Abstract
DNA methylation is a defining feature of mammalian cellular identity and essential for normal development1,2. Most cell types, except germ cells and pre-implantation embryos3–5, display relatively stable DNA methylation patterns with 70–80% of all CpGs being methylated6. Despite recent advances we still have a too limited understanding of when, where and how many CpGs participate in genomic regulation. Here we report the in depth analysis of 42 whole genome bisulfite sequencing (WGBS) data sets across 30 diverse human cell and tissue types. We observe dynamic regulation for only 21.8% of autosomal CpGs within a normal developmental context, a majority of which are distal to transcription start sites. These dynamic CpGs co-localize with gene regulatory elements, particularly enhancers and transcription factor binding sites (TFBS), which allow identification of key lineage specific regulators. In addition, differentially methylated regions (DMRs) often harbor SNPs associated with cell type related diseases as determined by GWAS. The results also highlight the general inefficiency of WGBS as 70–80% of the sequencing reads across these data sets provided little or no relevant information regarding CpG methylation. To further demonstrate the utility of our DMR set, we use it to classify unknown samples and identify representative signature regions that recapitulate major DNA methylation dynamics. In summary, although in theory every CpG can change its methylation state, our results suggest that only a fraction does so as part of coordinated regulatory programs. Therefore our selected DMRs can serve as a starting point to help guide novel, more effective reduced representation approaches to capture the most informative fraction of CpGs as well as further pinpoint putative regulatory elements.
Changes in DNA methylation (DNAme) patterns and the resulting differentially methylated regions (DMR) have been the focus of numerous studies in the context of normal development7 and disease8. These studies have characterized many different DMR classes including partially methylated domains (PMDs)9, condition specific10, cell type specific9,11–13 and tissue specific DMRs14,15 (tDMRs) as well as DMRs arising in disease such as cancer15,16. Due to the relatively small fraction of genomic CpGs assayed or small sample cohorts, the question of what fraction of genomic CpGs changes its methylation state in the context of normal development as well as their regulatory context remains underexplored.
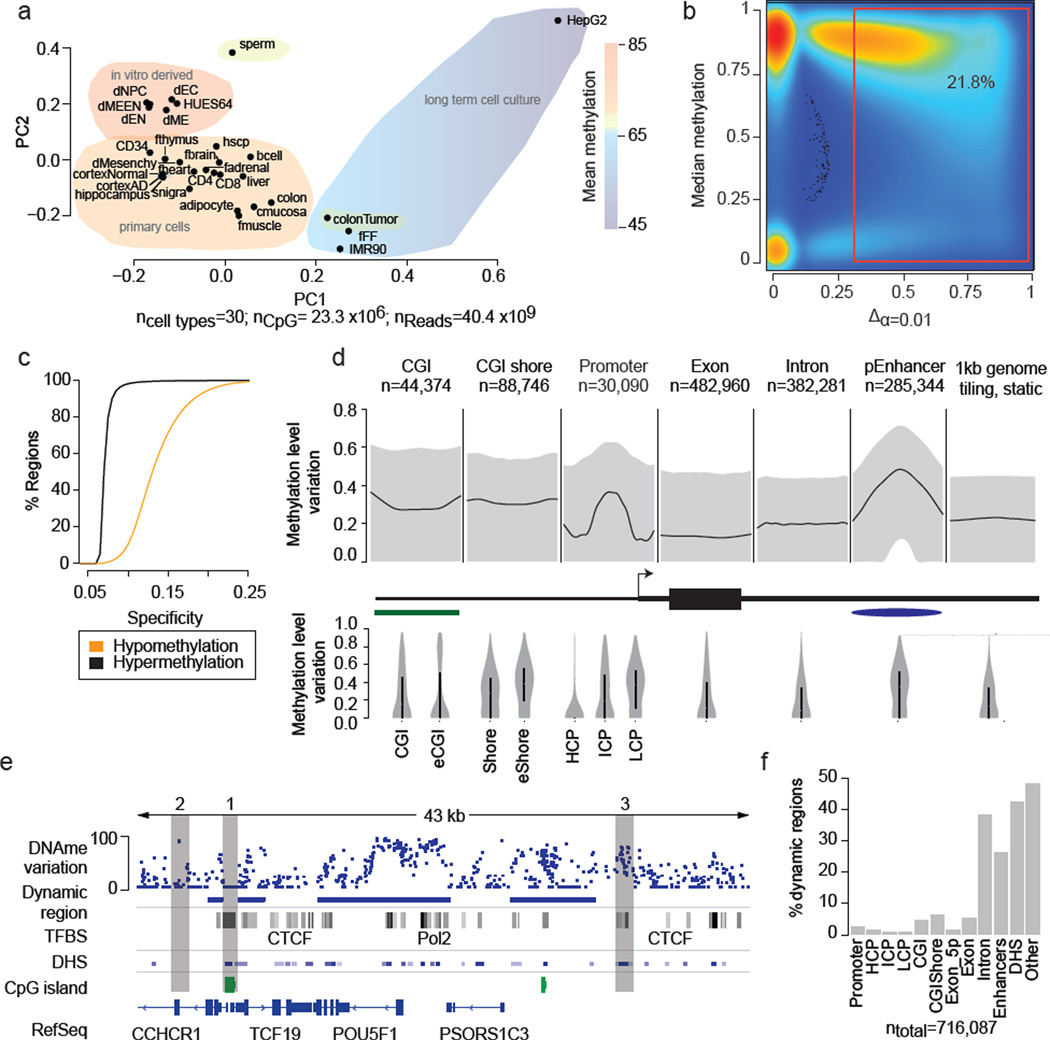
