Abstract
Ethanol is a modulator at the N-methyl-d-aspartate class of glutamate receptors in the brain. In animal studies the receptor adapts to sustained ethanol exposure through altered expression of the subunits that make up the receptor complex. We used real-time RT-PCR normalized to GAPDH to assay NR1, NR2A, and NR2B subunit mRNA in superior frontal and primary motor cortex tissue obtained at autopsy from chronic alcoholics with and without co-morbid cirrhosis of the liver, and from matched controls. The expression of all three subunits was significantly lower in both areas of cirrhotic alcoholics than in the corresponding areas in both controls and alcoholics without co-morbid disease, who did not differ significantly from each other. The decrease was area-dependent when cases were partitioned by the 5-HTTLPR allele. Thus, polymorphisms in one gene can have a significant effect on the expression of a second, unrelated, gene. The expression of the N-methyl-d-aspartate glutamate receptor complex is under multifactorial control.
Keywords: 5-HTTLPR, autopsy, glutamate, excitotoxicity, cerebral cortex, genotype–phenotype interactions
Ethanol has been used as a psychoactive substance for millennia; its misuse (alcohol dependence; alcoholism) is a major medical and social problem worldwide. There are an estimated 20 million alcoholics in the United States alone.1 It is a complex disease with specific neurologic effects that lead to brain damage. This manifests as generalized brain shrinkage, loss of white matter, and dendritic pruning. Loss of neurons also occurs, and is greater in the superior frontal cortex (SFC) than other cortical regions. The extent of neuronal loss is higher in subjects with co-morbid cirrhosis of the liver.2 The propensity to develop alcohol dependence is modulated by many genetic factors, and numerous polymorphisms associated with alcohol dependence have been identified. Genes include those involved in ethanol metabolism, such as alcohol and aldehyde dehydrogenases,3 and many that encode receptors and transporters for transmitters such as glutamate, γ-aminobutyrate (GABA), serotonin (5-HT), and dopamine.4-7
The N-methyl-d-aspartate class of glutamate receptors (NMDARs) is involved in several physiological and pathophysiological processes, including synaptic plasticity, ischemia, neurodegeneration, and convulsions. NMDARs also mediate, in part, the acute effects of ethanol. NMDARs are heteromeric ionotropic receptors that can be found postsynaptically to most glutamatergic neurons. The receptor consists of an essential NR1 subunit in combination with a number of NR2 subunits. Its stoichiometry has been contentious, although consensus opinion favors a tetramer of two NR1 and two NR2 subunits.8 The receptor forms an ion channel that is blocked by Mg2+. Binding of the co-agonists glutamate and glycine and removal of the Mg2+ block by plasma-membrane depolarization allows influx of K+ and Ca2+, which leads to the generation of an action potential. The influx of Ca2+ affects many Ca2+-dependent signaling pathways, including Ca2+/calmodulin-dependent kinase, protein phosphatases, proteases, and phospholipases.9
The NR1 subunit gene can give rise to eight splice variants. In contrast, four distinct genes encode NR2 subunits, yielding isoforms A–D. Differing combinations of NR1 splice variants and NR2 subunits in the receptor complex give rise to functional diversity. Chronic ethanol administration alters NMDAR subunit mRNA and protein expression in animal models.10 Up-regulation of NMDARs, changes in phosphorylation state, and variations in subunit combinations alter NMDAR function. This leads to tolerance to the effects of NMDAR modulators, including ethanol.10 Altered NMDAR function persists after ethanol intake ceases and may contribute to the symptoms of the alcohol withdrawal syndrome.10
Alcohol dependence is typical of many addictions in that (i) alcohol is consumed despite an awareness of its detrimental effects, both physiological and social, and (ii) relapse can occur even after an extended period of abstinence.11 Mesolimbic circuits utilizing monoamine neurotransmitters play a significant role in the addiction process.
Here we report the quantification of NM-DARs’ NR1, NR2A, and NR2B subunit mRNA in autopsy brain tissue from nonalcoholic, alcoholic, and cirrhotic-alcoholic subjects. We found decreases in all transcripts, and an area-dependent effect of the 5-HTTLPR genotype, in cirrhotic alcoholics. The data highlight the multifactorial nature of alcoholism.
