Abstract
We report the surprising finding that several transposable elements are highly active in Drosophila brain during normal aging. We also show that mutations in Drosophila Argonaute 2 (dAgo2) exhibit exacerbated transposon expression in brain, progressive and age-dependent memory impairment and shortened lifespan. These findings suggest that transposon activation may contribute to age-dependent loss of neuronal function.
Transposable elements (TEs) are highly abundant mobile genetic elements that constitute a large fraction of most eukaryotic genomes 1,2. Retrotransposons, which replicate through an RNA intermediate, represent approximately 40% and 30% of the human and Drosophila genomes respectively. Mounting evidence suggests that TEs can be active not only in the germline, but also in the brain. LINE-1 elements, for instance, are actively mobilized in normal mammalian brains during neurogenesis3–5, leading to genetic heterogeneity, which in principle could have a functional role in neurophysiology. However, because TEs are capable of replicating and inserting into new positions in the genome, they represent a massive reservoir of potential genomic instability as well as RNA-level toxicity. In fact, TE activation has been correlated with several neurodegenerative disorders6–13.
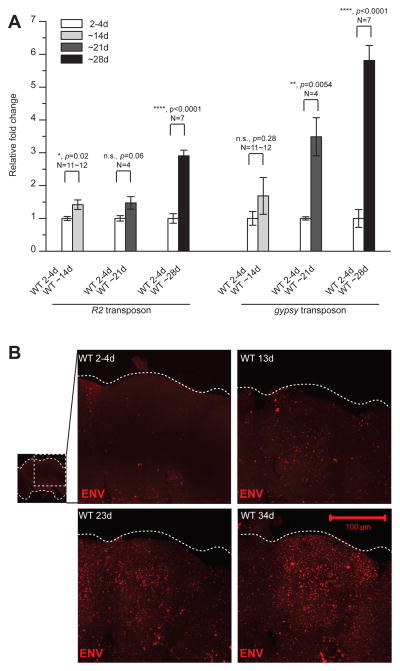
We examined TE expression in Drosophila, where it is feasible to manipulate the TE control mechanisms and to measure physiological effects on the nervous system. We first used Quantitative Real-Time PCR (QPCR) to measure levels of several TE transcripts in head tissues during normal aging by comparing transcript levels from 2–4-day old adult wild-type flies with that of ~14, ~21 and ~28-day old counterparts. Surprisingly, transcripts from R2, a LINE-like element and gypsy, an LTR element are dramatically elevated in aged relative to young animals (Fig. 1A). R1, a second LINE-like element also shows elevated expression with age (see below). Although we have not exhaustively examined expression of the TE families in the Drosophila genome, the age-dependent expression may impact certain TEs specifically because we do not see effects on gypsy4 or Zam (data not shown). In addition to the effects on transcripts from gypsy, R1 and R2, we detect age-dependent increase in expression of the gypsy membrane glycoprotein ENV, using immunohistochemical staining in whole mount brains (Fig. 1B). The ENV signal is most intense in the cortical regions that contain most of the cell bodies, but also is detected in neuropil, areas containing axons and dendrites (central brain projections shown in Fig 1B; see also individual confocal sections Figs 3B and S5B). This age-dependent de-repression of TEs is not due to loss of expression of either Dicer-2 or dAGO2, proteins required for TE silencing in somatic tissues 14 (Fig. S1).
Figure 1. Age dependent increases in expression of LINE-like and LTR retrotransposons in Drosophila brain.
(A) Levels of transcripts of R2 and gypsy quantified by QPCR from young (2–4-day) and aged (~14-day, ~21-day and ~28-day) wild type (WT) heads. Transcript levels normalized to Actin and shown as fold changes relative to WT (means ± SEM). (B) ENV immunofluorescence is elevated in brains from older animals (13-day, 23-day, 34-day) relative to ~2–4-day old animals. Projection through the central brain are shown. ENV signal is detected throughout the cortex layer that includes most of the cell bodies as well as in neuropil areas of axons and dendrites (see also individual confocal sections in Figs. 3B and S5B).