In this study, we systematically investigated the DNAme state of most human autosomal CpGs to determine those that show dynamic changes and hence may participate in genome regulation in a developmental context (dynamic CpGs). In total, we included 42 WGBS datasets comprising a range of human cell and tissue types (n=30). The combined 40.4 billion reads enabled us to assay 25.71 million autosomal CpGs (≥5× coverage in at least ≥50% of all samples; 96% of all hg19 autosomal CpGs) (Supplementary Table 1). We organized the samples into four classes; comprising human embryonic stem cells (hESCs), hESC derived cell populations, normal somatic tissues as well as disease conditions (Fig. 1a, Supplementary Table 1). On a global scale, hESC and their derivatives exhibit the highest DNAme levels, followed by primary tissues (~5% less), which is in sharp contrast to the global hypomethylation in colon cancer (~10–15% less) and long-term cultured cell lines (10–30% less).
Figure 1. Identification and characteristics of differentially methylated regions (DMRs) in the human genome.
a. Principal component analysis based on CpG methylation levels for 1kb tiles across 30 diverse human cell and tissue samples. Coloring indicates classification of samples into subgroups and group wise mean DNAme. Detailed sample annotations are listed in Supplementary Table 1. Gray area indicates Alzheimer’s disease (AD) samples..
b. Density scatterplot of CpG wise DNAme level differences (x-axis, p≤0.01) and CpG median methylation (y-axis) across the 24 developmental samples (excluding cancer and long-term culture). Coloring indicates CpG density from low (blue) to high (red). The red box highlights dynamic CpGs (≥0.3).
c. Cumulative distribution of DMR specificity. High hypo/hypermethylation specificity indicates that particular region is methylated/unmethylated in most tissues and deviates from this default state in only one or few cases.
d. Top: Composite plot of mean DNAme differences across various genomic features. Black line indicates the median of the average DNAme difference across each feature. Grey areas mark 25th and 75th percentile. Bottom: Distribution of mean DNAme difference for each genomic feature. Black bar indicates 25th and 75th percentile while white dot marks the median. For CGI islands, a smaller, experimentally determined set (eCGI; n=25,490) is shown as well. Promoters are broken down into high CpG content (HCP, n=24,899), intermediate CpG content (ICP, n=10,920) and low CpG content (LCP, n=7,946) regions (n=43,765 total).
e. Methylation level variation across the OCT4 locus (chr6:31,119,000–31,162,000) (top). Blue boxes indicate DMRs significant at p≤0.01 and exhibit a minimum difference ≥0.3 across the 24 developmental samples. For reference, ENCODE TFBS cluster track, DNAse I hypersensitive sites, CpG islands and RefSeq genes are shown.
f. Distribution of DMRs across various genomic features. Each region is assigned only to one of these genomic feature according the ranking promoter, CGI, CGI shore, exon, intron, putative enhancers, DNAse I hypersensitive site or other.
