 10 min, 12 000 rpm for 3 min) to prevent contamination by cellular nucleic acids. The diagnosis of aGVHD was based on clinical criteria and histologically confirmed by biopsy in the target organs. aGVHD was graded based on the severity of involvement of the target organs.21 All research samples were collected after obtaining written informed consent for participation in accordance with the Declaration of Helsinki through protocols approved by the Duke University Medical Center institutional review board. Patient characteristics are summarized in Table 1, and a diagram of sample collection and analysis is shown in supplemental Figure 1.
Table 1.
Clinical characteristics of patients recruited in the study
| Characteristic | Discovery and training phase | P | Validation and blinded testing phase | P | ||
|---|---|---|---|---|---|---|
| non-GVHD (n = 22) | aGVHD (n = 63) | non-GVHD (n = 41) | aGVHD (n = 70) | |||
| Median age (range) | 48.5 (24-72) | 50.5 (19-71) | 49 (19-72) | 58 (19-72) | ||
| Disease (%) | .431 | .833 | ||||
| Malignant* | 19 (86.4%) | 58 (92.1%) | 37 (90.2%) | 64 (91.4%) | ||
| Other† | 3 (13.6%) | 5 (7.90%) | 4 (9.80%) | 6 (8.60%) | ||
| Disease status at HCT‡ (%) | .344 | .588 | ||||
| Low/mediate risk | 12 (54.5%) | 27 (42.9%) | 25 (61.0%) | 39 (55.7%) | ||
| High risk | 10 (45.5%) | 36 (57.1%) | 16 (39.0%) | 31 (44.3%) | ||
| Donor type (%) | .281 | .246 | ||||
| Related | 12 (54.5%) | 26 (41.3%) | 24 (58.5%) | 33 (47.1%) | ||
| Unrelated | 10 (45.5%) | 37 (58.7%) | 17 (41.5%) | 37 (52.9%) | ||
| Regimen type (%) | .149 | .014 | ||||
| Nonmyeloablative | 13 (59.1%) | 26 (41.3%) | 13 (31.7%) | 39 (55.7%) | ||
| Myeloablative | 9 (40.9%) | 37 (58.7%) | 28 (68.3%) | 31 (44.3%) | ||
| Maximum GVHD grade (%) | ||||||
| 1-2 | 42 (66.7%) | 46 (65.7%) | ||||
| ≥3 | 21 (33.3%) | 24 (34.3%) | ||||
| Organ target at GVHD onset (%) | ||||||
| Skin | 23 (36.5%) | 16 (22.9%) | ||||
| Gut | 3 (4.76%) | 13 (18.6%) | ||||
| Liver | 2 (3.17%) | 2 (2.86%) | ||||
| Multiple organs | 35 (55.6%) | 39 (55.7%) | ||||
| GVHD diagnosis day after HCT | ||||||
| Before day 14 | 3 (4.80%) | 4 (5.70%) | ||||
| Between days 14 and 42 | 33 (52.4%) | 21 (30.0%) | ||||
| After day 42 | 27 (42.8%) | 45 (64.3%) | ||||
P values were calculated using a 2-sided χ2 test.
Malignant disease included acute myelogenous leukemia, acute lymphoblastic leukemia, chronic myeloid leukemia, chronic lymphocytic leukemia, non-Hodgkin lymphoma, Hodgkin disease, myelodysplastic syndrome, diffuse large B-Cell lymphoma, and multiple myeloma.
Other disease, which means nonmalignant disease, included severe aplastic anemia, sickle cell anemia, dyskeratosis congenita, myeloproliferative disorder, and myelofibrosis.
Low and high risk of disease status at BMT is according to CIBMTR guidelines.
RNA extraction from plasma
Total RNAs were extracted from 400 µL of plasma using the mirVana PARIS Kit (Ambion) according to the manufacturer’s instructions. Because no known endogenous miRNA in human plasma can be used as normalization control, we used synthetic Caenorhabditis elegans miRNA (cel-miR-39) as a spiked-in control. Two microliters of 5 nM synthetic cel-miR-39 (synthesized by Integrated DNA Technologies) was added to each denatured sample after combining the plasma sample with the denaturing solution. RNAs were eluted with 105 µL of preheated elution solution.
qRT-PCR analysis of plasma miRNAs
Plasma miRNAs were quantified using SYBR Green–based quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) as previously described.22 The detailed information is shown in the supplemental Methods.
Real-time qPCR-based high-throughput miRNA array
We used a real-time quantitative polymerase chain reaction (qPCR)-based high-throughput miRNA array that contains primers specific for 345 well-characterized human miRNAs as previously described23 to compare the plasma miRNA profiles of 4 aGVHD and 3 non-GVHD patients. Cel-miR-39 was used for normalization to obtain relative miRNA expression.
Enzyme-linked immunosorbent assay of plasma sIL-2Ra
Plasma soluble interleukin-2 receptor α (sIL-2Rα) was measured using a sIL-2Rα ELISA kit (R&D Systems) according to the manufacturer’s instructions. Four-fold dilutions of plasma samples and standards were measured in duplicate. Absorbance was measured with a Molecular Devices Emax plate reader, and the results were analyzed with SoftMax Pro 4.8 software (Molecular Devices).
Statistical analysis
Hierarchical clustering analysis was performed using Cluster 3.0. The Mann-Whitney test was used to compare the differences in plasma miRNA expression between aGVHD and non-GVHD groups. A 2-sided χ2 test was used to compare the differences in patient clinical characteristics. Receiver-operating characteristic (ROC) curves and the area under the ROC curve (AUC) were used to assess the sensitivity and specificity of miRNA biomarkers for the diagnosis of aGVHD. Logistic regression was used to develop a combined miRNA panel to predict the probability of developing aGVHD as previously described.23 Overall survival was analyzed using Kaplan-Meier curves. Cox regression was performed to determine the predictive value of the miRNA signature for diagnosis of aGVHD and survival prediction. Spearman correlation analysis was performed to test whether the expression levels of the 4 miRNAs are correlated with each other. All statistical analysis was performed using SPSS 16.0 software, and graphs were generated using GraphPad Prism 5.0 (GraphPad Software).
