Abstract
Ataxia-telangiectasia mutated (ATM) is a major regulator of the DNA damage response. ATM promotes the activation of BRCA1, CHK2, and p53 leading to the induction of response genes such as CDKN1A (p21), GADD45A and RRM2B that promote cell cycle arrest and DNA repair. The up-regulation of these response genes may contribute to resistance of cancer cells to genotoxic therapies. Here we show that histone deacetylases (HDACs) play a major role in mitigating the response of the ATM pathway to DNA damage. HDAC inhibition decreased ATM activation and expression, and attenuated the activation of p53 in vitro and in vivo. Select depletion of HDAC1 and HDAC2 was sufficient to modulate ATM activation, reduce GADD45A and RRM2B induction, and increase sensitivity to DNA strand breaks. The regulation of ATM by HDAC enzymes therefore suggests a vital role for HDAC1 and HDAC2 in the DNA damage response, and the potential use of the ATM pathway as a pharmacodynamic marker for combination therapies involving HDAC inhibitors.
Keywords: histone deacetylase, HDAC inhibitor, DNA damage signaling, ATM, p53
Introduction
Ataxia-telangiectasia mutated (ATM) is a major regulator of the DNA damage response. ATM is a member of the phosphatidylinositol-3 kinase-related kinases (PIKK) family, whose auto-phosphorylation is promoted by the MRN (MRE11, Rad50, and NBS1) complex in response to DNA double strand breaks (1). DNA damage signaling is then propagated by ATM through activation of BRCA1, CHK2, and p53, leading to the induction of response genes involved in growth arrest, DNA repair, and/or apoptosis (1-3). The repair of double strand breaks occurs primarily through homologous recombination (HR) and non-homologous end joining (NHEJ). Cells with defective ATM exhibit enhanced chromatin decondensation, increased genomic instability, and greater sensitivity to DNA damaging agents (4, 5). These features are similar to those observed in cells treated with histone deacetylase (HDAC) inhibitors (6-9). Emerging evidence suggests a role for HDACs in regulating ATM. ATM and HDAC1 associate in fibroblast cells, and their interaction increases after γ-irradiation (10). HDAC2 regulates the expression of chromatin remodeling genes including SMC1 (11), which is phosphorylated by ATM after the induction of double strand breaks (12).
HDACs catalyze the removal of acetyl groups from histone and non-histone proteins alike, altering gene expression and protein stability/function, respectively (13). The 18 human HDAC proteins are divided into 4 groups including the Zn+-dependent class I, IIa, IIb and IV, and the NAD+-dependent class III HDACs. HDAC1 and HDAC2 are members of class I HDACs. HDAC expression is frequently de-regulated in human cancers, and several pharmacological inhibitors of HDACs are undergoing clinical testing (14, 15). The HDAC inhibitors vorinostat and romidepsin have been approved by the US Food and Drug Administration for the treatment of refractory cutaneous T-cell lymphoma.
HDAC inhibitors sensitize cancer cells to DNA damaging therapies (e.g. irradiation and various chemotherapeutics) by altering chromatin structure and down-regulating DNA repair. HDAC inhibition reduced HR in several cell lines (16, 17). A clear synergistic effect has been demonstrated in vivo when combining HDAC inhibitors and anthracyclines (8, 18). Depletion of HDAC1 and HDAC2 by siRNA targeting also reduced HR, but had a greater effect on NHEJ (19). Furthermore, HDACs promote the stability and function of proteins involved in the DNA damage response such as Ku70 and p53 (20-22). Inhibition of class I/IIa HDACs by valproic acid attenuated the activation of the Mec1 (ataxia telangiectasia and Rad3-related (ATR) ortholog) pathway in the presence of DNA damage (23).
Clinical studies have demonstrated a benefit in some patients by adding HDAC inhibitors to therapeutic regimens that induce DNA damage (24). Epigenetic modulation is believed to play a role in therapy resistance, and these clinical trials have demonstrated responses in some patients who have previously progressed on treatment (24). The exact mechanisms governing HDAC inhibitor potentiation of DNA damage and their optimal use in the clinical setting are not yet fully understood (7, 25, 26). Therefore, we set out to investigate the role of HDACs in the response of cancer cells to chemotherapeutic induction of DNA damage.
Results from the present study demonstrate that treatment with an HDAC inhibitor caused reduced activation of ATM-mediated DNA damage signaling in various tumor cell types. ATM down-regulation via HDAC inhibition resulted in diminished DNA damage signaling and attenuated the induction of p53 response genes. The inability to initiate a robust DNA damage response was associated with increased sensitivity to DNA damaging agents and persistence of DNA damage. Select depletion of HDAC1 and HDAC2 (HDAC1/2) was sufficient to modulate ATM expression and confer sensitivity to DNA damage. Genetic depletion of ATM by siRNA mirrored the phenotypic effects of HDAC inhibition. Additionally, the results were recapitulated in vivo demonstrating an HDAC inhibitor-mediated reduction of DNA damage signaling. The relationship between ATM and HDAC1/2 supports further investigation of ATM-dependent DNA damage signaling in combination treatments including HDAC inhibitor treatment. The results suggest this HDAC inhibitor effect on DNA damage signaling may be applied to any DNA double strand break inducing therapy.
Materials and Methods
Chemicals
Entinostat (MS-275) was obtained from Selleck Chemicals LLC, epirubicin from Calbiochem (EMD Chemical), dimethyl sulfoxide (DMSO) from MP Biomedicals LLC. Vorinostat was provided by Aton Pharma Inc. All other chemicals were obtained from Sigma-Aldrich unless otherwise noted.
Cell Culture and Treatment
MCF-7, T-47D, SK-MEL-28, Saos-2, and A549 cell lines were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco’s Modified Eagle Medium high glucose (25 mM) supplemented with 10% fetal bovine serum, 4 mM L-glutamine, 100 units/ml penicillin and 100 mg/ml streptomycin in 5% CO2 at 37°C. Cell lines were authenticated by short tandem repeat profiling. For experiments, cells were treated for 48 hours with an HDAC inhibitor or vehicle (DMSO) before epirubicin (0.5 μM).