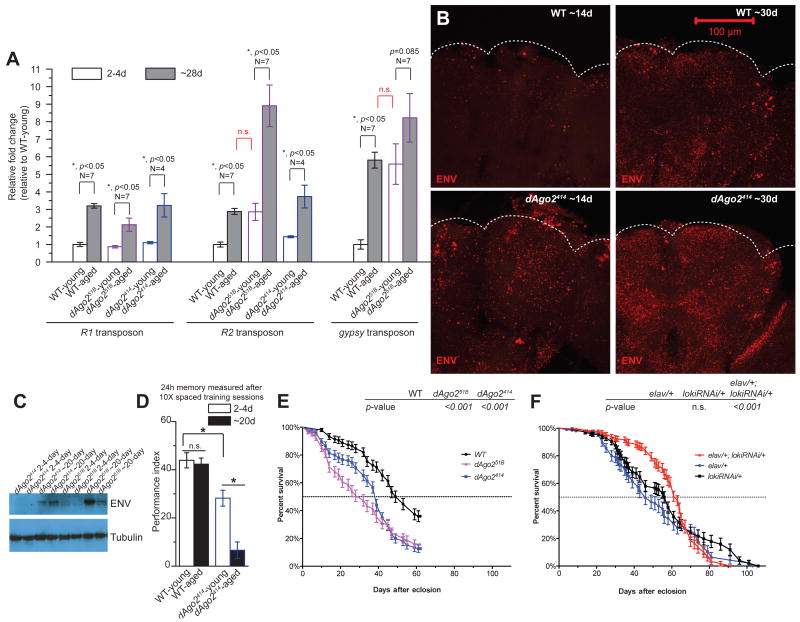
Figure 3. Age-dependent TE expression contributes to memory decline and age-dependent mortality.
(A) Levels of transcripts of R1, R2 and gypsy were quantified from young (2–4-day) and aged (~28-day) WT and dAgo2 mutant animal heads. Within all genotypes, aged animals have significantly elevated levels of each of the transposon transcripts (R1, R2, and gypsy), compared to young animals (*, p<0.05, N=4 for both young and aged dAgo2414 groups, N=7 for both young and aged WT and dAgo251B groups), except for the comparison between young and aged groups within dAgo251B (p=0.085) for gypsy, which also is elevated in young animals. For R2 and gypsy, transcript levels in dAgo251B young groups are as high as in WT aged groups. ~28-day old dAgo251B animals exhibit dramatically increased levels of R2 compared to aged WT group (*, p<0.05). For R2, the 5′ probe set was used in this experiment (see Online Methods) (B) ENV immunoreactivity is detected throughout the cortex layer that includes most of the somata as well as in neuropil (see also Figs. 1 and S5B). Central projections are shown for whole mount brains. Brains from dAgo2414 mutants exhibit higher levels of ENV immunolabeling in ~14-day old and ~30-day old animals, as also is observed with other dAgo2 alleles (Fig. S5B). (C) Western blot detection with ENV monoclonal antibody shows age-dependent accumulation in heads from dAgo2 mutant animals (see also Fig. S5A). Levels for dAgo251B appear increased although somewhat variable. (D) LTM performance (means ± SEM) shown for 2–4-day old and ~20-day old WT and dAgo2414 mutant animals. 2–4-day old dAgo2414 mutants exhibit significantly reduced LTM performance relative to 2–4-day old WT animals, and show a dramatic further reduction in performance in the 20-day old dAgo2414 mutant group (*, p<0.05 and N=15). (E) Lifespan is significantly shortened for dAgo2414 and dAgo251B animals relative to WT (log-rank test). (F) Knocking down loki gene expression with lokiRNAi in neurons significant delays mortality (Gehan-Breslow-Wilcoxon test) of the elav/+; lokiRNAi/+ animals compared to heterozygous controls for transgenes (elav/+ and lokiRNAi/+), as well as the onset of age-dependent memory decline (Fig. S7).