Material and Methods
Tissue
Unfixed frozen tissue samples were obtained from the Queensland Brain Bank and the NSW Tissue Resource Centre under the auspices of the Australian Brain Bank Network. Ethical clearance was obtained from the University of Queensland Medical Research Ethics Committee (HEC #2006000901).
RNA
Total RNA was extracted from tissue using Trizol reagent (Invitrogen, Mount Waverley, Victoria, Australia) following the manufacturer’s protocol. RNA was quantified spectrophotometrically and stored at −80°C. cDNA was made from 2 μg of total RNA using oligo-dT primers and Superscript III Reverse Transcriptase® (Invitrogen) following the manufacturer’s instructions. cDNA was diluted 50-fold for real-time RT-PCR
Real-Time RT-PCR
The levels of NR1, NR2A, NR2B, and GAPDH mRNA transcripts were determined using the following primer pairs: NR1-RTF, 5′-GTCCACCAGACTGAAGATTGTGAC-3′; NR1-RTR, 5′-CTCCTCCTTGCATGTCCCA-3′; NR2A-RTF, 5′-GCTCTTCTCCATCAGCAGGG-3′; NR2A-RTR, 5′-GGATCCCGTCAGATTGAAGTCT-3′; NR2B-RTF, 5′-GGTCTTCTCCATCAGCAGAGG-3′; NR2B-RTR, 5′-TGTTGTTCATGGTTGCGGT-3′; GAPDHF, 5′-GGCATGGACTGTGGTCATGAG-3′; GAPDHR, 5′-TGCACCACCAACTGCTTAGC-3′.
Primers were used at 300 nM final concentration. 2.5 μL of diluted cDNA was used with the appropriate primers plus 12.5 μL SYBR® GREEN PCR Master Mix (Applied Biosystems, Scoresby, Victoria, Australia) in a total volume of 25 μL. Real-time RT-PCR was carried out using an ABI Prism 7000 Sequence Detection System (Applied Biosystems) with cycling parameters of initial denaturation at 95°C for 10 min, then 45 cycles of 95°C for 15 s, and 60°C for 1 min. For each sample all four transcripts were measured in duplicate. Duplicates with cycle threshold (CT) values differing by more than 0.3 standard deviations (SD) were re-run until consistency was achieved. Expression values were normalized to GAPDH and expressed as 2−ΔCT, where ΔCT is the difference between the CT values of the target transcript and GAPDH from the same tissue sample.
Genotyping
The 5-HTTLPR genotype was determined for each case using PCR.12
Data Analysis
Initial assessment by normal probability plots (Fig. 1) showed that the 2−ΔCT data distribution was skewed, and required a logarithmic transformation.13 Since CT values are in essence a log2 transform of 2−CT values—the amount of PCR product doubles every cycle—all statistical tests were performed on ΔCT data. This gave colinear normal probability plots (Fig. 1). The ΔCT values were analyzed by ANOVA using the Statistica program (Statsoft, Inc., Tulsa, OK, USA) with Newman–Keuls post hoc tests for pair-wise comparisons.13 Statistical significance was accepted at P < 0.05. Means and SEMs calculated in ΔCT units were converted back to the 2−ΔCT scale for presentation.
Figure 1.
Normal probability plots of the distribution of NMDA receptor subunit mRNA expression data expressed on a 2−ΔCT scale (upper panel) or a ΔCT scale (lower panel). All the 2−ΔCT distributions differed significantly from normal by Shapiro–Wilks tests, P < 0.01 in each instance. Open circles, NR1 mRNA in SFC; open squares, NR1 in PMC; open triangles, NR2A in SFC; open diamonds, NR2A in PMC; solid circles, NR2B in SFC; solid squares, NR2B in PMC.