DNA Damage Detection Assay
Expression of γ-H2AX was detected using the Accuri 6 flow cytometer (BD Biosciences). Cells treated with vehicle or VPA (2mM) for 48 hours before the addition of epirubicin (0.5 μM) for 4 hours. Cells were either collected immediately, or washed to remove epirubicin, and allowed to recover for 12 hours. Collected cells were washed, fixed in 3% paraformaldehyde, permeablized (0.5% saponin, 10 mM HEPES, 0.14mM NaCl, and 2.5 mM CaCl2), and incubated with FITC-conjugated IgG or anti-phospho H2AX (Ser139) antibodies (Millipore).
Colony Forming Assay
Approximately 200 cells were seeded per well in a 12-well plate. Cells were then treated with vehicle or vorinostat (1.0 μM) and epirubicin (50 nM) for up to 14 days. Cells were fixed in methanol and stained with 2% crystal violet. Colonies measuring at least 50 cells were counted and normalized to plating efficiency (27).
Western Blotting
Protein was extracted in lysis buffer (0.1% SDS, 1% triton X-100, 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 10% glycerol), Halt protease and phosphatase inhibitor cocktail (Thermo Scientific), seprarated by SDS-PAGE, and transferred to Immobilon-P polyvinylidene fluoride microporous membranes (Milllipore). Membranes blocked in 5% nonfat milk were incubated with primary antibodies p53 and ATM (Abcam Inc.), phospho-p53 (S15), phospho-CHK2 (Thr68), and phospho-BRCA1 (S1423) (Cell Signaling Technology Inc.), p21 (Santa Cruz Biotechnology), HDAC1, acetylated-Tubulin, and phospho-ATM (S1981) (Millipore), E2F1 (BD pharmigen), and HDAC2, Acetyl-Histone H4, γ-H2AX, GAPDH (Upstate Biotechnology). Membranes were incubated with horseradish peroxidase linked secondary antibodies and visualized using the ECL Plus Western Blotting Detection System (GE Healthcare).
Immunofluorescence Microscopy
Cells were washed and fixed in 4% paraformaldehyde and permeabilized in 0.5% triton-X and blocked in 1% bovine serum albumin. Next they were incubated with primary antibodies (see above), washed, and incubated with FITC-labeled secondary antibodies. Cells were mounted in DAPI-supplemented media for imaging on the Zeiss Axio Imager 2 (Carl Zeiss MicroImaging, LLC).
Transfection
For siRNA experiments, cells were nucleofected using the Amaxa Cell Line Nucleofector kit V (Lonza Group Ltd) in buffer containing 1-2 μM of siRNA pools or Silencer negative control #2 (Ambion, Applied Biosystems). Pulsed cells were suspended in complete media without antibiotics, and experiments were performed 48 hours post-transfection. The pcDNA3.1(+) Flag-His-ATM-wt plasmid was obtained from Addgene and was originally created by the Kastan lab. Cells were nucleofected with 10ug of plasmid. The following day cells were treated with vehicle or vorinostat for 48 hours before addition of epirubicin for an additional 8 hours.
Quantitative Real-Time PCR
RNA was extracted from cells using the RNeasy kit (Qiagen). cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad Labs Inc.). Gene expression analysis was performed using TaqMan qPCR gene expression assays (MRE11, BRCA1, ATM, TP53, GADD45A, and RRM2B) on the ABI 7900 HT Thermocycler (Applied Biosystems), and normalized to β-glucuronidase (h.Gus).
In vivo tumor xenograft studies
Athymic, nude, female mice (Taconic Farms Inc.) with implanted estrogen pellets were injected with 1×107 MCF-7 breast cancer cells into the right flank. When mean tumor volumes reached 300 mm3, mice were randomized into 4 separate cohorts (n=5-6) to be treated with vehicle, vorinostat, epirubicin, or vorinostat-epirubicin. Mice were treated by intraperitoneal injection with vehicle (10% DMSO, 45% PEG400, 45% H2O) or vorinostat (150 mg/kg/day) for 2 days. On day 3 mice were treated with vehicle, vorinostat, and/or epirubicin (5 mg/kg). Tumors were harvested 8 hours after epirubicin administration and flash frozen in liquid N2. Care of animals was in accordance with institutional guidelines.
Statistics
Data are expressed as means ± S.E.M. A two-sided non-paired Student’s t test was used to determine differences between two groups with p < 0.05 considered statistically significant. For multiple groups the differences were measured by analysis of variance (ANOVA) using SPSS software.
Results
HDACs regulate p53-dependent DNA damage signaling
In the presence of DNA double strand breaks, DNA damage signaling includes p53 phosphorylation by ATM, BRCA1, and CHK2. Phosphorylation of p53 at various sites leads to enhanced protein stability, increased nuclear retention, and greater DNA binding (28). The subsequent induction of p53 response genes promotes cell cycle arrest to permit DNA repair, and their up-regulation may play a role in conferring chemotherapy resistance (29, 30). Because HDAC inhibitors sensitize tumor cells to DNA damage and may play a role in overcoming chemotherapy resistance, we investigated the effect of pharmacological HDAC inhibition on the DNA damage response.
MCF-7 breast cancer cells expressing wild-type p53 were exposed to 0.5 μM of the topoisomerase II inhibitor, epirubicin, to induce DNA double strand breaks. The damage caused by epirubicin triggered robust and time-dependent DNA damage signaling demonstrated by increased p53 Serine 15 (S15) phosphorylation and enhanced p53 protein stability (0-8 hours) (Figure 1A). In contrast, cells pre-treated with an HDAC inhibitor prior to the addition of epirubicin demonstrated a significantly altered DNA damage response. Exposure to therapeutic doses of the HDAC inhibitor vorinostat (1 μM) for 48 hours considerably reduced the activation of p53 (Figure 1A). Whereas epirubicin caused p53 activation within 4 hours of treatment, cells pre-treated with vorinostat had undetectable levels of phosphorylated p53 at the same time-point and significantly reduced p53 activation after 8 hours (Figure 1A). Additionally, vorinostat pre-treatment attenuated the stabilization of total p53 protein induced by epirubicin exposure. Reduced p53 activation was detected in cells pre-treated with vorinostat concentrations as low as 0.2 μM (Figure 1B), which is well within the range of therapeutic concentrations (31).