Focusing initially on our developmental sample set (n=24 total, hESCs, in vitro derived cell types and somatic tissues, Supplementary Table 1) we identified ~5.6 million dynamic CpGs (minimum methylation difference ≥0.3, FDR=10.4%, 21.8% of captured autosomal CpGs; Fig. 1b, Supplementary Fig. 1e, see Supplementary Information) distributed across 716,087 discrete differentially methylated genomic regions (DMRs, 19.2% of the mappable human genome). In addition to this moderately stringent cutoff, we also tested thresholds as low as 10% methylation difference that may account for DNAme changes arising from relevant small subpopulations in heterogeneous tissue samples or noise, but still only find 10.4 million CpGs to be dynamic.
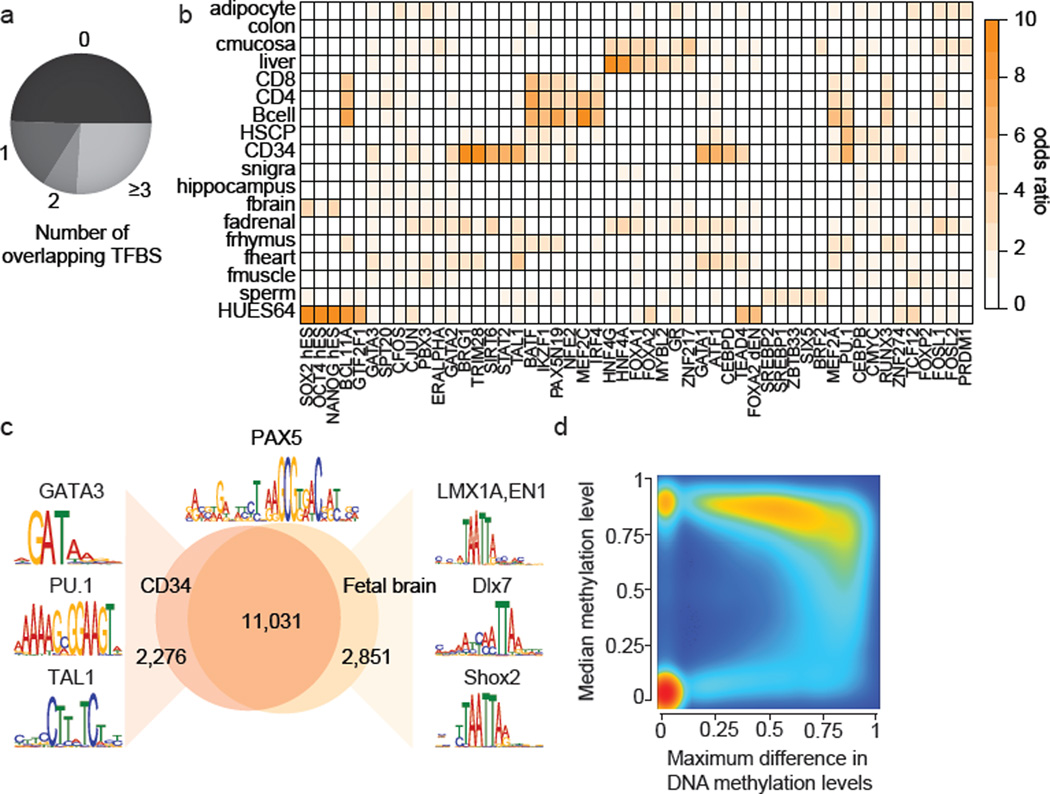
Focusing on the more stringent set (≥0.3 difference), we find approximately 70% are on average highly methylated (>75% methylation ratio) while less than 2% are on average unmethylated (<10% methylation ratio) (Supplementary Fig. 1h). In line with this observation, we find that hypomethylation of DMRs shows greater sample specificity than hypermethylation (Fig. 1c). Interestingly, most of the DMRs are small (>75% are smaller than 1kb, Supplementary Fig. 1i) and located distal to transcription start sites (TSS) (Supplementary Fig. 1j). However, the average variation in DNAme levels across all RefSeq promoters (n=30,090) does still exhibit a clear increase specifically at the TSS with most of this variation occurring at intermediate and low CpG density promoters (Fig. 1d). For CpG islands in general, we observe distinct dynamic regimes, highlighting that different classes of CpG islands are likely subject to different modes of regulation12,17,18 (Fig. 1d bottom). Consistent with previous reports15, we find CpG island shores to be among the most variable genomic regions (Supplementary Fig. 1o). These observations are exemplified at the OCT4 (POU5F1) locus, where the promoter and large parts of the gene body exhibit high DNAme dynamics, while the strong downstream CpG island as well as the surrounding CTCF binding sites remain static (Fig. 1e). Only 12.2% of our DMR set overlap with at least one of 568,430 annotated classic, gene centric genomic features (promoter, exon, CGI, CGI-shore) (Fig. 1f). To gain insights into the role of the remaining set, we first investigated their co-localization with DNase I HS sites across 92 distinct cell types19 as well as a catalog of putative enhancer elements for 31 cell and tissue types20. Strikingly, we found that 42.3% of our DMRs overlap with at least one DNAse I HS site (Fig. 1f) and 26.1% co-localize with enhancer like regions, which cover more than 50% of all H3K27ac regions in our catalog (n=285,344) and represents one of the most differentially methylated features (Fig. 1d). Next, we examined DMR overlap with transcription factor binding site (TFBS) clusters determined by the ENCODE project21 and uncovered a highly significant overlap of the two feature classes (jaccard=0.11, p-value<=0.1). Taking into account an even broader set of TFBS comprising 165 factors,, we find that more than 50% of all DMRs overlap with at least one and 25% with more than 3 TFBS accounting for an additional 13.0% of DMRs (Fig. 2a). Consistent with this we find dramatically increased variation in DNAme levels specifically across TFBS (Supplementary Fig. 2a). In summary, we were able to readily attribute 64.2% of all DMRs to at least one putative gene regulatory element or coding sequence (Supplementary Fig. 1e–h) suggesting they demarcate various classes of putative regulatory elements.