Results
Plasma miRNA profiling and signature in aGVHD patients
To identify a diagnostic plasma miRNA signature for aGVHD, we used a real-time PCR-based high-throughput miRNA array to compare the plasma miRNA profiles from 4 biopsy-confirmed aGVHD patients with the profiles from 3 non-GVHD patients 6 weeks after HCT. We identified a distinct miRNA signature including a set of 24 miRNAs that were differentially expressed in aGVHD patients compared with non-GVHD patients, including 8 upregulated and 16 downregulated miRNAs (supplemental Figure 2). Hierarchical clustering analysis showed that the plasma miRNA signature was able to distinguish aGVHD from non-GVHD subjects.
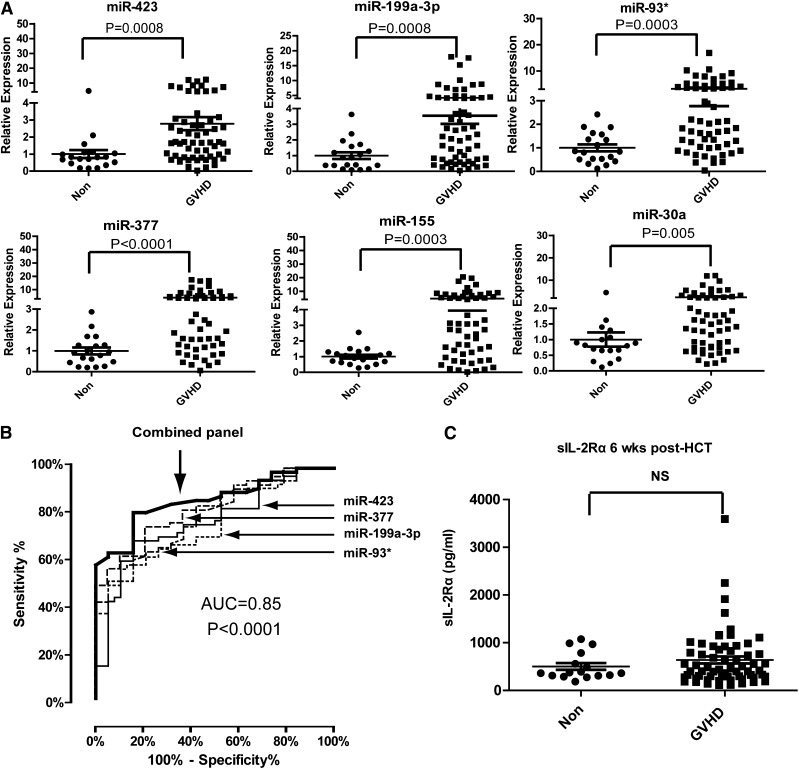
To verify the differentially expressed plasma miRNAs identified in the discovery phase, we performed miRNA qPCR in an independent training set consisting of 59 aGVHD patients and 19 non-GVHD controls. Among the 24 candidate miRNAs, 6 plasma miRNAs (miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a) were significantly upregulated in aGVHD patients compared with non-GVHD patients in the training phase (Figure 1A). The fold changes in the mean expression levels of these 6 miRNAs were 2.8 (miR-423), 3.5 (miR-199a-3p), 3.2 (miR-93), 4.0 (miR-377), 4.6 (miR-155), and 2.4 (miR-30a). We chose these 6 miRNAs for further analysis.
Figure 1.
Expression of plasma miRNA signature for aGVHD diagnosis in a training set. (A) Expression of miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a in plasma of aGVHD (n = 59) and non-GVHD patients (n = 19) 6 weeks after HCT. P values were calculated using the Mann-Whitney test. (B) ROC analysis for individual miRNAs and combined miRNA panel. Logistic regression demonstrated that a linear combination of values for miR-423, miR-199a-3p, miR-93*, and miR-377 produced the best model for aGVHD diagnosis; the equation was “Combined miRNA panel = 5.514 + 0.255 × miR-423 − 0.427 × miR-199a-3p + 0.522 × miR-93* − 1.161 × miR-377.” ROC curve of miRNA panel was generated based on the predicted probability (P) for each patient. Predicted probability = Exp (combined miRNA panel)/ [1+Exp (combined miRNA panel)]. (C) Comparison of sIL-2Rα protein levels in plasma between aGVHD and non-GVHD patients 6 weeks after HCT. P values were calculated using the Mann-Whitney test.
Development of a diagnostic panel containing 4 plasma miRNAs
The 6 altered miRNAs we identified are involved in inflammation, tissue damage, regulation of cell proliferation, and tissue repair,24-26 suggesting increased levels of these miRNAs in the plasma of aGVHD patients may be associated with the immune response and organ damage during aGVHD. We thus evaluated the sensitivity and specificity of these miRNAs in diagnosing aGVHD using ROC analysis. Next, we used logistic regression to determine the best combination of miRNAs to predict aGVHD. Because the expression of miR-155 was the lowest among the 6 miRNAs, we did not include miR-155 in the logistic regression model. We found that that a linear combination of the expression levels of miR-423, miR-199a-3p, miR-93*, and miR-377 produced the best model to predict aGVHD. Using a 4-miRNA panel significantly increased the AUC value when compared with using any of the miRNAs alone. The ROC curves had AUCs of 0.76 (95% confidence interval [CI], 0.64-0.87) for miR-423, 0.76 (95% CI, 0.65-0.87) for miR-199a-3p, 0.78 (95% CI, 0.67-0.89) for miR-93*, and 0.81 (95% CI, 0.71-0.91) for miR-377 (Figure 1B). The AUC for the combined miRNA biomarker panel of miR-423, miR-199a-3p, miR-93*, and miR377 was 0.85 (95% CI, 0.76-0.93) (P < .0001).