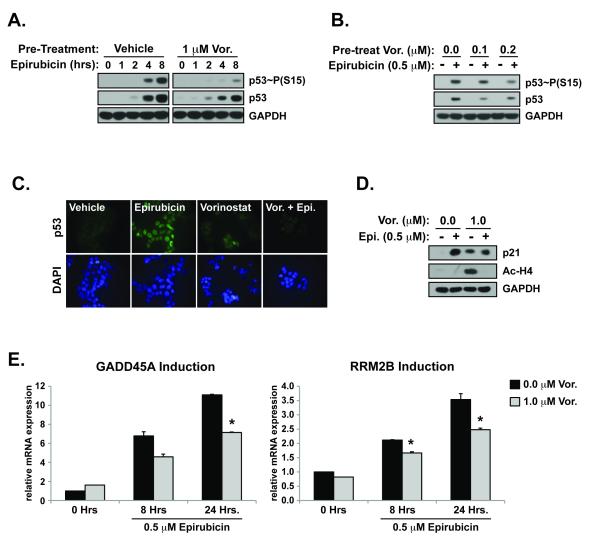
Figure 1. HDACs regulate the DNA damage response.
MCF-7 cells were treated with vehicle (DMSO) or vorinostat (Vor.) for 48 hours before adding epirubicin (0.5 μM) for an additional 0-8 hours. Total cell lysate was processed for western blotting examining the activation and stabilization of p53 due to increasing lengths of exposure to epirubicin (A.), and the concentration effect of vorinostat (B.). (C.) Cells grown on glass coverslips were treated as in (A.) before fixation, and immunofluorescence imaging of FITC-p53 and DAPI (D.). (E.) Cells treated as in (A.) were analyzed for induction of p21 and acetylation of histone H4 by western blotting. (E.) Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis for the induction of GADD45A and RRM2B normalized to β-glucuronidase (h.Gus) for total RNA in cells treated as in (A). (*p<0.05 vs. epirubicin treated)
The attenuation of p53 phosphorylation caused by HDAC inhibition in response to DNA damage was supported by immunofluorescence microscopy (Figure 1C). Nuclear accumulation of p53 significantly increased after exposure to epirubicin. Pre-treatment of cells with vorinostat, however, significantly inhibited this epirubicin-induced nuclear localization of p53 (Figure 1C). Activation and nuclear stabilization of p53 caused by DNA damage leads to induction of its response genes that contribute to cell cycle arrest and/or DNA repair. The cell cycle checkpoint regulator CDKN1A (p21) can be induced in a p53-dependent and independent manner. p21 was induced by exposure to vorinostat and to a greater extent by epirubicin (Figure 1D). Interestingly, treatment of cells with the combination resulted in lower p21 induction compared to treatment with epirubicin alone. The acetylation of histone H4 induced by vorinsotat was strongly inhibited by the addition of epirubicin (Figure 1D). This is likely due to inhibition of N-terminal lysine acetylation caused by binding of epirubicin to histone proteins (32).
The reduced activation of p53 caused by HDAC inhibition attenuated the induction of p53 response genes, GADD45A and RRM2B. Exposure to epirubicin increased the expression of GADD45A and RRM2B mRNA in a time-dependent manner (Figure 1E). Pre-treatment with vorinostat, however, partially abrogated DNA damage-induced expression of GADD45A and RRM2B (Figure 1E). This suggests an HDAC inhibitor-dependent attenuation of DNA damage signaling in the presence of genotoxic stress.
HDAC inhibitors attenuate ATM-pathway activation
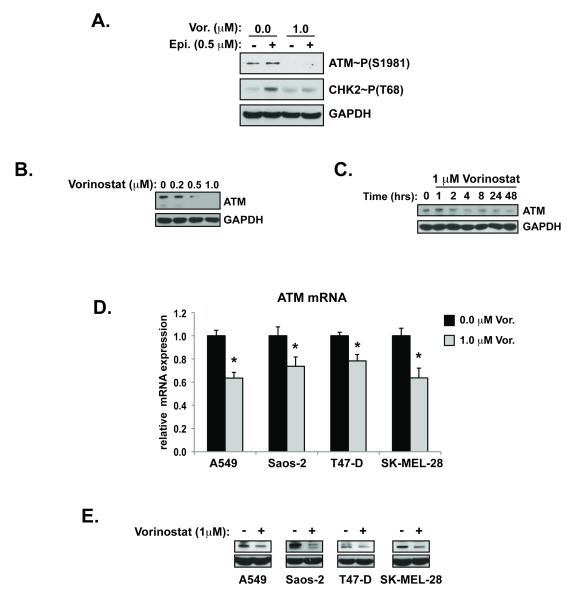
In order to identify the factors upstream of p53 that may be influenced by HDACs, we investigated the activation of ATM, BRCA1, and CHK2 in the presence of vorinostat. Pre-treatment with therapeutic doses of vorinostat reduced the activation of ATM in the presence of DNA double strand breaks (Figure 2A). Phosphorylation of BRCA1 at S1423 and CHK2 at T68 was also reduced in the presence of epirubicin after pre-treatment with an HDAC inhibitor (Supplemental Figure S1A and Figure 2A). These sites are modified by ATM in the presence of double strand breaks. ATM expression was reduced by HDAC inhibitor treatment in a dose (Figure 2B) and time dependent (Figure 2C and Supplemental Figure S1B, S1C) manner. The effect of vorinostat on ATM mRNA and protein expression was also observed in lung adenocarcinoma (A549), melanoma (SK-MEL-28), breast adenocarcinoma (T-47D) and osteosarcoma (Saos-2) cells (Figure 2D and 2E). Reduced expression of ATM in T-47D breast cancer cells also inhibited DNA damage induced activation of p53 (Supplemental Figure S1D).
Figure 2. HDAC inhibitors attenuate ATM pathway activation.