Figure 2. Dynamic CpG methylation regions frequently co-localize with transcription factor binding sites (TFBS).
a. Overlap of DMRs with ENCODE TFBS.
b. Enrichment of the top four TFBSs significantly overrepresented (p<0.01, empirical test) in DMRs specific to the cell type indicated (specificity >0.15). Color code quantifies median enrichment odds ratio compared to size matched random control regions.
c. Overlap of PAX5 motifs (±100bp top) unmethylated in CD34 cells or fetal brain across the entire human genome. Regions specifically unmethylated in CD34 or fetal brain were subjected to motif analysis and top differentially co-occurring motifs are highlighted on the left for CD34 and on the right for fetal brain.
d. Density scatterplot of maximum DNAme difference across 24 developmental samples for TFBS cluster track (n=2.7 million) and median methylation level across all samples. Color code indicates density of TFBS from low (blue) to high (red)..
We determined all cell type specific hypomethylated regions (n=430,250, see Supplementary Information) and investigated the enrichment for 161 ENCODE factors (excluding MBD4, SETDB1, POL2P, HDAC2 from the prior set). Strikingly, we observe significant enrichment of cell type specific TFs that are known to be involved in the regulation of the respective cellular states (Fig. 2b). For instance, the top three factors bound in HUES64 specific DMRs are OCT4, SOX2, and NANOG (Fig. 2b). Similarly, PU.1 and TAL1 are highly enriched in CD34 cells and HNF factors in adult liver (Fig. 2b). In further support of this, motif enrichment analysis revealed many more interesting cell-type-TF-associations such as enrichment of distinct NKX factors in fetal heart and fetal brain as well as ESRRG in fetal adrenal cells (Supplementary Fig. 2b, Supplementary Table 3). Moreover, we tested whether the DMR set can be used to gain insights into the combinatorial control of cellular states by TFs. To that end, we determined all unmethylated (<10% methylation) PAX5 motif instances (±100bp) across the human genome in CD34 or fetal brain cells (Fig. 2c). While, both footprint sets show a large overlap (11,031 sites), regions exclusively unmethylated in CD34 or fetal brain are enriched for distinct sets of other known lineage specific TF motifs; such as PU.1 in CD34 and LMX1A or EN1 in fetal brain (Fig. 2c). Taken together, these findings highlight that cell type specific DNAme patterns can be used to detect footprints and infer potentially regulatory TFs. In fact, more than 60% of all ENCODE TFBS are hypermethylated in most samples, but become hypomethylated very specifically in only one or two cell types (Fig. 2d), while 25% are constitutively unmethylated and never change (Fig. 2d).