Results and Discussion
Data Pre-Processing and Confounding Factors
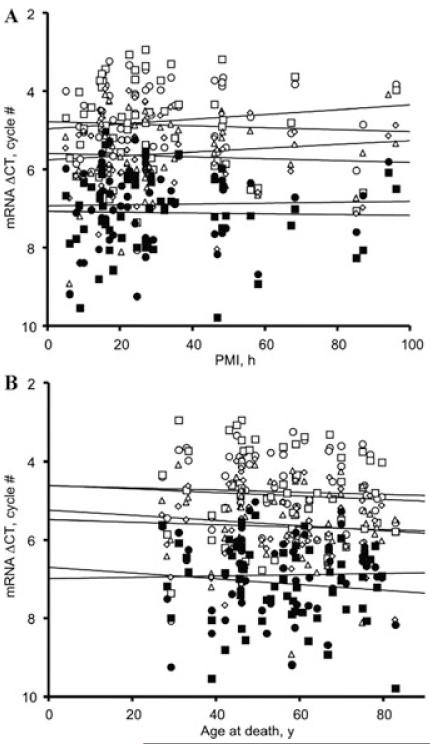
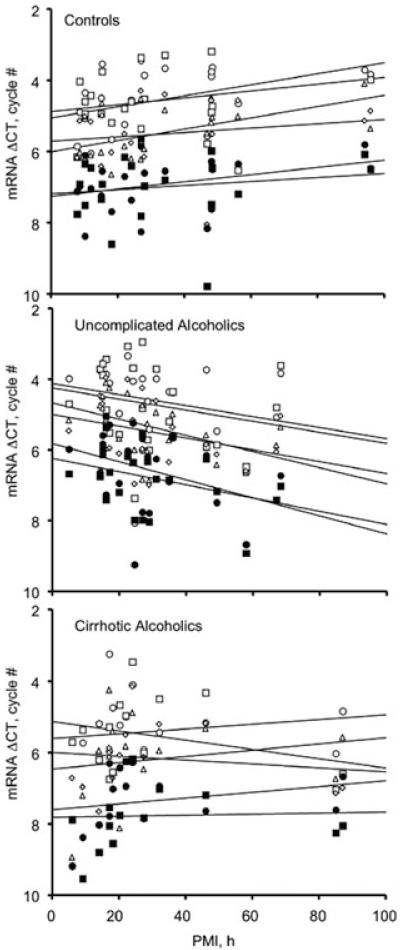
The relative levels of NR1, NR2A, and NR2B transcripts were quantified in control, alcoholic, and alcoholic-cirrhotic samples in an ‘affected’ area (SFC) and a ‘spared’ area (primary motor cortex, PMC), using GAPDH expression as a housekeeper. In a series of studies we have found that GAPDH expression does not differ between controls and alcoholics with and without co-morbid disease over a range of cortical areas, ages, and postmortem intervals, and the same was true here (not shown). Receptor transcripts and proteins are generally quite stable post mortem in human brain, although case-numbers have generally not been very high in earlier work.14 All three NMDAR subunit mRNA transcripts were strikingly stable up to at least 100 hours post mortem (Fig. 2). Transcript expression also did not vary significantly in subjects ranging between about 30 and 80 years of age (Fig. 2). Our large data set (55 cases and controls) allowed us to examine whether these regressions differed between case-groups. Although the Group × PMI interaction fell short of statistical significance, there was a significant tendency for the expression of all three transcripts, in both cortical areas, to trend in opposite senses with increasing postmortem delay in controls and alcoholics with-out co-morbid disease (Fig. 3). There was no trend in cirrhotic alcoholics (Fig. 3). To guard against possible confounds, the analyses presented below were repeated using a heterogeneous slopes model with PMI as the covariate. In all instances tested, this did not alter the key statistical indices, or the calculated within-cell mean and SEM values, to any but the most minor extent, primarily because the case groups were closely matched on PMI. The regressions on age did not differ significantly between groups (Fig. 2 [legend]).
Figure 2.
Regression of mRNA expression on postmortem interval (A) and age at death (B). The overall regressions were not statistically significant: PMI, F1,54 = 0.03, P = 0.87; age, F1,54 0.12, P = 0.073. The group × age interaction was = also not significant, F2,52 = 1.054, P = 0.36. Key as for Figure 1.
Figure 3.