(A.) Cells pre-treated with vehicle or vorinostat were exposed to epirubicin for 8 hours before collection and analysis of whole cell lysate by western blot for ATM and CHK2 phoshporylation. (B.) MCF-7 cells treated were treated with increasing concentrations (48 hours) and time-points of vorinostat (1μM) (C.). ATM expression in cell lines treated with vehicle or vorinostat were determined by qRT-PCR for mRNA normalized to β-glucuronidase (h.Gus) (D.) and for total protein by western blot (E.). (*p<0.05 vs. 0.0 μM vorinostat)
HDAC inhibitors potentiate the effects of epirubicin and reduce DNA repair
Next we determined if the HDAC inhibitor-mediated reduction of DNA damage signaling led to increased sensitivity to DNA damage. In agreement with previous results, exposure to an HDAC inhibitor significantly enhanced the cell-killing effects of topoisomerase II inhibition (20, 24, 25). Cells pre-treated with vorinostat prior to epirubicin exhibited decreased cell survival and increased apoptosis compared to cells treated with epirubicin alone (Figure 3A-3C). Vorinostat alone had little effect on cell viability at therapeutically relevant concentrations (1 μM) (Figure 3A and 3C).
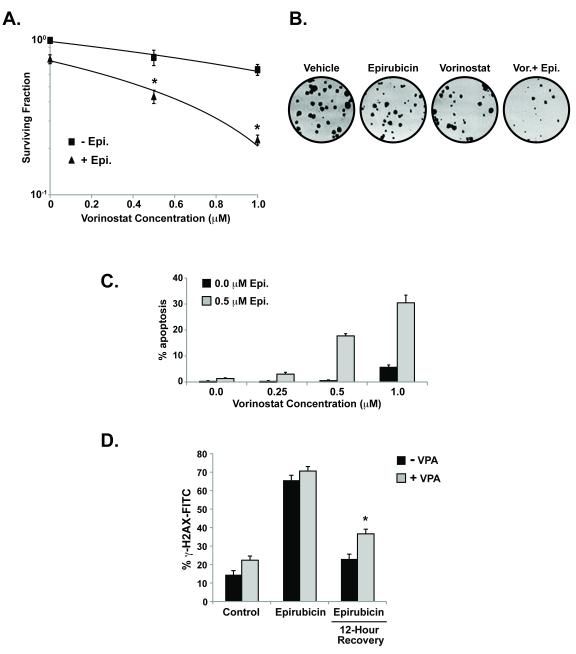
Figure 3. HDAC inhibitors potentiate the effects of epirubicin and down-regulate DNA repair.
(A) MCF-7 cells were treated with vorinostat (1 μM) and/or epirubicin (50 nM). After 10-14 days colonies (>50 cells) were counted and normalized to controls. Surviving fractions were calculated based on colony forming capacity. (B.) Images of colonies from (A.). (C.) Cells were treated with vehicle or vorinostat before the addition of epirubicin and scored by microscopy for apoptotic nuclei counting at least 100 cells per condition. (D.) Induction of γ-H2A-FITC expression as determined by flow cytometry for cells pre-treated with vehicle or valproic acid (4mM) before the addition of epirubicin (4 hours). Cells were either collected immediately and fixed, or washed to remove epirubicin to permit recovery for 12 hours at 37°. Cells were gated based on control IgG-FITC treated cells analyzing at least 0.8×105-1×105 per condition (*p<0.05 vs. epirubicin treated)
Vorinostat can itself affect chromatin stability and induce DNA damage in breast cancer cells by inhibition of HDAC3 (Supplemental Figure S2A and S2B and (33)). To separate the effects of HDAC inhibitor-mediated induction of DNA damage from its effect on DNA repair, cells were treated with valproic acid that causes only minimal DNA damage (34). With a small increase in detectable DNA strand breaks after treatment with valproic acid alone, cells treated with epirubicin (0.5 μM) for 4 hours exhibited a significant induction of γ-H2AX expression (Figure 3D). When epirubicin was removed to permit DNA repair, DNA damage persisted to a significantly greater extent in cells that were treated with valproic acid compared to vehicle treated cells (Figure 3D). Importantly, γ-H2AX expression can occur in the absence of ATM due to the overlapping function of DNA-PK (35).
HDAC1/2 regulate the DNA damage response
To identify the HDAC enzymes that regulate the expression of DNA repair genes, cells were treated with pan- or class-specific HDAC inhibitors. Breast cancer cells were exposed to DNA damage after 48 hours of HDAC inhibitor pre-treatment and evaluated for DNA damage signaling. At therapeutic doses, vorinostat acts as a pan-HDAC inhibitor targeting classes I, II, and IV (HDACs 1-11), whereas valproic acid inhibits classes I and IIa (HDACs 1-5 and HDAC8), and entinostat (MS-275) inhibits class I HDACs (HDAC1-3), as well as HDAC9 (36). Epirubicin treatment elicited robust p53 phosphorylation (Figure 4A) that was attenuated in the presence of all three HDAC inhibitors (Figure 4A). Entinostat pre-treatment was sufficient to delay epirubicin-induced p53 phosphorylation, suggesting the involvement of class I HDACs in DNA damage signaling (Figure 4A).
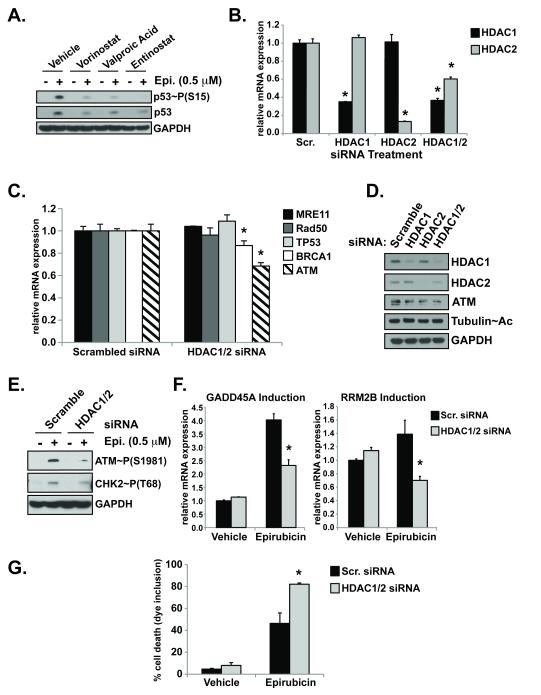
Figure 4. HDAC1/2 regulate ATM expression.