Breaking down this distribution of TFBS reveals distinct patterns of variation for different types of TFs (Supplementary Fig. 2e). More generally, we find that DNAme variation across TFBS is strongly correlated with its median methylation level and therefore the (hypo-) methylation specificity (Supplementary Fig. 2c), as well as the TFs specificity of expression22 (Supplementary Fig. 2d). These observations support the notion23 that selective TF binding creates spatially highly constrained hypomethylated regions and confers cell type specificity.
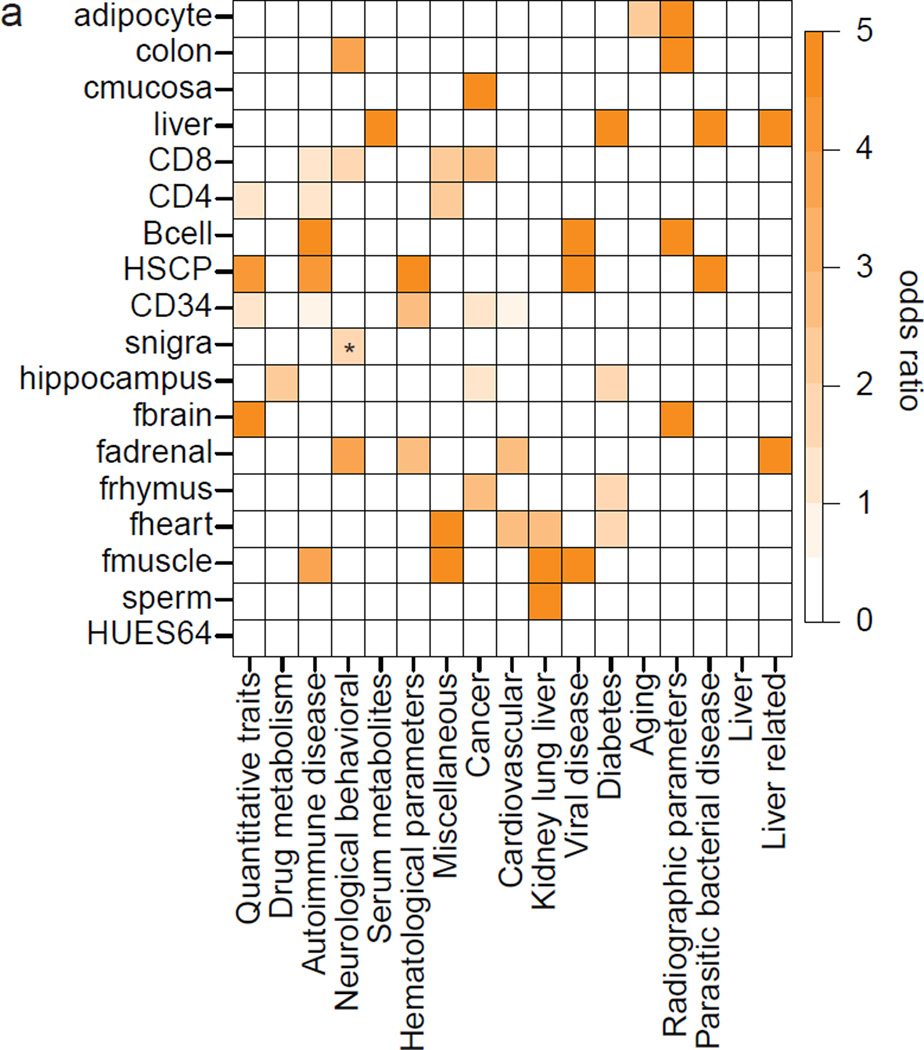
Based on these findings and previous reports24 we asked whether DMRs are more susceptible to point mutations that are functionally consequential. Even with strict filtering criteria, we found a significant enrichment of SNPs in DMRs compared to genomic background as well as different sets of random control regions (odds ratio 1.06, p-value<10−16, binomial test, Supplementary Information). We then determined the overlap of DMRs with recently evolved human specific CpGs, termed CpG beacons25, which shows a striking enrichment (odds ratio 1.37–1.6 compared to genomic background and random control regions, p-value<10−16). This suggests overall higher genetic intra-species variability specifically at regions that change their DNAme state. In concordance with the increased SNP frequency, DMRs are also significantly enriched for GWAS SNPs from the GWAS catalog26 (odds ratio 1.16, p-value=3.27×10−10, binomial test). Similar to our observations on TFBS, GWAS SNPs exhibit a non-random enrichment distribution across cell type specific DMRs (Fig. 3a). For instance, we find DMRs specific to adult liver to be enriched for liver and serum metabolite related GWAS SNPs, fetal heart for cardiovascular and many of our blood cell types for autoimmune diseases and hematological parameters.