Within-group regressions on PMI. The group × PMI interaction approached statistical significance, F2,52 = 2.986, P = 0.059. Note the consistent upward tend with increasing PMI in controls, and the opposite tendency in alcoholics without co-morbid disease. Although neither within-group regression considered separately reached significance (controls: F1,19 = 3.178, P = 0.091; alcoholics with-out co-morbid disease: F1,21 = 3.135, P = 0.091), the between-group comparison restricted only to these subjects did (F1,38 = 6.189, P = 0.017). There was no discernible trend in cirrhotic-alcoholic subjects (F1,12 = 0.014, P = 0.91). Key as for Figure 1.
Uncomplicated Alcoholics
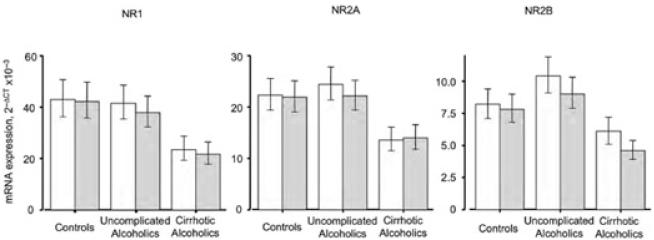
For alcoholics without co-morbid disease, NR2B mRNA expression in the SFC trended higher than that in the PMC (Fig. 4), but this regional difference only reached statistical significance in cirrhotic alcoholics (Fig. 4). A number of techniques have been used to quantify the level of NMDAR and its subunits in animal models of alcohol misuse. In mouse hippocampus, binding of the NMDAR-specific ligand MK801 is significantly increased.15 A similar increase is seen in rat forebrain.16,17 Notably, however, the increased binding in mouse brain was not accompanied by increased NR1 or NR2A subunit mRNA expression, even though the corresponding protein isoforms were more abundant.18 In electrophysiological studies in vitro there is increased NMDAR density in cultured cerebellar granule cells exposed to ethanol.19 These data suggest that NM-DAR functionality may be broadly enhanced in response to alcohol to compensate for its effects.
Figure 4.
Relative subunit mRNA expression in frontal and motor cortex. SFC: open columns; PMC: shaded columns. The main effect for group was significant, F2,56 = 4.824, P = 0.012; post hoc Newman–Keuls testing showed that overall mean subunit mRNA expression in cirrhotic alcoholics differed from that in both controls (P = 0.0087) and non-cirrhotic alcoholics (P = 0.011), who did not differ from each other. For the individual transcripts averaged across the two areas, Newman–Keuls tests showed that values in cirrhotic alcoholics differed from those in controls (NR1: P = 0.012; NR2A: P = 0.021; NR2B, P = 0.041); from those in non-cirrhotic alcoholics (NR1: P = 0.018; NR2A, P = 0.054; = NR2B, P = 0.041); and from values averaged across controls and non–co-morbid alcoholics (NR1: P = 0.0057; NR2A, P = 0.025; NR2B, P = 0.0078). In no instance did controls differ from alcoholics without co-morbid disease on these tests. For NR2B transcripts in cirrhotic alcoholics, SFC expression differed from that in the PMC, P = 0.025 by Newman–Keuls test; no other regional difference was significant for any transcript in any group. Mean ΔCT values were converted to 2−CT values for presentation; error bars represent SEM.
A general NMDAR upregulation was not reflected in the mRNA expression measured here, where only the NR2B transcript in the alcoholic SFC trended nonsignificantly higher than in control SFC. In whole-animal and cell-culture models, mRNA expression is highly dependent on the ethanol-exposure paradigm used and the brain area assayed. For example, chronic ethanol exposure led to lower NR2B mRNA expression in caudate-putamen and the hippocampal CA region in rats,20 whereas NR2B mRNA levels were unchanged under a low-dose paradigm, and significantly reduced in a high-dose paradigm, in a cell-culture model.21 The NR2B transcript showed a regional trend here, with the level in SFC higher than that in PMC. This suggests that the NR2B subunit may play a role in susceptibility to alcohol-induced neuronal loss. However, there was a general lack of regional difference in mRNA expression between alcoholics and controls.