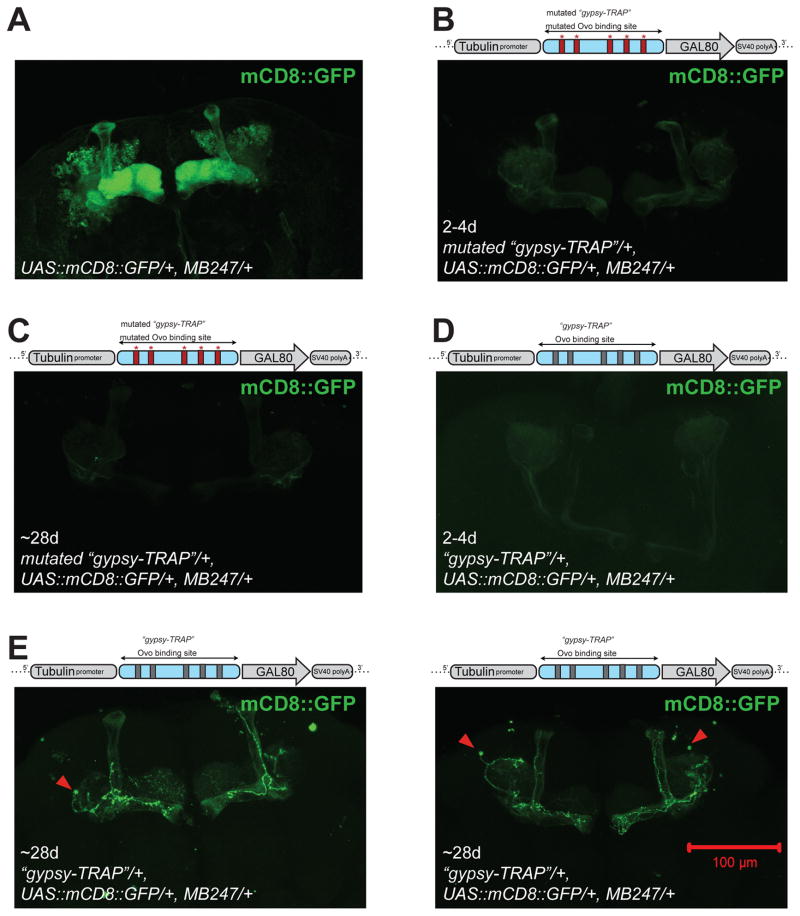
To determine if expression of gypsy in older animals is associated with physical transposition, we designed a “gypsy-TRAP” reporter system to detect de novo gypsy integration events. We adapted an elegant reporter system that was previously established for detecting gypsy integration in the germline15. To achieve this, we expressed GAL80, which is an effective repressor of GAL4, under control of the ubiquitous α-tubulin promoter. In the presence of GAL80 protein, GAL4-mediated expression of GFP is effectively silenced. We placed a ~500bp fragment from the ovo regulatory region between the promoter and GAL80 in order to attract gypsy insertional mutations (Fig. S2A). This fragment contains 5 Ovo binding sites to which the Ovo protein normally binds in its own regulatory region. In the germline, these Ovo binding sites are necessary and sufficient to attract de novo gypsy insertions15. In our reporter system, somatic integration of gypsy downstream of the promoter or within the GAL80 transcription unit disrupts expression of GAL80, permitting activation of GFP by GAL4 (Fig. 2B).
Figure 2. “Gypsy-TRAP” reporter detects de novo integration in neurons in aged animals.
A ~500bp fragment from the ovo regulatory region containing 5 Ovo binding sites is inserted between Tub promoter and GAL80 gene. A mutated “gypsy-TRAP” construct contains mutations that disrupt each of the 5 Ovo binding sites. In the absence of gypsy insertions, GAL80 expression suppresses GAL4, and UAS::mCD8::GFP is not expressed. In the presence of gypsy integration into the “gypsy-TRAP”, GAL80 expression is blocked, and UAS::mCD8::GFP is turned on (see Fig. S2). (A) Approximately 800 mushroom body Kenyon cell neurons per brain hemisphere are labeled by MB247-GAL4-driven UAS::mCD8::GFP. (B) An example brain from 2–4-day old mutated “gypsy-TRAP”; UAS::mCD8::GFP/+; MB247/+. No GFP labeled neurons seen. (C) An example brain from ~28-day old mutated “gypsy-TRAP”;UAS::mCD8::GFP/+; MB247/+. No GFP labeled neurons seen. (D) An example brain from ~2–4-day old “gypsy-TRAP”; UAS::mCD8::GFP/+; MB247/+. No GFP labeled neurons seen. (E) Example brains from ~28-day old “gypsy-TRAP”; UAS::mCD8::GFP/+; MB247/+. Several GFP-labeled MB neurons seen in each brain. See Table S1 and Fig. S2 for statistical summary and additional example images.