Previous studies suggested sIL-2Rα as a potential biomarker for aGVHD, with the highest AUC value achieved by a panel of protein biomarkers including sIL-2Rα, interleukin-8, hepatocyte growth factor, and tumor necrosis factor receptor 1.27-29 Despite these findings, we found no significant difference in the plasma levels of sIL-2Rα between aGVHD and non-GVHD patients 6 weeks after transplantation (Figure 1C). Taken together, our data suggest that the plasma miRNA panel may be a potential noninvasive biomarker for aGVHD.
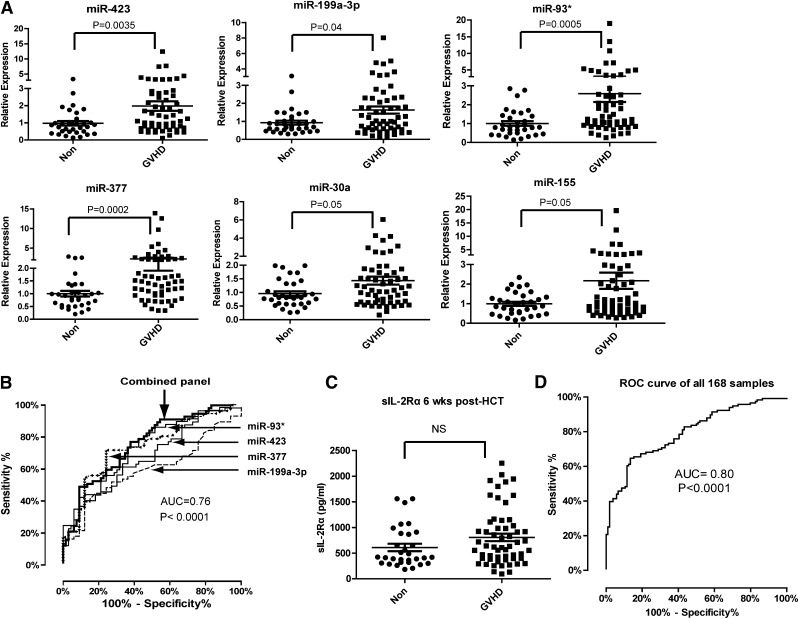
Validation and blinded testing of the miRNA signature for aGVHD
We further validated the expression levels of the 6 miRNAs in an independent validation study consisting of 57 aGVHD patients and 33 non-GVHD control subjects. The upregulation of the 6 miRNAs in the plasma of aGVHD patients was validated. The AUC for the 4-miRNA biomarker panel including miR-423, miR-199a-3p, miR-93*, and miR-377 was 0.76 (95% CI, 0.65-0.86) (P < .0001) (Figure 2B). No significant difference in plasma sIL-2Rα levels was found between the aGVHD and non-GVHD patients 6 weeks after transplantation (Figure 2C). Analysis of the data from all 168 samples in training and validation sets produced an AUC value of 0.80 (Figure 2D). To determine whether plasma miRNA levels differed between the aGVHD and non-GVHD groups prior to HCT, we analyzed the levels of miRNAs in plasma samples taken before transplantation. No significant difference was found in the expression of the 6 miRNAs in plasma of aGVHD and non-GVHD patients before HCT (supplemental Figure 3), suggesting that the elevated miRNA levels in aGVHD plasma were not due to the presence of elevated miRNA in the patients before transplantation.
Figure 2.
Expression of plasma miRNA signature for aGVHD diagnosis in a validation set. (A) Expression of miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a in plasma of aGVHD (n = 57) and non-GVHD patients (n = 33) 6 weeks after HCT. (B) ROC analysis for individual miRNAs and combined miRNA signature including miR-423, miR-199a-3p, miR-93*, and miR-377 as described in the legend of Figure 1B. The equation of logistic regression in validation set was “Combined miRNA panel = 11.059 − 0.19 × miR-423 + 0.175 × miR-199a-3p-0.04 × miR-93* − 0.89 × miR-377.” (C) Comparison of sIL-2Rα protein levels in plasma between aGVHD and non-GVHD patients 6 weeks after HCT. (D) ROC curve of the 4-miRNA panel generated by analyzing all 168 samples from training and validation sets.
We next tested the predictive value of the miRNA signature in a blinded study including 13 aGVHD patients and 8 non-GVHD patients 6 weeks after HCT. In this study, we were blinded to the patients’ clinical data when measuring miRNA levels. Based on the model developed in the validation phase, a signature composed of the 4 miRNAs (miR-423, miR-199a-3p, miR-93, and miR-377) correctly discriminated 12 of 13 aGVHD samples (92% sensitivity) and 5 of 8 non-aGVHD samples. Taken together, our data suggest that the 4-miRNA panel is a potential noninvasive biomarker for aGVHD.