Cirrhotic Alcoholics
All three transcripts, in both areas, showed reduced expression in cirrhotic alcoholics. This reduction was significant for mean NMDA receptor subunit mRNA expression, as well as for the three transcripts considered separately (Fig. 4). This is not consistent with increased NMDAR functionality in response to ethanol. However, liver failure in an animal model led to a significant reduction in NMDA-sensitive l-glutamate binding sites in several brain regions, including cortex.22 The mechanism involved is unresolved, but liver failure and consequent hyperammonemia alter brain glutamatergic systems.23 High concentrations of ammonia induce astrocytes to increase glutamate release, reduce expression of the transporters EAAT1 and EAAT2, and decrease glutamine synthetase activity through tyrosine nitration.23,24 The overall effect is an increase in extracellular glutamate levels, which would be expected to downregulate glutamate receptors. The lower levels of all three transcripts in both areas in cirrhotic alcoholics could arise from such a mechanism, and thereby more than counteract the increase in NMDARs in response to ethanol. It must also be noted that development of alcoholic cirrhosis may depend on drinking patterns, lifetime ethanol consumption, and beverage preference.25 Thus, the striking effects seen in cirrhotic alcoholics may be due to cirrhosis in combination with a number of other factors.
Sex Effects
The expression of NMDAR subunit mRNA was analyzed using sex as a categorical predictor (Fig. 5). The four-factor interaction showed a strong trend but Newman–Keuls post hoc testing did not reveal any significant pair-wise differences. The less-stringent Fisher’s LSD test did detect a trend toward lower NR1 levels in PMC in cirrhotic-alcoholic females than in the same area in female controls and uncomplicated alcoholics (Fig. 5). In our previous work female cirrhotic alcoholics showed higher [3H]MK801 binding capacity, but lower glutamate-mediated enhancement of [3H]MK801 binding, than other subjects, indicative of a possible subunit switch,26 although case-numbers were small. Post-translational mechanisms, such as receptor assembly, trafficking, and survival, may also play significant roles in determining receptor density and functionality.
Figure 5.
NMDA receptor subunit transcript expression in male and female subjects. Controls: open columns, n = 20; alcoholics without co-morbid disease: lightly shaded columns, n = 22; cirrhotic alcoholics: darkly shaded columns, n = 13. The transcript × area × group × sex interaction showed a trend toward differing patterns across the groups, F4,104 = 2.018, P = 0.097. No pair-wise difference was significant by Newman–Keuls post hoc test, although in female PMCS, NR1 expression in cirrhotic alcoholics showed a tendency to differ from that in both controls (P = 0.035) and combined controls plus non-co-morbid alcoholics (P 0.039) on Fisher’s LSD test.
5-HTTLPR Genotype
The cases were genotyped on a number of markers reportedly associated with human alcoholism, including the 5-HTTLPR polymorphism. 5-HT plays roles in the control of mood, appetite, and sleep, among other functions. The 5-HT transporter (5-HTT) mediates 5-HT up-take into the synaptic ending to terminate serotonergic transmission. 5-HTT is the site of action of some antidepressants, and of cocaine and amphetamines.
A functional polymorphism has been identified in the promoter of the 5-HTT gene. This contains a varying number 20–23-bp imperfect repeats, which can occur as the long (L) allele (16 repeats) or the short (S) allele (14 repeats). This polymorphism, 5-HTTLPR, affects the transcription of the 5-HTT gene: transcription levels are 3-fold higher in cells carrying the L allele.27
The 5-HTTLPR genotyping data for the subjects studied here is summarized in Table 1. The distribution of the genotypes within each group conformed to Hardy–Weinberg equilibrium. χ2 analysis showed no significant group differences, but the sample size is too small for statistical validity. Comparing all others to alcoholics, or all others to cirrhotic alcoholics, failed to reveal significant differences. The S allele is reportedly significantly associated with alcohol dependence4 and with increased alcohol consumption in social drinkers.28 However, several studies dispute this, and find no association of 5-HTTLPR with alcoholism.12,29,30 The association is further complicated by the identification of 10 additional polymorphisms within the 5-HTT promoter region,31 one of which (an A→G substitution in the L allele) gives similar expression levels to the S allele in cell culture.32
TABLE 1. Genotype and Allele Frequencies of the 5HTTLPR Polymorphism.