(A.) After pre-treatment with the HDAC inhibitors vorinostat (1 μM), valproic acid (1 mM), or entinostat (1 μM), cells were treated with epirubicin (Epi.) and analyzed by western blot for activation and stabilization of p53. (B.) Cells nucleofected with siRNA pools targeting HDAC1, HDAC2, both HDAC1/2 or control scrambled sequences for 48 hours were examined by qRT-PCR for gene mRNA expression normalized to h.Gus examining HDAC (B.) and DNA repair genes (*p<0.05 vs. scramble) (D.). Western blot of HDACs, ATM, and acetylated tubulin expression from cells in (B). Cells transfected as in (B.) were treated with vehicle or 0.5 μM epirubicin (8 hours) and examined for activation of ATM and CHK2 by western blot (E.) and p53 response genes induction by qRT-PCR (*p<0.05 vs. epirubicin treated) (F.). (G.) Cell death was assayed by trypan blue inclusion in cells with depleted HDAC expression and treated with epirubicin. (*p<0.05 vs. epirubicin treated)
The DNA damage response activated by DNA double strand breaks is promoted by the MRN complex (MRE11, Rad50, NBS1), ATM, BRCA1, and p53. To determine if class I HDACs affect the expression of these genes, MCF-7 cells were transfected with siRNAs targeting HDAC1 and HDAC2, alone or simultaneously due to some functional redundancy (37). When HDAC1 and HDAC2 were simultaneously depleted (HDAC1/2), there was a minor but reproducible, compensatory effect on HDAC2 compared to knock-down of HDAC2 alone (Figure 4B).
Previous studies have demonstrated reductions in MRE11 and Rad50 protein after vorinostat treatment in LNCaP and A549 cells (38). Similarly we observed decreased MRE11 and Rad50 protein in MCF-7 breast cancer cells following vorinostat treatment (Supplemental Figure S3A). Vorinostat treatment only caused a minimal decrease in MRE11 mRNA levels and had no effect on Rad50 mRNA (Supplemental Figure S3B). Select depletion of HDAC1/2 by RNAi had no effect on MRE11, Rad50, or TP53 mRNA (Figure 4C). In contrast, both ATM mRNA and protein levels were both affected by HDAC1/2 depletion (Figure 4C and 4D). ATM mRNA was decreased approximately 32% after HDAC1/2 knock-down. BRCA1 mRNA was only slightly reduced (~13%) (Figure 4C). Depletion of HDAC3, another class I HDAC, did not affect the expression of any of the DNA damage response genes examined (data not shown). The reduction of ATM mRNA by pharmacological HDAC inhibition was comparable to that detected after select depletion of HDAC1/2 by RNAi (Figure 4C and Supplemental Figure S1C).
Next we tested whether the reduced ATM expression caused by HDAC1/2 siRNA led to abrogated DNA damage signaling. In fact, cells transfected with scrambled siRNAs and exposed to epirubicin demonstrated significantly higher levels of activated ATM and CHK2 compared to those treated with HDAC1/2 siRNA pools (Figure 4E). In addition, depletion of HDAC1/2 also reduced the induction of p53 response genes GADD45A and RRM2B (Figure 4F) and increased sensitivity to DNA damage (Figure 4G).
Rescue of diminished DNA damage signaling by ATM over-expression
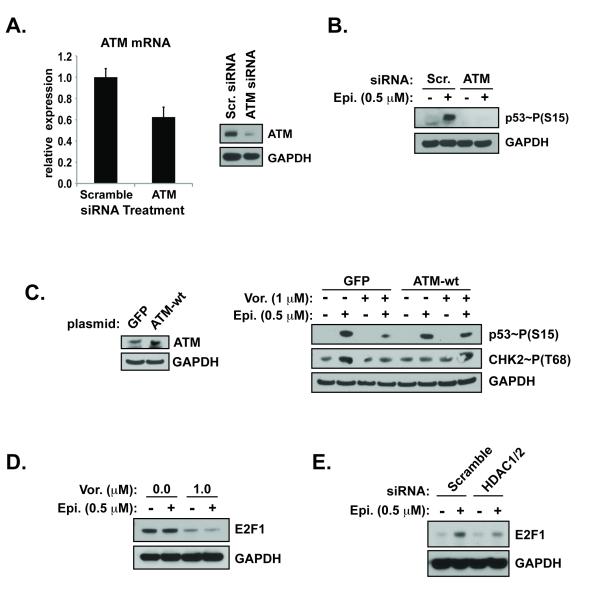
To provide mechanistic insight into the HDAC inhibitor-mediated reduction of DNA damage signaling, p53 phosphorylation was evaluated after siRNA-mediated depletion of ATM. Concentrations of siRNAs were utilized that reduced ATM to levels comparable to those detected with HDAC inhibitor treatment (Figures 5A). Induction of DNA damage by epirubicin was associated with significant phosphorylation of p53 in cells transfected with scrambled siRNA sequences, but not in those treated with siRNAs against ATM (Figure 5B).
Figure 5. Rescue of diminished DNA Damage Signaling by ATM Over-Expression.
(A.) Nucleofection of cells with siRNA pools against ATM or scrambled control sequences were examined by qRT-PCR for ATM/h.Gus expression (left) and by western blot (right). (B.) DNA damage signaling in the presence of epirubicin. (C.) Cells were transfected with plasmid DNA encoding constitutively expressed green fluorescent protein (GFP) or wild-type ATM (left). The following day cells were treated with vehicle or vorinostat before the addition of epirubicin (right). (E.) Cells treated with vorinsotat or siRNAs against HDAC1/2 were compared to those treated with vehicle or scrambled sequences, respectively, for DNA damage signaling.