miRNA signature as a predictive factor for aGVHD
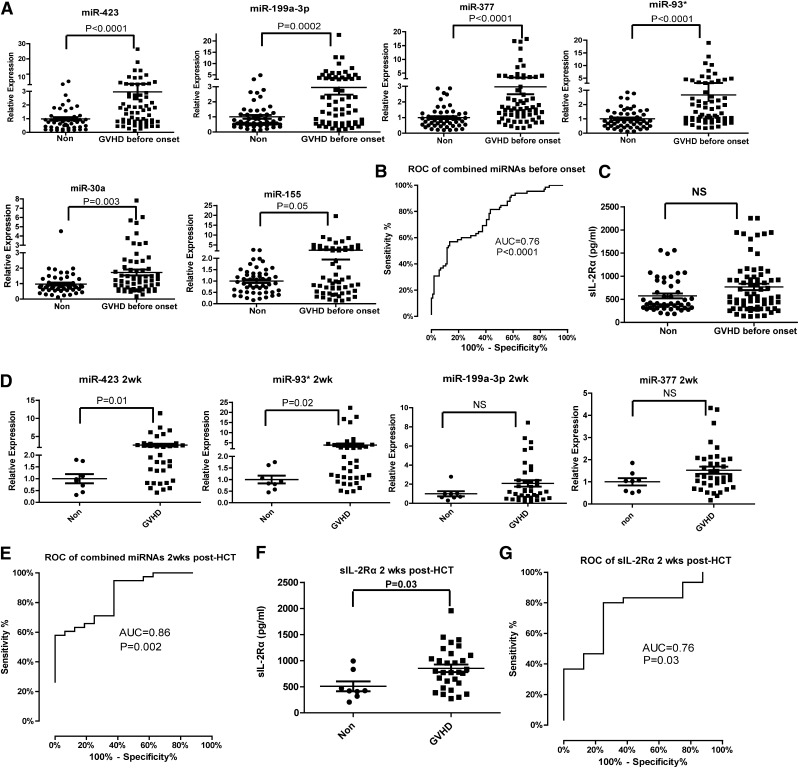
An ideal miRNA biomarker should be detectable before the onset of aGVHD. We analyzed the miRNA panel in plasma samples collected from aGVHD patients before diagnosis. Among all aGVHD samples at 6 weeks after HCT, we selected the aGVHD patients who did not have the symptoms 6 weeks after transplantation but developed aGVHD 1 to 78 (median = 16) days later into the analysis. We observed significantly elevated expression of the 6 miRNAs in the prediagnosis plasma samples of aGVHD patients (n = 65) compared with non-GVHD controls (n = 52) (Figure 3A). When we used miR-423, miR-199a-3p, miR-93*, and miR-377 as the miRNA signature to predict aGVHD, the AUC was 0.76 (95% CI, 0.68-0.85) (P < .0001) (Figure 3B). In contrast, sIL-2Rα levels did not differ significantly between prediagnosis plasma samples from aGVHD patients and samples from non-GVHD patients (Figure 3C).
Figure 3.
Predictive value of the miRNA signature for aGVHD. (A) The expression of miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a in the plasma of aGVHD patients prior to aGVHD diagnosis (median = 16 days before diagnosis). (B) ROC curve of the 4-miRNA panel (miR-423, miR-199a-3p, miR-93*, miR-377) 6 weeks after HCT for prediction of aGVHD before onset. (C) Plasma concentration of sIL-2Rα 6 weeks after HCT in non-GVHD and aGVHD patients before aGVHD diagnosis. (D) The expression of the 4-miRNA panel 2 weeks after HCT in plasma of aGVHD and non-GVHD patients. (E) ROC curve of the 4-miRNA panel 2 weeks after HCT. (F) Plasma concentration of sIL-2Rα 2 weeks after HCT. (G) ROC curve of sIL-2Rα 2 weeks after HCT.
To diagnose the onset of aGVHD development as early as possible, we further analyzed the expression of the miRNA panel in plasma samples taken 2 weeks after HCT. The levels of miR-423 and miR-93* were significantly upregulated in the plasma of aGVHD patients compared with non-GVHD patients. The levels of miR-199a-3p and miR-377 were also increased but failed to reach statistical significance (Figure 3D). Consistent with published results,27,28 the plasma levels of sIL-2Rα 2 weeks after HCT were significantly higher in aGVHD patients than in non-GVHD patients (Figure 3F). However, the AUC for the miRNA signature including miR-423, miR-199a-3p, miR-93*, and miR-377 was higher than the AUC for sIL-2Rα 2 weeks after HCT (0.86 vs 0.76) (Figure 3E,G). This result suggests that use of the miRNA signature will increase the accuracy of diagnosis for aGVHD.
Subsequently, we performed the Cox regression analysis using the diagnosis of aGVHD as the end point to evaluate whether the 4-miRNA panel can predict the probability of a patient developing aGVHD. Univariate Cox analysis with the 4-miRNA panel and clinical factors (age, malignancy, risk score, donor type, and regimen type) revealed that the 4-miRNA panel had predictive value for aGVHD in the training and validation sets (P = .001, hazard ratio 1.534 in training phase; P < .001, hazard ratio 1.481 in validation phase; Table 2). Furthermore, multivariate Cox regression analysis of each of these parameters suggests that the expression of the 4-miRNA panel was an independent predictor for aGVHD after adjusting for age, malignancy, risk score, donor type, and regimen type (P = .004, hazard ratio 1.524 in training phase; P < .001, hazard ratio 1.478 in validation phase; Table 2).
Table 2.
Cox regression analysis for the estimation of risk prediction of aGVHD in the training phase and validation phase
| Training phase | Validation phase | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| Hazard ratio | P | Hazard ratio | P | Hazard ratio | P | Hazard ratio | P | |
| Age | 1.004 | .720 | 1.007 | .590 | 1.022 | .083 | 1.013 | .327 |
| Malignant (yes/no) | 0.342 | .137 | 0.590 | .516 | 1.069 | .869 | 1.065 | .894 |
| Risk score (low/high) | 0.495 | .019 | 0.670 | .209 | 1.014 | .960 | 1.300 | .384 |
| Donor type (related/unrelated) | 0.630 | .134 | 0.609 | .126 | 0.835 | .504 | 0.971 | .919 |
| Regimen type (nonmyeloablative/myeloablative) | 0.585 | .073 | 0.772 | .469 | 1.385 | .229 | 1.236 | .494 |
| miRNA signature | 1.534 | .001 | 1.524 | .004 | 1.481 | <.001 | 1.478 | <.001 |
For Cox regression analysis, the reference group is the group that is coded as “0.” For example, for the variable of malignant (Y/N), we code “malignant patients (yes)” as “1” and “no malignant patients (no) as “0.” The group with lower classification is the reference group. “Risk score (low), donor type (related), regimen type (nonmyeloablative)” are reference groups for their variables.