We used this system to screen for de novo gypsy integration events in the brain by focusing on neurons of the mushroom body (MB) for which highly specific and strongly expressing GAL4 lines exist. We used the MB247 GAL4 line16, which is known to label about 800 out of ~2000–2500 mushroom body Kenyon cell neurons per brain hemisphere (Fig. 2A). GAL80 expression from our “gypsy-TRAP” (Tubp-OvoSite-GAL80) transformant lines is sufficient to silence GFP (Figs. 2 and S2). In fact, we do not observe any labeled neurons in 2–4-day old animals containing this construct (0/26 brains from 2–4day old animals, Figs. 2B, 2C, S2 and Table S1). However, we observe sparse GFP-labeled MB Kenyon cells at later ages in each of two transformant lines containing “gypsy-TRAP” (Tubp-OvoSite-GAL80), often in multiple neurons (14/39 brains labeled from 28–35 day old animals, Figs. 2E, S2 and Table S1). This effect of age was statistically significant (Chi-square Analysis, p<0.01). The labeling appears to be stochastic because we see both intra and inter-hemisphere variation. The accumulation of GFP positive neurons also requires the 5 Ovo binding sites, as is true for gypsy insertions in the germline, because we do not observe any GFP labeled cells in control transformant lines containing a “gypsy-TRAP” with an ovo fragment in which the binding sites are mutated (Tubp-MutatedOvoSite-GAL80) (Figs. 2B; S2; Table S1; Chi-square Analysis, p<0.001). Our results using this reporter system strongly support the conclusion that gypsy not only is expressed in neurons of aging animals, but also is actively mobile in an age dependent manner.
We next used genetic manipulations of dAgo2 to create a situation in which transposons are unleashed prematurely in young animals. In both animals and plants, TE control is mediated by Argonaute proteins guided by small regulatory RNAs14. Germline tissues are protected against TEs by the concerted action of Argonaute proteins of the PIWI clade and their small RNA partners, the piRNAs14. While control of TEs in somatic tissues in Drosophila is dependent on dAGO2 guided by endogenous small interfering RNAs, a different Argonaute protein in flies, dAGO1, preferentially loads the microRNAs that target cellular mRNAs, but has no known impact on TEs. Therefore, using dAgo2 mutants, we can create a condition in which we selectively disrupt the somatic TE control mechanism.
Although dAgo2 mutants have been shown to exhibit elevated TE expression in somatic tissue, the phenotypic consequences of such mutations on aging are not known. We found that transcripts from R2 and gypsy are significantly elevated in head tissue of young dAgo2414 and dAgo251B mutant animals, as well as in trans-heterozygous dAgo2414/51B animals (Figs S4A and 3A). In addition, the age-dependent elevation of both R2 and gypsy expression is accelerated in the dAgo2 mutants such that transcript levels of both R2 and gypsy in 2–4-day old mutant animals are comparable to that seen in ~28-day old wild type animals. At the protein level we observe an accelerated age dependent increase in ENV in dAgo2 mutants (the dAgo2414 and dAgo251B hypomorphic alleles and the dAgo2454 null allele) both in whole mount brains (Figs. 3B and S5B) and on western blots from adult heads (Figs. 3C and S5A). Furthermore, elevated expression of gypsy in dAgo2 mutants also is associated with de novo insertions into the ovo locus, as detected by genomic PCR and sequencing (Fig. S3).