To determine if the attenuated DNA damage signaling caused by HDAC inhibitors could be rescued, cells were transfected with wild-type ATM DNA whose constitutive expression was driven by the cytomegalovirus (CMV) promoter. The over-expression of ATM was sufficient to rescue the reduction in epirubicin-induced DNA damage signaling detected after pre-treatment with an HDAC inhibitor (Figure 5C). Unexpectedly, in the ATM over-expressing cells, the induction of CHK2 activation was reduced in the presence of epirubicin despite robust phosphorylation of p53. The abundant ATM levels may have increased direct activation of p53, reducing the dependency on CHK2. Loss of DNA repair gene expression caused by HDAC inhibitors is due in part to reduced E2F1 dependent transcription (16, 39). Finally, treatment with vorinostat (Figure 5D) and HDAC1/2 depletion (Figure 5E) significantly reduced the expression of the E2F1 transcription factor in the presence of damage. E2F1 promotes the expression of ATM (40). Thus the expression of ATM was necessary and sufficient to promote robust DNA damage signaling in the context of combination therapy treatment.
HDAC inhibition mitigates DNA damage response activation in vivo
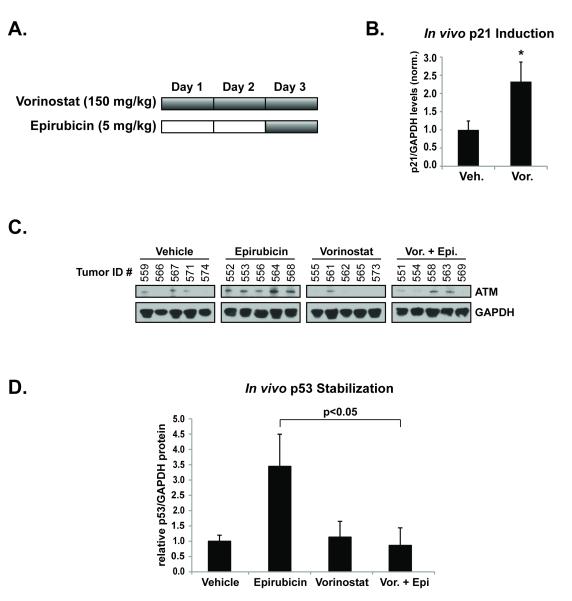
To confirm the regulation of DNA damage signaling by HDACs in vivo, tumors from an MCF-7 breast cancer xenograft model were examined for their response to epirubicin in the presence or absence of an HDAC inhibitor (Figure 6A). Tumor bearing mice were treated for 2 days with 150 mg/kg of vorinostat or vehicle. On the third day, mice were treated with vehicle, vorinostat, and/or epirubicin (5 mg/kg). Expression of p21 protein, an indicator of HDAC inhibition (41), was increased 2.3-fold in the tumors of mice treated with vorinostat, versus those treated with vehicle (Figure 6B). The level of ATM expression was lower in the group treated with vorinostat that the control treated group (Figure 6C). Vorinostat pre-treatment significantly attenuated p53 stabilization in the presence of epirubicin (Figure 6D). In fact, tumors from mice treated with the vorinostat-epirubicin combination had levels of p53 similar to those of control treated mice (Figure 6D). Vorinostat treatment alone did not increase p53 activation in vivo. These results emphasize the in vitro findings that the ATM-mediated DNA damage response is targeted by HDAC inhibition in the presence of genotoxic stress.
Figure 6. Vorinostat inhibits DNA Damage Signaling In Vivo.
(A.) Using an MCF-7 breast cancer mouse xenograft model, animals with similar mean tumor volumes were randomized and separated into groups to be treated for 2 days with vehicle or vorinostat (150 mg/kg), before treatment with vehicle, HDAC inhibitor, and/or epirubicin (5 mg/kg) on day 3. Tumors were harvested and prepared for analysis. (B.) Mean levels of p21/GAPDH protein levels in tumors from mice in the vehicle vs. vorinostat treated groups. (C.) The expression of ATM in tumors determined by western blot. (D.) Mean levels of stabilized p53 protein in tumors quantified relative to GAPDH.
Discussion
Acquired and de novo resistance of tumor cells to DNA damaging modalities (e.g. irradiation, chemotherapeutics) can be the result of alterations to DNA damage signaling and repair. Treatments targeting components of these pathways such as poly (ADP-ribose) polymerase (PARP) and the CHK1/CHK2 proteins have been explored extensively (42, 43). Pre-clinical and clinical studies have demonstrated that HDAC inhibitors potentiate the effects of DNA damage inducing therapies and may contribute to overcoming therapy resistance (20, 44). As single agents, however, HDAC inhibitors have limited therapeutic efficacy against solid tumors, despite their effects on chromatin stability and transcription. Baseline HDAC2 expression correlates with response to HDAC inhibitor-anthracycline based regimens and plays an important role in chromatin regulation (11, 31). The mechanism of potentiation by HDAC inhibitors is not yet fully understood. Furthermore, the lack of predictive markers in the clinical setting has hindered identification of optimal drug combinations and patient populations for novel treatment regimens.
Results from the present study demonstrated that inhibition of HDACs significantly reduced the initiation of DNA damage signaling following epirubicin treatment in several tumor types. We found that HDAC inhibition diminished the phosphorylation of ATM, BRCA1, CHK2 and p53 in response to DNA damage (Figure 2). The reduced activation of these proteins occurred at early time points (0-6 hours) before expected contribution of ATR (45). Pharmacological or siRNA-mediated inhibition of HDAC1/2 caused a significant deficiency in the induction of crucial p53 response genes that regulate cell cycle arrest and promote DNA repair (i.e. GADD45A and RRM2B) (Figure 4). This prevented cells from sufficiently inducing late-acting ATM-mediated DNA repair pathways in the presence of chemotherapeutically-induced genotoxic stress. RRM2B encodes p53R2, an enzyme that catalyzes the creation of deoxyribonucleoside diphosphates (dNTP precursors) required for DNA synthesis and repair (46). Consistent with other reports, down-regulation of RRM2B in MCF-7 cells coincided with reduced DNA repair and increased sensitivity to DNA damage (29).