Association of the miRNA biomarker panel with aGVHD severity
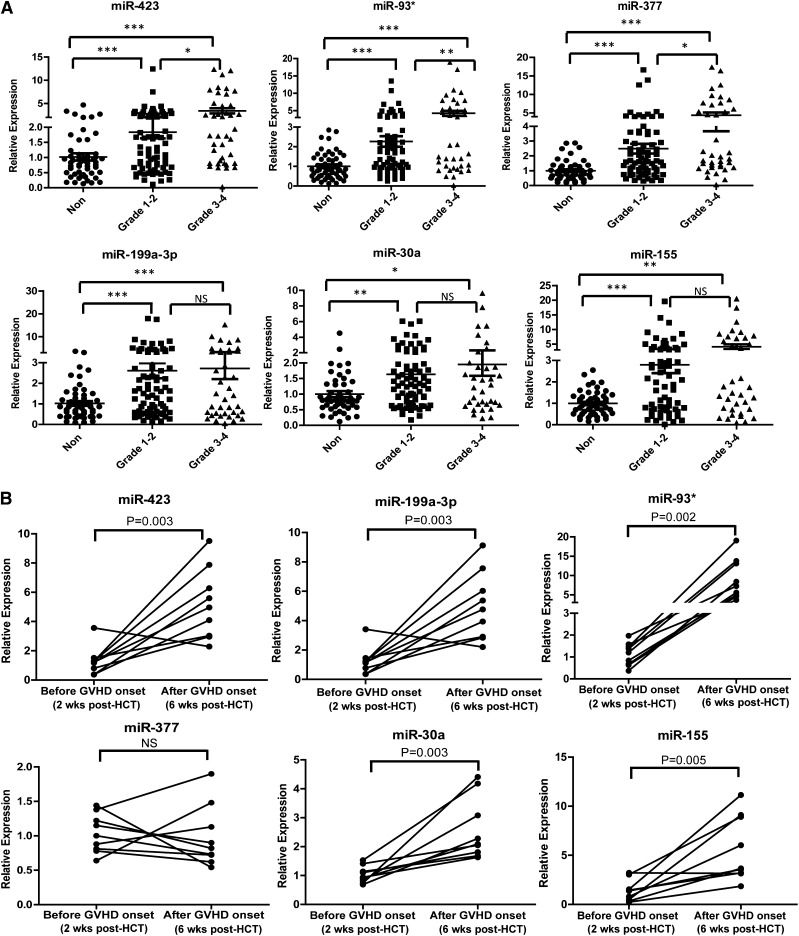
We examined whether the levels of the plasma miRNA biomarker panel were associated with the severity of aGVHD (grade 1-4). All aGVHD patients were divided into 2 groups based on max aGVHD grade: low grade (1-2) and high grade (3-4). The expression of miR-423, miR-93*, and miR-377 was significantly elevated in high-grade aGVHD patients compared with low-grade aGVHD patients, whereas the expression of miR-199a-3p, miR-155, and miR-30a did not differ in plasma samples from high-grade and low-grade aGVHD patients (Figure 4A). In addition, we found significantly elevated expression of the 6 miRNAs in all aGVHD patients (n = 168) compared with non-aGVHD patients. Moreover, the levels of miR-423, miR-199a-3p, miR-377, miR-93*, miR-155 were significantly upregulated in more severe aGVHD patients (grade 2-4), compared with non-aGVHD patients (grade 0) and less severe aGVHD patients (grade 1) (supplemental Figure 4). The above data suggest that the miRNA panel is associated with aGVHD severity. To investigate whether miRNA biomarker expression is associated with the development of aGVHD, the levels of the miRNA signature were measured in a set of 9 patients before disease onset (2 weeks after HCT) and after onset (6 weeks after HCT). The levels of miR-423, miR-199a, and miR-93* were significantly increased in the postdiagnosis samples compared with the prediagnosis samples (Figure 4B).
Figure 4.
Association of the miRNA signature with aGVHD severity. (A) Expression levels of miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a in aGVHD patients with different grades of disease (grade 1-2 vs grade 3-4). P values were calculated using the Mann-Whitney test. *P < .05; **P < .01; ***P < .001. (B) The kinetic expression of the miRNA signature in a set of 9 patients before disease onset (2 weeks after HCT) and after onset (6 weeks after HCT). We selected 9 aGVHD patients in the analysis according to the following criteria: they had similar diagnosis time (median day 20, range: 18 to 23) and didn’t develop aGVHD 2 weeks after HCT but had symptoms 6 weeks after HCT. P values were calculated using paired t tests.
Association of the miRNA biomarker panel with overall survival in aGVHD patients
We evaluated whether the 4-miRNA panel can be used as a prognostic marker in aGVHD patients. In the training set, 44 aGVHD patients with updated follow-up information were divided into 2 groups (high risk and low risk) based on their predicted probabilities for developing aGVHD using the 4-miRNA panel from the logistic model in the training set. Patients were designated as high risk if the probability of a single patient developing aGVHD was higher than the median probability of all the patients. Kaplan-Meier survival analysis showed that patients designated as high risk based on miR-423, miR-199a-3p, miR-93*, and miR-377 levels had poor survival compared with low-risk patients (P = .02) (supplemental Figure 5). Subsequently, multivariate Cox regression analysis with the 4-miRNA panel and clinical characteristics including age, malignancy, risk score, donor type, and regimen type indicated that the 4-miRNA panel was an independent unfavorable prognostic factor for aGVHD overall survival (P = .01, hazard ratio 2.110; supplemental Table 1).