To investigate the correlation between age-dependent neuronal decline and TE activation, we used a robust and sensitive Pavlovian learning and memory assay that is well established in Drosophila17. We compared 24-hour LTM performance in animals that were trained when they were young (2–4-day) or trained when they were at an intermediate age (~20-day). dAgo2 mutants already exhibit a partial reduction in memory at 2–4-days old (Figs. S4C, S4D, 3D), an effect which can be rescued by neuronal expression of a dAgo2 transgene (Fig. S4E). The mild defect seen in young animals becomes dramatically worse in 20-day old adults (Fig. 3D). In contrast, wild-type animals trained at the ~20-days age exhibit normal robust levels of LTM that are equivalent to that seen in 2–4-day old wild type animals (Fig. 3D), only developing impairment at ~28-day of age (Fig. S7B).
We also examined the effects of dAgo2 mutations on longevity and found that dAgo2414, dAgo251B and dAgo2454 mutants exhibit significantly shorter lifespans than their wild type counterparts (Figs. 3E and S5C). This finding is consistent with reports that mutations in Dicer-218 and loquacious19, other components of the somatic small RNA-dependent TE silencing pathway, also exhibit short lifespan. Although dAgo2 mutants are susceptible to exogenous viruses, viral infections do not contribute to the age-dependent decline in these mutants and do not cause the observed induction of transposons (Fig. S6).
TE activation in the germline is sufficient to cause sterility, at least in part by triggering Checkpoint kinase 2 (Chk2)-mediated DNA damage-induced apoptosis. In fact, disruption of Chk2 in the germline prevents cells from undergoing programmed cell death, which is sufficient to suppress TE dependent sterility20. To test whether DNA-damage leading to Chk2 signaling also contributs to age dependent mortality in wild type animals, we used an RNAi transgene to target loki, the Drosophila ortholog of chk2. Remarkably, we found that disrupting loki function exclusively in neurons by expressing the lokiRNAi under control of the pan-neuronal elav-GAL4 can significantly delay mortality (Figs. S7A and 3F). This disruption of loki function also yields a modest but significant delay in age-dependent memory impairment (Fig. S7B and S7C). Although a causal role for TE activation and TE-induced DNA damage in age-related neuronal decline remains to be demonstrated, the effects with disruption of Chk2 signaling are at least consistent with this interpretation.
There is accumulating evidence that TEs actively mobilize in neurons3,4. This phenomenology has lead to the suggestion that regulated TE jumping provides a source of somatic mosaicism that may contribute to normal brain physiology, although such functional effects remain to be established. On the other hand, LINE, SINE and LTR elements are de-repressed in a variety of neurodegenerative diseases6–13, suggesting the possibility that misregulation of TEs is detrimental. The results reported here, together with the above literature, suggest the novel hypothesis that TE activation with age or disease may contribute to neuronal decline.
ONLINE METHODS
Fly Stocks
The wild type flies utilized in this study were w1118 (isoCJ1), a Canton-S derivative21. The dAgo2 mutants and UAS::dAgo2 transgenic strains were backcrossed to the above wild type strain for at least five generations. Flies were cultured in standard fly food and laboratory room temperature (22.5°C). The “gypsy-TRAP” transgenic flies were made by cloning a ~500bp Ovo binding site15 into the NotI site between Tubulin promoter and GAL80 gene in the Tubp-GAL80 in pCaSpeR4 plasmid. The resulting construct was injected into w1118 (isoCJ1) recipient embryos and transformant lines were isolated by standard procedures at the BestGene, Inc. The mutated “gypsy-TRAP” transgenic flies were made by injecting a similar construct bearing mutations in Ovo binding sites15. The MB247, Repo and Elav -Gal4 lines are as reported previously 22.