The reduced DNA damage response caused by HDAC inhibition appeared to be HDAC6 and HDAC8-independent since class I HDAC inhibitor, entinostat (MS-275), treatment was sufficient to mitigate epirubicin-induced p53 activation in the presence of damage (Figure 4A). Furthermore, select HDAC1/2 depletion attenuated ATM activation and GADD45A and RRM2B induction. The effect on p53 response genes in these cells is likely indirect as HDAC1/2 knockdown did not significantly alter their expression in the absence of DNA damage (Figure 4F).
Although vorinostat reduced MRE11 and Rad50 protein levels, select HDAC1/2 depletion had no effect on their mRNA expression (Figure 4C). Only ATM mRNA and protein were reduced by pharmacological and siRNA-mediated inhibition of HDAC1/2 in these cells. HDAC inhibitor treatment affected BRCA1 expression, but HDAC1/2 knock-down had only a minor effect on its mRNA levels. HDAC inhibition reduced ATM levels, and potentiated the effects of DNA damaging agents. Importantly, HDAC inhibitor treatment did not have a significant impact on cell viability at the therapeutically relevant concentrations (Figure 2A and 2C). The loss of ATM was deleterious to cell survival only after induction of DNA damage. These findings are consistent with the clinical observations that HDAC inhibitor mono-therapy is not an effective treatment against solid tumors (47, 48).
Results from our group and others suggest that HDAC inhibition down-regulates DNA repair by modulating the expression of DNA repair genes (16, 25, 49). Other reports have demonstrated that HDAC inhibitors can activate the DNA damage response within 2-4 hours of treatment by causing replication and transcription-associated damage involving HDAC3 (33, 50). Prolonged pharmacologic and siRNA-mediated inhibition of HDAC1/2, however, down-regulates the expression of ATM (Figure 2C), which is consistent with reports demonstrating reduced expression of DNA repair and chromatin remodeling genes at comparable time points (8, 11, 38). This may explain the need for sequence specific administration of HDAC inhibitors prior to DNA damaging therapies in order to achieve synergistic cell-death (25). Reduced expression of DNA repair genes by HDAC inhibitors has been reported to be due to diminished recruitment of E2F1 to promoter regions (16, 39). Importantly, E2F1 also promotes the expression of ATM (40) and we demonstrate a clear reduction of E2F1 by vorinsotat and HDAC1/2 knock-down in the presence of genotoxic stress (Figure 5D and 5E).
Taken together, these results demonstrate that HDAC inhibition in vitro and in vivo attenuates ATM-dependent DNA damage signaling in response to induction of DNA strand breaks (Supplemental Figure S4). Treatment with an HDAC inhibitor targets E2F1 and HDAC1/2 reducing the expression of ATM and other DNA repair genes. Subsequent treatments inducing DNA double strand breaks then induce an insufficient DNA damage response, resulting in sustained DNA damage, and increased cell death. This suggests the need for further exploration into ATM pathway activation as a potential pharmacodynamic marker for identifying patients most likely to benefit novel therapeutic approaches combining HDAC inhibitors with DNA damage inducing modalities. Furthermore, this underlying mechanism suggests that the therapeutic addition of HDAC inhibition should not be limited to topoisomerase II inhibitors, and should be explored with other agents that induce DNA double strand breaks.
Supplementary Material
Acknowledgements
The authors would like to thank the University of California, San Francisco Genome and Laboratory for Cell Analysis Cores for their assistance, and also Stephanie Chen and Nevetha Ganesan for their help.
Financial Support: This work was supported by grants from the National Institutes of Health (R01 CA122657-01) awarded to P.N. Munster. Work performed by K.T. Thurn, S. Thomas, P. Raha, and I. Qureshi were supported by the R01 CA122657 grant.
Footnotes
Conflict of Interest: The authors have no conflicts of interests to disclose.
References
- 1.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–6. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 2.Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, Weber B, et al. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 2000;60:3299–304. [PubMed] [Google Scholar]
- 3.Kurz EU, Douglas P, Lees-Miller SP. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J Biol Chem. 2004;279:53272–81. doi: 10.1074/jbc.M406879200. [DOI] [PubMed] [Google Scholar]
- 4.Hittelman WN, Pandita TK. Possible role of chromatin alteration in the radiosensitivity of ataxia-telangiectasia. Int J Radiat Biol. 1994;66:S109–13. [PubMed] [Google Scholar]
- 5.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 6.Eot-Houllier G, Fulcrand G, Magnaghi-Jaulin L, Jaulin C. Histone deacetylase inhibitors and genomic instability. Cancer letters. 2009;274:169–76. doi: 10.1016/j.canlet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Munshi A, Kurland JF, Nishikawa T, Tanaka T, Hobbs ML, Tucker SL, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res. 2005;11:4912–22. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 8.Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005;65:3815–22. doi: 10.1158/0008-5472.CAN-04-2478. [DOI] [PubMed] [Google Scholar]
- 9.Ji P, Yeh V, Ramirez T, Murata-Hori M, Lodish HF. Histone deacetylase 2 is required for chromatin condensation and subsequent enucleation of cultured mouse fetal erythroblasts. Haematologica. 2010;95:2013–21. doi: 10.3324/haematol.2010.029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim GD, Choi YH, Dimtchev A, Jeong SJ, Dritschilo A, Jung M. Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J Biol Chem. 1999;274:31127–30. doi: 10.1074/jbc.274.44.31127. [DOI] [PubMed] [Google Scholar]
- 11.Marchion DC, Bicaku E, Turner JG, Schmitt ML, Morelli DR, Munster PN. HDAC2 regulates chromatin plasticity and enhances DNA vulnerability. Mol Cancer Ther. 2009;8:794–801. doi: 10.1158/1535-7163.MCT-08-0985. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–38. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thurn KT, Thomas S, Moore A, Munster PN. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Future Oncol. 2011;7:263–83. doi: 10.2217/fon.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer. 2004;112:26–32. doi: 10.1002/ijc.20395. [DOI] [PubMed] [Google Scholar]