Expression correlation within the 4-miRNA panel
Multivariate logistic regression analysis showed that use of the 4-miRNA panel improved the distinction between aGVHD and non-GVHD patients over the use of individual miRNAs. We performed Spearman correlation analysis for all of the samples 6 weeks after HCT in training and validation sets to test whether the expression levels of the 4 miRNAs were correlated with each other. Significant positive correlations were found between miR-423 and miR-199a-3p and between miR-93* and miR-377 plasma levels, with R2 values of 0.822 and 0.932, respectively (P < .001; supplemental Figure 6). In addition, Spearman correlation analysis showed that the expression levels of the 4 miRNAs in the panel (miR-423, miR-199a-3p, miR-93*, and miR-377) were significantly positively correlated with each other, with all P values < .001 (supplemental Table 2). These results further support the overlapping diagnostic values of these 4 miRNAs. However, none of the individual miRNA levels were correlated with sIL-2Rα levels, indicating that the miRNA panel is an independent biomarker for aGVHD (supplemental Table 3).
Disease specificity of the 6 miRNAs
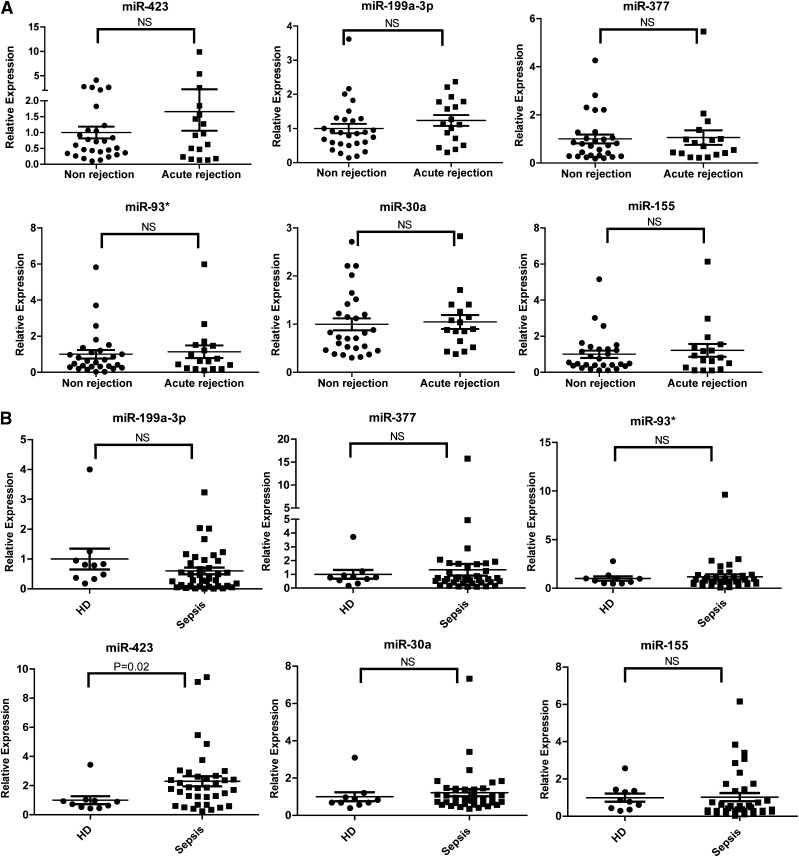
To determine the disease specificity of the 6 miRNAs identified in aGVHD patients, we enrolled patients with or without acute rejection after lung transplantation and patients with or without sepsis as non-HCT disease controls. No significant differences were found in the expression of the 6 miRNAs in plasma samples from acute rejection (n = 17) and nonrejection (n = 28) lung transplant patients (Figure 5A). Sepsis is a posttransplantation complication associated with a significant increase in inflammation-related markers, including sIL-2Rα.28 We compared the expression of the 6 miRNAs in the plasma of 38 sepsis patients and 10 healthy donors. The expression of 5 miRNAs (miR-199a-3p, miR-93*, miR-377, miR-30a, and miR-155) was comparable between sepsis patients and healthy controls; however, the level of miR-423 was higher in sepsis patients than in healthy donors (Figure 5B). These data suggest that these 5 miRNAs are related to the development of aGVHD and that miR-423 may be involved in the regulation of inflammation.
Figure 5.
Expression of the aGVHD miRNA signature in sepsis or lung transplant patients. (A) Expression levels of the aGVHD miRNA signature including miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a in plasma of lung transplant patients with (n = 17) or without (n = 28) acute rejection. (B) Expression levels of the aGVHD miRNA signature in plasma of sepsis patients (n = 38) and healthy donors (n = 10). HD, healthy donors; NS, nonsignificant.
Discussion
Recent publications demonstrate that circulating miRNAs may serve as noninvasive biomarkers for many different diseases.12-20 Here, we show that plasma miRNAs can be used as biomarkers for aGVHD. We found that the plasma levels of 6 miRNAs (miR-423, miR-199a-3p, miR-93*, miR-377, miR-155, and miR-30a) were significantly elevated in the plasma of aGVHD patients when compared with non-GVHD patients. We have developed a model including 4 miRNAs to diagnose aGVHD with an AUC of 0.80. The miRNA panel can improve the diagnosis sensitivity and specificity for aGVHD when compared with a single miRNA biomarker (supplemental Table 4). The levels of the miRNA biomarkers were positively associated with aGVHD severity. Moreover, the elevated miRNAs were detected before the onset of aGVHD and were not found in lung transplant patients or patients with sepsis. In addition, we found that high expression of a 4-miRNA panel was associated with poor survival. Taken together, our data show that the plasma miRNA signature is an independent predictive, diagnostic, and prognostic factor for aGVHD.