| Genotype Frequency |
Allele Frequency |
H–WE P value |
|||||
|---|---|---|---|---|---|---|---|
| Group | n | LL | LS | SS | L | S | |
| Control | 20 | 0.200 | 0.450 | 0.350 | 0.425 | 0.575 | 0.939 |
| Alcoholic | 22 | 0.364 | 0.455 | 0.182 | 0.591 | 0.409 | 0.961 |
| Cirrhotic alcoholic | 13 | 0.231 | 0.538 | 0.231 | 0.500 | 0.500 | 0.962 |
ANOVA on the expression data was carried out using 5-HTTLPR genotype as a categorical predictor. The analysis was simplified by comparing SS homozygotes to all others (Fig. 6). There was a significant four-factor interaction on the 2−ΔCT scale, but not on the CT scale (Fig. 4). In the SFC, the SS genotype was associated with lower NMDAR subunit expression in cirrhotic alcoholics than in controls and non-co-morbid alcoholics, most markedly for the NR1 subunit (Fig. 6). The SS genotype was associated with higher NR1 and NR2A levels in PMC in non-co-morbid alcoholics (Fig. 6).
Figure 6.
Effect of 5-HTTLPR genotype on NMDA receptor subunit mRNA expression. The genotype was simplified to a comparison of SS homozygotes with LS heterozygotes plus LL homozygotes. Although the four-factor interaction term (transcript × area × group × genotype) was significant on the 2−ΔCT scale (F4,100 = 2.599, P = 0.041), this finding must be treated with caution as it did not achieve significance on the ΔCT scale (F4,100 = 1.275, P= 0.29). Newman–Keuls testing showed there was =a difference in mean mRNA expression averaged across the three transcripts: in SFC, expression in SS cirrhotic alcoholics was significantly lower than that in combined controls plus non-co-morbid alcoholics, P = 0.023; in PMC, expression in LL LS cirrhotic alcoholics was significantly lower than that in combined controls plus non-co-morbid alcoholics, P = 0.014. For the transcripts considered separately, no pair-wise comparison reached significance on this test, but NR1 expression in SS cirrhotic alcoholics was lower by Fisher LSD test than that in SS controls (P = 0.033), SS uncomplicated alcoholics (P = 0.036), and combined SS controls plus non-co-morbid alcoholics (P = 0.019). NR1 expression in LL + LS cirrhotic alcoholics was also lower than that in LL + LS controls on this criterion (P = 0.023). Key as for + Figure 5.
The serotonergic system has been associated with alcohol consumption and dependence. Animal studies have supported its role in alcohol intake, and by inference in the development of alcohol misuse in human subjects. Specific 5-HT antagonists, serotonergic neurotoxins, and 5-HT uptake enhancers can all reduce the level of brain 5-HT and increase ethanol intake. In contrast, the direct or systemic application of 5-HT, or the application of 5-HT-releasing compounds or selective serotonin reuptake inhibitors (SSRIs), can lead to an increase in brain 5-HT and a concomitantly reduced ethanol in-take.33 Clearance of 5-HT from the brain is inhibited by ethanol via a mechanism independent of 5-HTT.34 It is possible that individuals with the SS genotype, with reduced 5-HTT activity, may have a compensatory decrease in 5-HT release to counteract this. This may in turn lead to altered ethanol consumption behavior to maintain higher levels of 5HT. These individuals may drink to maintain a constant blood alcohol level and hence higher overall NMDAR density, as in some cell-culture and animal models.
Conclusions
From the data presented here, alcohol misuse had a significant regional effect on the levels of NMDAR subunit mRNA. Liver cirrhosis modulated the levels of all subunit transcripts in both SFC and PMC. An interaction between neurotransmitter systems was shown by the influence of the 5-HTTLPR polymorphism on NMDAR subunit mRNA expression. The analysis of autopsy brain tissue is a useful way to understand the effect of chronic human alcoholism and highlights the need for an integrated approach in which the influence of co-morbid conditions and genotype are considered.