- 15.Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–10. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kachhap SK, Rosmus N, Collis SJ, Kortenhorst MS, Wissing MD, Hedayati M, et al. Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS One. 2010;5:e11208. doi: 10.1371/journal.pone.0011208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adimoolam S, Sirisawad M, Chen J, Thiemann P, Ford JM, Buggy JJ. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proc Natl Acad Sci U S A. 2007;104:19482–7. doi: 10.1073/pnas.0707828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. In vivo synergy between topoisomerase II and histone deacetylase inhibitors: predictive correlates. Mol Cancer Ther. 2005;4:1993–2000. doi: 10.1158/1535-7163.MCT-05-0194. [DOI] [PubMed] [Google Scholar]
- 19.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–51. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CS, Wang YC, Yang HC, Huang PH, Kulp SK, Yang CC, et al. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007;67:5318–27. doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- 21.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell death and differentiation. 2011 doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harms KL, Chen X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res. 2007;67:3145–52. doi: 10.1158/0008-5472.CAN-06-4397. [DOI] [PubMed] [Google Scholar]
- 23.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–9. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munster P, Marchion D, Bicaku E, Schmitt M, Lee JH, DeConti R, et al. Phase I trial of histone deacetylase inhibition by valproic acid followed by the topoisomerase II inhibitor epirubicin in advanced solid tumors: a clinical and translational study. J Clin Oncol. 2007;25:1979–85. doi: 10.1200/JCO.2006.08.6165. [DOI] [PubMed] [Google Scholar]
- 25.Marchion DC, Bicaku E, Daud AI, Richon V, Sullivan DM, Munster PN. Sequence-specific potentiation of topoisomerase II inhibitors by the histone deacetylase inhibitor suberoylanilide hydroxamic acid. J Cell Biochem. 2004;92:223–37. doi: 10.1002/jcb.20045. [DOI] [PubMed] [Google Scholar]
- 26.Singh S, Le H, Shih SJ, Ho B, Vaughan AT. Suberoylanilide hydroxyamic acid modification of chromatin architecture affects DNA break formation and repair. Int J Radiat Oncol Biol Phys. 2010;76:566–73. doi: 10.1016/j.ijrobp.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 28.Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. The EMBO journal. 1999;18:7002–10. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devlin HL, Mack PC, Burich RA, Gumerlock PH, Kung HJ, Mudryj M, et al. Impairment of the DNA repair and growth arrest pathways by p53R2 silencing enhances DNA damage-induced apoptosis in a p53-dependent manner in prostate cancer cells. Molecular cancer research : MCR. 2008;6:808–18. doi: 10.1158/1541-7786.MCR-07-2027. [DOI] [PubMed] [Google Scholar]
- 30.Sato J, Kimura T, Saito T, Anazawa T, Kenjo A, Sato Y, et al. Gene expression analysis for predicting gemcitabine resistance in human cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2011;18:700–11. doi: 10.1007/s00534-011-0376-7. [DOI] [PubMed] [Google Scholar]
- 31.Munster PN, Marchion D, Thomas S, Egorin M, Minton S, Springett G, et al. Phase I trial of vorinostat and doxorubicin in solid tumours: histone deacetylase 2 expression as a predictive marker. Br J Cancer. 2009;101:1044–50. doi: 10.1038/sj.bjc.6605293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan SN, Danishuddin M, Varshney B, Lal SK, Khan AU. Inhibition of N-terminal lysines acetylation and transcription factor assembly by epirubicin induced deranged cell homeostasis. PloS one. 2012;7:e51850. doi: 10.1371/journal.pone.0051850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–80. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nature cell biology. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNAPK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 36.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–9. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 37.Jurkin J, Zupkovitz G, Lagger S, Grausenburger R, Hagelkruys A, Kenner L, et al. Distinct and redundant functions of histone deacetylases HDAC1 and HDAC2 in proliferation and tumorigenesis. Cell Cycle. 2011;10:406–12. doi: 10.4161/cc.10.3.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci U S A. 2010;107:14639–44. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez G, Liu J, Ren W, Wei W, Wang S, Lahat G, et al. Combining PCI-24781, a novel histone deacetylase inhibitor, with chemotherapy for the treatment of soft tissue sarcoma. Clin Cancer Res. 2009;15:3472–83. doi: 10.1158/1078-0432.CCR-08-2714. [DOI] [PubMed] [Google Scholar]
- 40.Berkovich E, Ginsberg D. ATM is a target for positive regulation by E2F-1. Oncogene. 2003;22:161–7. doi: 10.1038/sj.onc.1206144. [DOI] [PubMed] [Google Scholar]
- 41.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A. 2004;101:1241–6. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 43.Ashwell S, Zabludoff S. DNA damage detection and repair pathways--recent advances with inhibitors of checkpoint kinases in cancer therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:4032–7. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
- 44.Nome RV, Bratland A, Harman G, Fodstad O, Andersson Y, Ree AH. Cell cycle checkpoint signaling involved in histone deacetylase inhibition and radiation-induced cell death. Mol Cancer Ther. 2005;4:1231–8. doi: 10.1158/1535-7163.MCT-04-0304. [DOI] [PubMed] [Google Scholar]
- 45.Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–81. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura T, Takeda S, Sagiya Y, Gotoh M, Nakamura Y, Arakawa H. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nature genetics. 2003;34:440–5. doi: 10.1038/ng1212. [DOI] [PubMed] [Google Scholar]
- 47.Blumenschein GR, Jr., Kies MS, Papadimitrakopoulou VA, Lu C, Kumar AJ, Ricker JL, et al. Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest New Drugs. 2008;26:81–7. doi: 10.1007/s10637-007-9075-2. [DOI] [PubMed] [Google Scholar]
- 48.Luu TH, Morgan RJ, Leong L, Lim D, McNamara M, Portnow J, et al. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res. 2008;14:7138–42. doi: 10.1158/1078-0432.CCR-08-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Carr T, Dimtchev A, Zaer N, Dritschilo A, Jung M. Attenuated DNA damage repair by trichostatin A through BRCA1 suppression. Radiat Res. 2007;168:115–24. doi: 10.1667/RR0811.1. [DOI] [PubMed] [Google Scholar]
- 50.Lee JS. Functional link between DNA damage responses and transcriptional regulation by ATM in response to a histone deacetylase inhibitor TSA. Cancer Res Treat. 2007;39:116–24. doi: 10.4143/crt.2007.39.3.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.