Behavioral Assays
Aversive Pavlovian olfactory task was performed by training flies in a T-maze apparatus using a Pavlovian conditioning paradigm. Approximately 50–100 flies were loaded into an electrifiable training grid. For a single training session, flies were exposed sequentially to one odor (the conditioned stimulus, CS+), which was paired with a 60-volt electric shock and then a second odor (the unconditioned stimulus, CS−) without shock. 2 minutes after this training session, the flies were tested and allowed to choose between the two odors. A half performance index was calculated by dividing the number of flies that chose correctly, minus the flies that chose incorrectly by the total number of flies in the experiment. The same protocol was then performed with another group of 50–100 flies and reciprocal odor presentation. The final PI was calculated by averaging both reciprocal half PIs. The long-term-memory (LTM) experiment was an adaptation of this training protocol. Flies were subjected to ten such training sessions in robotic trainers spaced out with a 15-minute rest interval between each. Flies then were transferred into food vials and incubated at 18 °C until being tested 24 hours after the training. All genotypes were trained and tested in parallel, and rotated between all the robotic trainers to ensure a balanced experiment. Odor pairs and concentrations used for these behavior paradigms are: 3-Octanol (1.5×10−3 v/v) and 4-Methylcyclohexanol (1×10−3 v/v), or, 3-Octanol (1.5×10−3 v/v) and Benzaldehyde (0.5×10−3 v/v). Pure odors were purchased from Sigma and delivered as the stated concentrations with air flow at 750ml/min. In all cases, behavior experiments within a figure were performed in parallel. Behavioral data are normally distributed and are shown as means ± SEM. One-Way ANOVA and post-hoc analyses were performed.
Lifespan
Lifespan were measured with ~50–150 animals/genotype. Equal numbers of male and female flies were used for each genotype. Survival analyses were performed with the Kaplan-Meier Method. Log-rank test and Gehan-Breslow-Wilcoxon test were used to compare survival curves. Pair-wise comparisons were made with Bonferroni corrections.
QPCR
The QPCR was performed according to the assay manual. In brief, massive numbers of fly heads were collected for each genotype and total RNA was purified with Trizol (Invitrogen) and treated by DNaseI (Promega). Total RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer. For the reverse transcription (RT) reaction, each 20μl RT reaction was performed with 2μg total RNA using the High capacity RNA-to-cDNA kit (Applied Biosystems). The QPCR reactions for each assay were carried out in duplicate, and each 20μl reaction mixture included 1μl previous RT products. The QPCR reaction was carried out and analyzed in an Applied Biosystems 7900HT Fast Real-Time PCR System in 96-well plates at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Linearity tests were performed on all custom designed primers and probes to ensure linearity.
Custom TaqMan Probes
All TaqMan® Gene Expression Assays (Applied Biosystems) utilized the FAM Reporter and MGB Quencher. TaqMan® probes for each transcript were designed following the vendor’s custom assay design service manual. The customized gene-specific Taqman probes and inventoried Taqman probes had the following sequences and Assay IDs.
R1-ORF2 (Assay ID AID1TD0, FBgn0003908):
probe: 5′-ACATACGCCATAATCTG-3′
Blood-ORF2 (assay ID AIFARJ8, FBgn0000199)
Probe: 5′-TCGGTGCATAACTTAGTTAGTTCA-3′
GypsyORF2 (Assay ID AI5IO6V, FBgn0001167)
Probe: 5′-AAGCATTTGTGTTTGATTTC-3′
Gypsy4-ORF2 (AID1TGU, FBgn0063433)