Figure 3. DMRs exhibit elevated SNP frequency and show non-random GWAS SNP enrichment.
a. Odds ratio of significantly overrepresented (p<0.05, empirical test, see Supplementary Information) GWAS SNPs grouped into 16 categories in regions specifically hypomethylated within the sample indicated on the left. Asterisk indicates p-value <0.1.
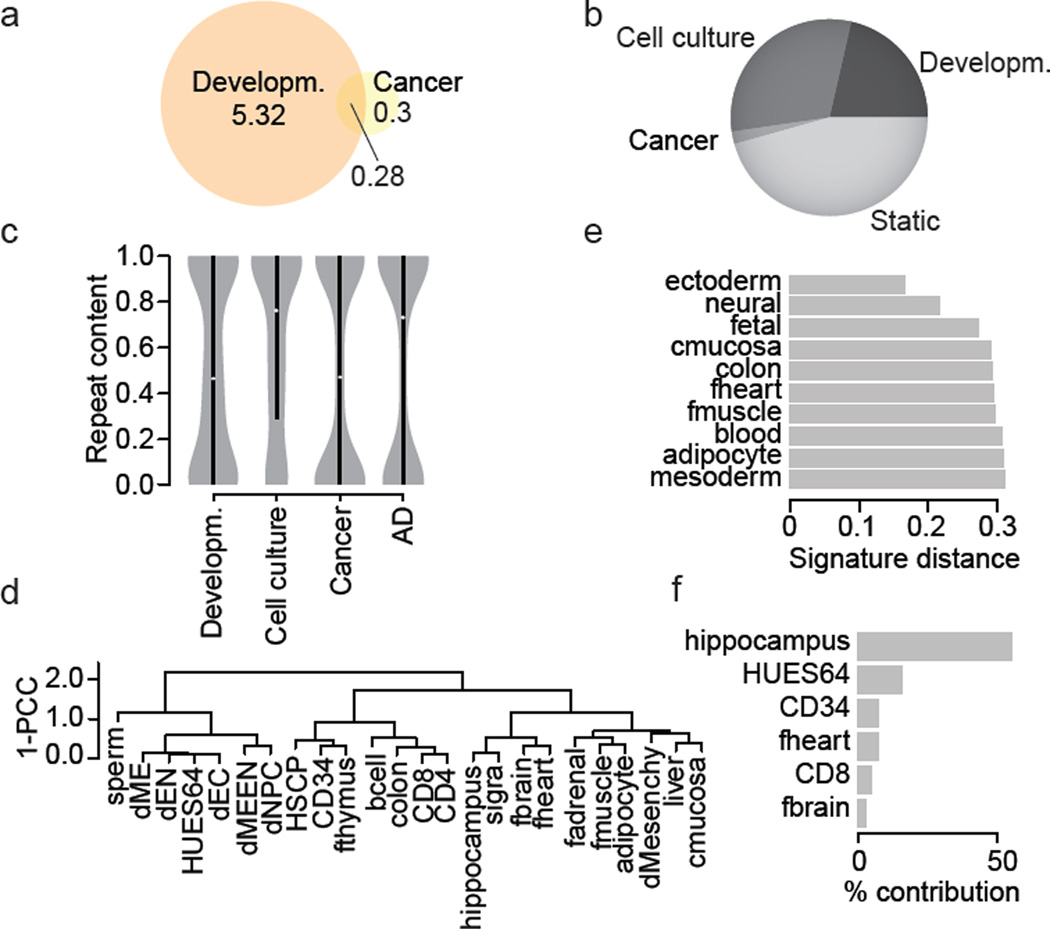
It is well known that many cancers exhibit dramatic DNAme changes27, we therefore compared a colon cancer to a matched control and found 532,665 differentially methylated CpGs. 40% of these overlapped with the previously identified developmental dynamic set (Fig. 4a). Similarly, 36% of differentially methylated CpGs found in Alzheimer Disease (AD) samples compared to normal controls (n=12,408) overlapped with our previous set of developmental CpGs. The most dramatic change in the number of dynamic CpGs occurs when comparing our developmental sample cohort to the long-term cell culture cohort, leading to the identification of 8.4 million additional dynamic CpGs (Fig. 4b). Importantly, this expanded set differs notably in terms of their sequence features, with cancer and AD dynamic CpGs residing in less conserved regions that also exhibit lower motif complexity compared to the developmental and cell culture (Supplementary Fig 4a,b). The cell culture specific CpGs exhibit elevated repeat content relative to developmental CpGs, a feature that is shared with AD (Fig. 4c). While the disease samples clearly add more dynamic CpGs, our analysis suggests a notable overlap with our prior set for CpGs that may participate in actual regulatory events.
Figure 4. Effective classification and sample deconvolution using only the DMR set.
a. Overlap of dynamic CpGs (p≤0.01 Δ≥0.3) in normal samples and between colon cancer and matching control CpG numbers (in million).
b. Distribution of autosomal CpGs across three conditions. Class name indicates sample group where a CpG was observed dynamic (developmental (n=24), cell culture (n=3), cancer (n=2)) or remained unchanged over the entire sample set (n=30).
c. Repeat content distribution of DMRs (sets as in b).
d. Hierarchical clustering using pearson correlation coefficient (PCC) of the DMR values across the entire sample set (n=30).
e. Distance of the fetal brain sample to different sets of signature regions defined for sample classes or individual samples, but excluding regions identified by means of the fetal brain sample.
f. Contribution of individual sample signature region sets to an in silico generated hybrid sample (HUES64 and hippocampus).