Our data suggest that the plasma miRNA panel has excellent sensitivity and specificity as a biomarker for aGVHD. Extensive studies have been devoted to identifying blood biomarkers for aGVHD to improve diagnosis and individualized treatment; however, the majority of the work has focused on whole proteins or polypeptides.4,5,30 To date, there is no widely accepted protein biomarker for aGVHD. The reported potential protein biomarkers for aGVHD have several drawbacks. First, these biomarkers are often elevated during inflammation such as that caused by bacterial infection and veno-occlusive disease4,28 and are thus not specific for aGVHD. Published analyses testing these protein biomarkers often excluded patients with other inflammatory conditions such as sepsis, idiopathic pneumonia syndrome, and veno-occlusive disease.29,31 Second, compared with qPCR-based assays, the methods for protein detection either have a relatively low sensitivity or are too complicated for clinical use.4 Third, most reported protein biomarkers for aGVHD could aid in diagnosis, but their predictive and prognostic values remain to be established.4 Recent findings suggest that circulating miRNAs are an excellent class of biomarkers. Serum or plasma miRNAs are highly stable32 and tissue-specific.33-35 Our results show that compared with sIL-2Rα, a promising biomarker with the highest AUC value in a panel of protein biomarkers for aGVHD,29 the plasma miRNA signature has obvious advantages. The levels of sIL-2Rα peak 2 to 3 weeks after HCT and decline rapidly to baseline levels 6 weeks after HCT.27,28 Furthermore, the levels of sIL-2Rα are also elevated during infection.28 Thus, it is not clear whether the elevated sIL-2Rα level is due to infection or aGVHD. Our measurement of the expression levels of the miRNA signature for aGVHD clearly demonstrate that it is elevated in the plasma of the aGVHD patients 2 to 6 weeks after HCT but is not detected in other conditions, such as sepsis and lung transplantation.
Our results show that the miRNA panel can predict aGVHD. We found that the levels of the plasma miRNA signature were elevated in aGVHD patients at a median of 16 days before the diagnosis of aGVHD. In contrast, the plasma levels of sIL-2Rα in these samples were not significantly elevated. The low relative abundance of protein in the plasma may render its detection difficult. However, the change in the expression levels of the miRNA signature prior to aGVHD diagnosis can be detected by qPCR. Cox regression analysis indicated that the miRNA signature containing 4 miRNAs (miR-423, miR-199a-3p, miR-93*, and miR-377) had the best predictive value for aGVHD. Predicting aGVHD development is essential for the early identification of high-risk individuals who need individualized treatment. In future studies, we can identify new patients in whom early intervention might be warranted according to the following criteria. First, we will use a miRNA model developed from large-scale validation study, such as the following equation in our study: combined miRNA panel = 11.059 − 0.19 × miR-423 + 0.175 × miR-199a-3p-0.04 × miR-93* − 0.89 × miR-377. Second, we will measure the expression of the 4-miRNA panel for each patient and calculate the predicted probability of aGVHD based on the following equation: predicted probability (P) = Exp (combined miRNA panel) / [1 + Exp (combined miRNA panel)]. Third, we will determine if the patient has developed aGVHD based on the threshold that was set in the model. For example, we consider the patient as aGVHD if the predicted probability (P) > 0.66 (sensitivity 60%, specificity 76%). We have successfully predicted the occurrence of aGVHD prior to onset by using the miRNA model in a blinded test. We correctly discriminated 12 of 13 aGVHD samples (92% sensitivity) and 5 of 8 non-aGVHD samples (62% specificity; false-positive rate, 38%) by using the 4-miRNAs panel (data not shown).
Our data suggest that the plasma miRNA signature is associated with poor survival in aGVHD patients and is a prognostic factor for predicting the outcome of aGVHD patients. The severity and outcome of aGVHD is heterogeneous. Growing evidence shows that diverse clinical factors such as age, HLA matching, the intensity of conditioning, and the intensity of posttransplant immunosuppression may be associated with aGVHD36; however, prognostic factors that can be identified at the onset of aGVHD remain largely elusive. If patients who are likely to have poor outcomes can be predicted at the onset of aGVHD, early administration of tailored therapy may improve the outcome of these patients. The skin, liver, and gastrointestinal tract are the 3 main target organs of aGVHD, and particular miRNAs may be associated with particular aGVHD target organs. However, we have not performed organ-specific association studies because most of the patients enrolled in this study had damage to multiple organs.
The 6 miRNAs identified in the plasma of aGVHD patients are involved in inflammation, tissue damage, regulation of cell proliferation, and tissue repair. miR-423 may serve as a potential circulating biomarker for heart failure.24 miR-155 plays a key role in inflammation regulation and immune responses.37 Interestingly, a recent report showed that miR-155 is upregulated in the effector T cells of mice with aGVHD and that miR-155 expression in lymphocytes is essential for lethal aGVHD in mice.26 miR-199a-3p was reported to play a role in hepatocyte injury and hepatocellular carcinoma.25,38 miR-93* regulates cell proliferation,39 whereas miR-30a is associated with autophagy and apoptosis.40,41 Spearman analysis in our study demonstrated a high correlation between several miRNAs in the panel, suggesting that the above miRNAs have a similar function in the development of aGVHD. Although the mechanism underlying the secretion of the plasma miRNAs remains to be investigated, our hypothesis is that the miRNA panel may play a potential role in the donor T cell’s attacking process or in the response to damage in target cells during aGVHD. The upregulated cellular miRNAs will be selectively included into multivesicular bodies or exosomes and then released into the bloodstream.42 However, recent work indicates that circulating miRNAs can be transferred in an exosome-independent manner, and miRNAs can be stably exported in conjunction with RNA-binding proteins, such as nucleophosmin 1,43 Argonaute 2,44 and high-density lipoprotein.45 Therefore, the origin of the plasma miRNA signature of aGVHD requires further investigation.
Supplementary Material
Acknowledgments
The authors thank Dr Claire Gordy for critically reading this manuscript.