Probe: 5′-CCCGATCTGGGTTGTC-3′
ZAM-ORF2 (Assay IDAICSVAM, FBgn0023131)
Probe: 5′-CCCCATGATTAGTCTTTACTG-3′
1731-ORF2 (Assay ID AICSU7S, FBgn0000007)
Probe: 5′-AAGCTGAAGACTGATTTATG-3′
297-ORF2 (Assay ID AI70LJB, FBgn0000005)
Probe: 5′-TTGATCAAACATACAAATTAATTAC-3′
R25′ (Assay ID AJ0IV12, FBgn0003909)
Probe: 5′-GAATGCCATTCCAAATGGAGAGCCC-3′
R23′ (Assay ID AJY9XVU, FBgn0003909)
Probe: 5′-TAGAAAAATATTGGGCGAACAAGTT-3′
DBV (Assay ID AIV13YJ)
Probe: 5′-CCTATTAGTGATCCGCTCGCG-3′
DTRV (Assay ID AIS07L3)
Probe: 5′-CTTCGATCCGAGGTATGC-3′
DAV (Assay ID AIX00AZ)
Probe: 5′-AAGGTAGTAGGTTACATTTGTC-3′
Sigma V (Assay ID AIWR14R)
Probe: 5′-CCGTAGTCCGATGGTTCC-3′
Nora V (Assay ID AIQJA9N)
Probe: 5′-CTGAGGCTTCTCTTGTTTAAT-3′
DCV (Assay ID AIPAC3F)
Probe: 5′-TTGTCGACGCAATTCTT-3′
DXV (Assay ID AIRR9FV)
Probe: 5′-TCATAGATGATGTCAAATTT-3′
ANV (Assay ID AIT95SB)
Probe: 5′-CAGACAATTTCTCAGAATCAT-3′
Act5C (Assay ID Dm02361909_s1)
Loki (Assay ID Dm01811114_g1)
Ago2 (Assay ID Dm01805432_g1 and Dm01805433_g1)
Dcr-2 (Assay ID Dm01821537_g1 and Dm01821540_g1)
Western Blots
~15 adult fly heads per sample were homogenized in 20ul Nupage@ sample loading buffer, heated to 95 °C for 5 min and 10 μl loaded onto Nupage@ 4–12% Bis-Tris gels, then transferred to PVDF membrane (Invitrogen) and blotted by standard protocols. Primary antibodies used were anti-tubulin (1:10,000, E7, Developmental Studies Hybridoma Bank), anti-ENV (1:5000). The WesternBreeze® Chemiluminescent Kit–Anti-Mouse system was used to visualize the blotted bands on films.
Bleach Treatment of Embryos
In order to remove virus infection in fly stocks, 2hr embryos from wild type controls and dAgo2 mutants were collected and treated with 50% bleach 2 times for 20 minutes each. Treated embryos were then grown in a virus free clean room equipped with UV lamps to sterilize surfaces. Expanded fly stocks after bleach treatment are proven to be virus-free. All strains also were grown on a rotating set of 6 antibiotics.
Immunohistochemistry and GFP Imaging
Dissection, fixation, immunolabeling and confocal imaging acquisition were performed as previously described23. Ascites containing anti-gypsy ENV monoclonal antibody (mAb) was prepared from the anti-gypsy ENV 7B3 hybridoma cell line24. A 1:100 dilution of ENV primary mAb and a 1: 200 dilution of secondary antibody of Cy3-conjugated goat anti-mouse IgG were used. We used 2μM DilC18(5)-DS lipophilic dye solution (Molecular Probes) to label cell membranes throughout the brain as counterstaining. For Env immunolabeling, we imaged multiple brains of each genotype and age. Representative images are shown in figures. Totals numbers imaged for wild type were: 6 (0–4day), 14 (14day), 16 (21–28 day), 4 (70 day). For dAgo2414, total number imaged were: 8 (14 day), 7 (21–28 day). For dAgo2-51b, total number imaged were: 5 (0–4 day), 6 (14 day), 6 (21–28 day), 3 (70 day). For dAgo2454, total number imaged were 9 (0–4 day), 13 (14 day), 6 (21–28 day).
Nested PCR
DNA was extracted from ~300 fly heads of the indicated ages. Standard PCR was preformed in nested fashion with the first round of PCR utilizing primer 1 and 3 followed by a second round of pcr with primer 2 and 4. Primer sequences are listed below. Nested PCR was then run on .9% agarose gel (Sigma) and size was estimated according to 1 kb plus DNA ladder (Invitrogen). The PCR product was then gel purified using illustra GFX PCR DNA and Gel Band Purification Kit from GE Healthcare. The fragment was cloned using the TOPO TA Cloning Kit for sequencing from Invitrogen and sequenced by ELIM BIOPHARM using the Sanger sequencing method. MacVector was used to display sequencing results.