Finally, we investigated the utility and power of the reduced region set to accurately classify unknown samples or help deconvolute a mixture of samples. We first clustered our developmental sample set based on the DMRs only (Fig. 4d) and found the result to be in excellent agreement with genome-wide 1kb tiling based clustering (Supplementary Fig. 5a). To probe the potential of our DMR set to accurately classify unknown samples, we derived signature region sets for different sample groups. These signature regions turned out to be excellent classifiers of an unseen sample (fetal brain, Fig. 4e). Next, we tested as a proof of principle whether it is possible to utilize our DMR set to infer the different cell populations present within a heterogeneous sample. To that end, we deconvoluted an in silico mixture of HUES64 and hippocampus WGBS libraries using our DNAme signatures. Notably, the two top hits after application of a very simple deconvolution algorithm indeed proved to be hippocampus and HUES64 (Fig. 4f).
Our study highlights and defines a relatively small subset of all genomic CpGs that change their DNA methylation state across a large number of representative cell types. Although we expect that number to somewhat increase with more diverse cell types as more WGBS data sets becoming available, our analysis suggests that the rate of newly discovered regulatory CpGs will drop rapidly once all major cell and tissue types have been mapped, mostly owed to the fact that between tissue variability exceeds within tissue variability by one order of magnitude (Supplementary Fig. 3a,b). Future studies are likely to fine map dynamics occurring in more specific subpopulations, giving rise to smaller changes in DNAme that we were unable to detect or include because of power constraints. Extreme conditions in vitro or in vivo such as loss or misregulation of DNMT1 may affect a larger subset including many intergenic CpGs that are generally static, but most of these additional CpGs are unlikely to overlap with functional elements such as TFBS or enhancers. In combination with the fact that sequencing of WGBS libraries is very inefficient, as about 65% of all 101bp reads in our set did not even contain any CpGs to begin with, this amounts to an approximate, combined loss of around 80% of sequencing depth on non-informative reads and static regions. Finally, once defined, it will likely be sufficient in most cases to profile only a representative subset of CpGs across a comprehensive set of DMRs using an array28 or hybrid capture29 based technology to recover representative dynamics and measure regulatory events. Using these results as a guiding principle, we expect further improved efficiencies in mapping DNAme and enhance its applicability as a marker for various regulatory dynamics in normal and disease phenotypes.