This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases grant AI074754 (to Y.-W.H.). E.L.T. was supported by a career development award from the Durham Veterans Administration Medical Center. The enrollment of healthy and sepsis controls was supported by funding from the Defense Advanced Research Projects Agency and the National Institutes of Heath, National Institute of Allergy and Infectious Diseases Research Project Cooperative Agreement (U01 AI066569). N.J.C. was supported by National Institutes of Health, National Cancer Institute Research Program grant 2P01 CA47741.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.X. and Y.-W.H. designed the study; B.X. and Y.W. performed the experiments; B.X., W.L., and Z.L. contributed to the statistical analysis; M.B., K.C., E.-L.T., S.-M.P., C.-W.W., and N.-J.C. provided the GVHD, lung transplant, and sepsis samples; B.X., J.G., Q.-J.L., N.-J.C., and Y.-W.H. analyzed the data; and B.X. and Y.-W.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare competing financial interests. The reported data have been filed for patent invention by Duke University Office of Technology and Licensing.
Correspondence: You-Wen He, Box 3010, Department of Immunology, DUMC, Durham, NC 27710; e-mail: youwen.he@duke.edu; and Nelson J. Chao, Department of Medicine, DUMC, Durham, NC 27710; e-mail: nelson.chao@duke.edu.
References
- 1.Grasedieck S, Sorrentino A, Langer C, et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121(25):4977–4984. doi: 10.1182/blood-2013-01-480079. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012;12(6):443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57(2):840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 5.Paczesny S. Discovery and validation of graft-versus-host disease biomarkers. Blood. 2013 doi: 10.1182/blood-2012-08-355990. 121(4)585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Lint MT, Milone G, Leotta S, et al. Treatment of acute graft-versus-host disease with prednisolone: significant survival advantage for day +5 responders and no advantage for nonresponders receiving anti-thymocyte globulin. Blood. 2006;107(10):4177–4181. doi: 10.1182/blood-2005-12-4851. [DOI] [PubMed] [Google Scholar]
- 7.Deeg HJ. How I treat refractory acute GVHD. Blood. 2007;109(10):4119–4126. doi: 10.1182/blood-2006-12-041889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10):2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Z, Zhao Q, Warrick J, et al. Circulating microRNA miR-323-3p as a biomarker of ectopic pregnancy. Clin Chem. 2012;58(5):896–905. doi: 10.1373/clinchem.2011.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li LM, Hu ZB, Zhou ZX, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70(23):9798–9807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 15.Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 16.Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwabara Y, Ono K, Horie T, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4(4):446–454. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 18.Vasilescu C, Rossi S, Shimizu M, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS ONE. 2009;4(10):e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mas VR, Dumur CI, Scian MJ, Gehrau RC, Maluf DG. MicroRNAs as biomarkers in solid organ transplantation. Am J Transplant. 2013;13(1):11-19. [DOI] [PMC free article] [PubMed]
- 20.Starkey Lewis PJ, Dear J, Platt V, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54(5):1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 21.Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, Thomas ED. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995;15(6):825–828. [PubMed] [Google Scholar]
- 22.Jiang S, Li C, Olive V, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118(20):5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang ZN, Xu JJ, Fu YJ, et al. Transcriptomic Analysis of Peripheral Blood Mononuclear Cells in Rapid Progressors in Early HIV Infection Identifies a Signature Closely Correlated with Disease Progression. Clin Chem. 2013;59(8):1175–1186. doi: 10.1373/clinchem.2012.197335. [DOI] [PubMed] [Google Scholar]
- 24.Tijsen AJ, Creemers EE, Moerland PD, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106(6):1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 25.Lee CG, Kim YW, Kim EH, et al. Farnesoid X receptor protects hepatocytes from injury by repressing miR-199a-3p, which increases levels of LKB1. Gastroenterology. 2012;142(5):1206-1217.e7. [DOI] [PMC free article] [PubMed]
- 26.Ranganathan P, Heaphy CE, Costinean S, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012;119(20):4786–4797. doi: 10.1182/blood-2011-10-387522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm J, Zeller W, Zander AR. Soluble interleukin-2 receptor serum levels after allogeneic bone marrow transplantations as a marker for GVHD. Bone Marrow Transplant. 1998;21(1):29–32. doi: 10.1038/sj.bmt.1701041. [DOI] [PubMed] [Google Scholar]
- 28.Foley R, Couban S, Walker I, et al. Monitoring soluble interleukin-2 receptor levels in related and unrelated donor allogenic bone marrow transplantation. Bone Marrow Transplant. 1998;21(8):769–773. doi: 10.1038/sj.bmt.1701163. [DOI] [PubMed] [Google Scholar]
- 29.Paczesny S, Krijanovski OI, Braun TM, et al. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113(2):273–278. doi: 10.1182/blood-2008-07-167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hori T, Naishiro Y, Sohma H, et al. CCL8 is a potential molecular candidate for the diagnosis of graft-versus-host disease. Blood. 2008;111(8):4403–4412. doi: 10.1182/blood-2007-06-097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasan R, Daniels J, Fusaro V, et al. Accurate diagnosis of acute graft-versus-host disease using serum proteomic pattern analysis. Exp Hematol. 2006;34(6):796–801. doi: 10.1016/j.exphem.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 33.Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82(16):8215–8223. doi: 10.1128/JVI.02575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55(11):1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 36.Harris AC, Ferrara JL, Levine JE. Advances in predicting acute GVHD. Br J Haematol. 2013;160(3):288–302. doi: 10.1111/bjh.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28(5):264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 38.Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Liu S, Patel SH, Ginestier C, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8(6):e1002751. doi: 10.1371/journal.pgen.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou Z, Wu L, Ding H, et al. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem. 2012;287(6):4148–4156. doi: 10.1074/jbc.M111.307405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, et al. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis. 2012;33(4):732–739. doi: 10.1093/carcin/bgs020. [DOI] [PubMed] [Google Scholar]
- 42.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 43.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38(20):7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.