Acknowledgments
We are grateful to the neuropathologists from the Queensland Brain Bank, SMMS, University of Queensland, and from the NSW Tissue Resource Centre, for providing tissue samples from alcoholic cases and controls; and to the next of kin for providing informed written consent for the studies. The tissue banks are part of the NHMRC-supported Australian Brain Bank Network. The NSW Centre and Australian Brain Donor Program are supported by the University of Sydney, NHMRC, Schizophrenia Research Institute, NIAAA, and NSW Department of Health. Financial support was provided by the NHMRC (Australia) under Grant No. 401551.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.WHO . Global Status Report on Alcohol. Department of Mental Health and Substance Abuse, WHO; Geneva: 2004. [Google Scholar]
- 2.Charness ME. Brain lesions in alcoholics. Alcohol. Clin. Exp. Res. 1993;17:2–11. doi: 10.1111/j.1530-0277.1993.tb00718.x. [DOI] [PubMed] [Google Scholar]
- 3.Edenberg HJ, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum. Mol. Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 4.Feinn R, et al. Meta-analysis of the association of a functional serotonin transporter promoter polymorphism with alcohol dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;133:79–84. doi: 10.1002/ajmg.b.30132. [DOI] [PubMed] [Google Scholar]
- 5.Wernicke C, et al. Polymorphisms in the N-methyl-D-aspartate receptor 1 and 2B subunits are associated with alcoholism-related traits. Biol. Psychiatry. 2003;54:922–928. doi: 10.1016/s0006-3223(03)00072-6. [DOI] [PubMed] [Google Scholar]
- 6.Samochowiec J, et al. Family-based and case-control study of DRD2, DAT, 5HTT, COMT genes polymorphisms in alcohol dependence. Neurosci. Lett. 2006;410:1–5. doi: 10.1016/j.neulet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Loh E-W, et al. Association between variants at the GABAAβ2, GABAAα6 and NMDA subunit mRNA in alcoholics GABAAγ2 gene cluster and alcohol dependence in a Scottish population. Mol. Psychiatry. 1999;4:539–544. doi: 10.1038/sj.mp.4000554. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa H, et al. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 9.Lerea LS, et al. N-methyl-D-aspartate receptors activate transcription of c-fos and NGFI-A by distinct phospholipase A2-requiring intracellular signaling pathways. Mol. Pharmacol. 1995;47:1119–1125. [PubMed] [Google Scholar]
- 10.Dodd PR, et al. Glutamate-mediated transmission, alcohol, and alcoholism. Neurochem. Int. 2000;37:509–533. doi: 10.1016/s0197-0186(00)00061-9. [DOI] [PubMed] [Google Scholar]
- 11.Schaffer A, Naranjo CA. Recommended drug treatment strategies for the alcoholic patient. Drugs. 1998;56:571–585. doi: 10.2165/00003495-199856040-00005. [DOI] [PubMed] [Google Scholar]
- 12.Foley PF, et al. Association studies of neurotransmitter gene polymorphisms in alcoholic Caucasians. Ann. N.Y. Acad. Sci. 2004;1025:39–46. doi: 10.1196/annals.1316.005. [DOI] [PubMed] [Google Scholar]
- 13.Winer BJ, et al. Statistical Principles in Experimental Design. McGraw-Hill; Sydney, NSW: 1991. [Google Scholar]
- 14.Dodd PR, Lewohl JM. Cell death mediated by amino acid transmitter receptors in human alcoholic brain damage: conflicts in the evidence. Ann. N.Y. Acad. Sci. 1998;844:50–58. [PubMed] [Google Scholar]
- 15.Snell LD, et al. Radioligand binding to the N-methyl-D-aspartate receptor/ionophore complex: alterations by ethanol in vitro and by chronic in vivo ethanol ingestion. Brain Res. 1993;602:91–98. doi: 10.1016/0006-8993(93)90246-j. [DOI] [PubMed] [Google Scholar]
- 16.Gulya K, et al. Brain regional specificity and time-course of changes in the NMDA receptor-ionophore complex during ethanol withdrawal. Brain Res. 1991;547:129–134. [PubMed] [Google Scholar]