Methods Summary
Biological materials and sequencing libraries
Genomic DNA was fragmented to 100–500 bp using a Covaris S2 sonicator. DNA fragments were cleaned-up, end-repaired, A-tailed, and ligated with methylated paired-end adapters (purchased from ATDBio). See Supplementary Information for details.
Data processing and analysis
In house generated WGBS libraries were aligned using MAQ30 in bisulfite mode to the hg19/GRCh37 reference assembly. Subsequently, CpG methylation calls were made using custom software, excluding duplicate, low quality reads as well as reads with more than 10% mismatches. Methylation ratios of individual CpGs were modeled using a beta-binomial model estimating parameters from the number of methylated and total reads overlapping a given CpG, incorporating replicates. Only CpGs covered by ≥5× reads were considered for further analysis. Differential methylation values of individual CpGs were estimated using the beta-difference distributions. CpG cluster differential methylation was determined by pooling CpG level methylation differences using a random effects model. CpG cluster methylation specificity was determined using the Jensen-Shannon divergence of a CpG cluster’s methylation level distribution across all samples and a reference distribution representing either of the two extremes: completely unmethylated or fully methylated. In silico identified CpG islands were defined by genomic regions of at least 700bp length, an CpG observed vs. expected ratio of greater than 0.6 and a GC content greater or equal than 0.5. For the SNP analysis, we obtained the CEPH SNP set from USCS. GWAS SNPs were retrieved from the GWAS catalog, while most of the GWAS SNP grouping was taken from Maurano et al.24. For TFBS analysis, we retrieved peak files from the ENCODE projects and collapsed replicates. Motif analysis was carried out using FIMO.
Supplementary Material
Acknowledgements
We would like to thank K. Clement, P. Samavarchi-Tehrani, Z. Smith, M. Chan and R. Karnik for helpful discussions and feedback. We would also like to thank F. Kelley, T. Durham, Chuck Epstein, Noam Shoresh, G. Lauwers and the MGH tissue repository for assisting in sample and data management. E.D. Rosen is supported by the NIH Roadmap Project (ES017690). AM is supported by the Pew Charitable Trusts and is a New York Stem Cell Foundation, Robertson Investigator. This work was funded by NIH grants (U01ES017155 and P01GM099117) and The New York Stem Cell Foundation.
Footnotes
WGBS data are deposited at the Gene Expression Omnibus (see Supplementary Table 1 for the specific accession numbers).
The authors declare competing financial interests due to the filing of a patent application on the selected regions.
Author contributions
M.Z. and A.M. conceived the study and interpreted the results. M.Z. designed the statistical framework, analysis strategy and analyzed the data. H.G. performed all in house WGBS library production, F.M. contributed bioinformatics tools and J.D. performed cell culture experiments. L.T-Y.T. and E.D.R. provided adipocyte nuclei for WGBS profiling while P.L.D. and D.A.B. made adult brain and AD samples available. O.K. provided support on analysis strategy and statistical methods. B.E.B. and A.M. organized samples as part of the NIH Roadmap Epigenomics Project. H.G., A.G. and A.M established the WGBS at the Broad. A.M. supervised the project. M.Z. and A.M. wrote the paper with assistance from the other authors.
References
- 1.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 3.Seisenberger S, et al. The Dynamics of Genome-wide DNA Methylation Reprogramming in Mouse Primordial Germ Cells. Molecular cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackett JA, Surani MA. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110328. doi: 10.1098/rstb.2011.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 7.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 8.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 9.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nazor KL, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 12.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varley KE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman BP, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen NM, Kenigsberg E, Tanay A. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell. 2011;145:773–786. doi: 10.1016/j.cell.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Lienert F, et al. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 19.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 24.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell CG, et al. Human-specific CpG "beacons" identify loci associated with human-specific traits and disease. Epigenetics. 2012;7:1188–1199. doi: 10.4161/epi.22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dedeurwaerder S, et al. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 29.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.