Primers
Primer 1 - CAACTCTGCACCCACGACTA
Primer 3 - CAGCGGAAAGCTGACACTTC
Primer 2 - CACACACCCATGGAATTGAA
Primer 4 - GGCTCATTGCCGTTAAACAT
Statistical Testing
Behavioral data from the Pavlovian memory task are normally distributed 21 and are shown in all figures as means ± SEM. For these data, one-Way ANOVA and post-hoc analyses were performed. For the life-span curves, survival analyses were performed with the Kaplan-Meier Method. Log-rank test and Gehan-Breslow-Wilcoxon test were used to compare survival curves. Pair-wise comparisons were made with Bonferroni corrections.
Supplementary Material
Acknowledgments
We thank Benjamin Czech and Gregory Hannon for the dAgo2414 and lokiRNAi fly line, Fen-Biao Gao for the dAgo251B fly line, Barry Dickson for the UAS::dAgo2 transgenic fly line, Michael Welte for the dAgo2454 fly line, Tzumin Lee for the pTub-GAL80 in Casper4 plasmid, Jeff Boek for the 7B3 hybridoma cell line and Carmelita Bautista at the CSHL shared resources for ascites production. We also are grateful to Scott Waddell, Jennifer Beshel, Michael Cressy, Benjamin Czech, Gregory Hannon, Kyle Honegger, Josh Huang, Maurice Kernan, Rob Martienssen, Hongtao Qin, Carmen Sandoval, Yichun Shuai, Glenn Turner, Tony Zador and Yi Zhong for helpful discussions or comments on the manuscript. This work was supported by NIH TR01(5R01NS067690-03) and DART neuroscience LLC awarded to JD. SG received additional support from the Shakespeare Fellowship and the Ernst Göhner Foundation.
Footnotes
Author Contributions:
W.L and L.P contributed equally. W.L, L.P and J. D conceived and designed the project and analyzed the experiments. The behavior experiments and western blots were performed by W.L. The QPCR and lifespan analyses were performed by L.P with assistance from W. L, C.S, L.K. and T.D. Imaging experiments were performed by L.P, N.C. and S.G. The manuscript was written by W.L, L.P. and J.D with comments from the other authors.
Competing interests: This work was funded in part by DART LLC via a research grant to J.Dubnau
References
- 1.Belancio VP, Hedges DJ, Deininger P. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 2.Goodier JL, Kazazian HH., Jr Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Muotri AR, et al. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 4.Coufal NG, et al. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baillie JK, et al. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lathe R, Harris A. J Mol Biol. 2009;392:813–822. doi: 10.1016/j.jmb.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 7.Muotri AR, et al. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong BH, Lee YJ, Carp RI, Kim YS. J Clin Virol. 2010;47:136–142. doi: 10.1016/j.jcv.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Coufal NG, et al. Proc Natl Acad Sci U S A. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douville R, Liu J, Rothstein J, Nath A. Ann Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko H, et al. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan H, et al. Hum Mol Genet. 2012;21:57–65. doi: 10.1093/hmg/ddr437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Jin Y, Prazak L, Hammell M, Dubnau J. PLoS One. 2012;7:e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czech B, Hannon GJ. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labrador M, Sha K, Li A, Corces VG. Genetics. 2008;180:1367–1378. doi: 10.1534/genetics.108.094318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwaerzel M, Heisenberg M, Zars T. Neuron. 2002;35:951–960. doi: 10.1016/s0896-6273(02)00832-2. [DOI] [PubMed] [Google Scholar]
- 17.Dubnau J, Chiang AS. Curr Opin Neurobiol. 2012 doi: 10.1016/j.conb.2012.09.006. [DOI] [Google Scholar]
- 18.Lim DH, et al. FEBS Lett. 2011;585:3079–3085. doi: 10.1016/j.febslet.2011.08.034. [DOI] [PubMed] [Google Scholar]
- 19.Liu N, et al. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Pane A, Schupbach T. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tully T, Preat T, Boynton SC, Del Vecchio M. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 22.Qin H, et al. Curr Biol. 2012;22:608–614. doi: 10.1016/j.cub.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, et al. PLoS Comput Biol. 2008;4:e1000026. doi: 10.1371/journal.pcbi.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.