- 17.Haugbol SR, et al. Upregulation of glutamate receptor subtypes during alcohol withdrawal in rats. Alcohol Alcohol. 2005;40:89–95. doi: 10.1093/alcalc/agh117. [DOI] [PubMed] [Google Scholar]
- 18.Snell LD, et al. Regional and subunit specific changes in NMDA receptor mRNA and immunore-activity in mouse brain following chronic ethanol ingestion. Brain Res. Mol. 1996;40:71–78. doi: 10.1016/0169-328x(96)00038-1. [DOI] [PubMed] [Google Scholar]
- 19.Iorio KR, et al. Chronic exposure of cerebellar granule cells to ethanol results in increased N-methyl-D-aspartate receptor function. Mol. Pharmacol. 1992;41:1142–1148. [PubMed] [Google Scholar]
- 20.Darstein MB, et al. Changes in NMDA receptor subunit gene expression in the rat brain following withdrawal from forced long-term ethanol intake. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2000;361:206–213. doi: 10.1007/s002109900180. [DOI] [PubMed] [Google Scholar]
- 21.Kalev-Zylinska ML, During MJ. Paradoxical facilitatory effect of low-dose alcohol consumption on memory mediated by NMDA receptors. J. Neurosci. 2007;27:10456–10467. doi: 10.1523/JNEUROSCI.2789-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson C, et al. Selective loss of N-methyl-D-aspartate-sensitive L-[3H]glutamate binding sites in rat brain following portacaval anastomosis. J. Neurochem. 1990;55:386–390. doi: 10.1111/j.1471-4159.1990.tb04149.x. [DOI] [PubMed] [Google Scholar]
- 23.Vaquero J, Butterworth RF. The brain glutamate system in liver failure. J. Neurochem. 2006;98:661–669. doi: 10.1111/j.1471-4159.2006.03918.x. [DOI] [PubMed] [Google Scholar]
- 24.Chan H, Butterworth RF. Cell-selective effects of ammonia on glutamate transporter and receptor function in the mammalian brain. Neurochem. Int. 2003;43:525–532. doi: 10.1016/s0197-0186(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 25.Mann RE, et al. The epidemiology of alcoholic liver disease. Alcohol Res. Health. 2003;27:209–219. [PMC free article] [PubMed] [Google Scholar]
- 26.Dodd PR, et al. Genes and gene expression in the brains of human alcoholics. Ann. N.Y. Acad. Sci. 2006;1074:104–115. doi: 10.1196/annals.1369.010. [DOI] [PubMed] [Google Scholar]
- 27.Heils A, et al. Allelic variation of human serotonin transporter gene expression. J. Neurochem. 1996;66:2261–2264. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 28.Munafo MR, et al. Association between the serotonin transporter gene and alcohol consumption in social drinkers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;135:10–14. doi: 10.1002/ajmg.b.30162. [DOI] [PubMed] [Google Scholar]
- 29.Dick DM, et al. Association analyses of the serotonin transporter gene with lifetime depression and alcohol dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Spy chiatr. Genet. 2007;17:35–38. doi: 10.1097/YPG.0b013e328011188b. [DOI] [PubMed] [Google Scholar]
- 30.Kohnke MD, et al. The serotonin transporter promotor polymorphism 5-HTTLPR is not associated with alcoholism or severe forms of alcohol withdrawal in a German sample. Psychiatr. Genet. 2006;16:227–228. doi: 10.1097/01.ypg.0000218629.11375.b0. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura M, et al. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol. Psychiatry. 2000;5:32–38. doi: 10.1038/sj.mp.4000698. [DOI] [PubMed] [Google Scholar]
- 32.Hu XZ, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am. J. Hum. Genet. 2006;78:815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeMarquand D, et al. Serotonin and alcohol intake, abuse, and dependence: findings of animal studies. Biol. Psychiatry. 1994;36:395–421. doi: 10.1016/0006-3223(94)91215-7. [DOI] [PubMed] [Google Scholar]
- 34.Daws LC, et al. Ethanol inhibits clearance of brain serotonin by a serotonin transporter-independent mechanism. J. Neurosci. 2006;26:6431–6438. doi: 10.1523/JNEUROSCI.4050-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]