Abstract
Non-linear optical microscopy methods can characterize over time multiple functional properties of engineered tissues during development. Here, we demonstrate how the combined use of third-harmonic generation (THG) and two-photon excited fluorescence (2PEF) imaging can provide direct quantitative biomarkers of adipogenic stem cell differentiation and metabolic state, respectively. Specifically, we imaged over nine weeks silk scaffolds embedded with human mesenchymal stem cells and exposed to either propagation (PM) or adipogenic differentiation media (AM). THG was employed to visualize the formation of lipid droplets. 2PEF was used to assess the metabolic state of the cells through the redox ratio defined based on the endogenous FAD and NADH fluorescence. The redox ratio of cells in the AM scaffold was significantly lower than that in the PM scaffold during week 5 and 9, and correlated with significant increases in lipid-to-cell volume ratio, and number and size of lipid droplets in the AM scaffold. These findings indicate that the decrease in redox ratio during adipogenic differentiation is associated with fatty acid synthesis and lipid accumulation. Our methods therefore enabled us to identify and measure dynamic correlations between lipid droplet formation and cell metabolic state, while providing insight on the spatial heterogeneity of the observed signals.
Keywords: Lipid droplets, Stem cell metabolism, Adipogenic differentiation, Third-harmonic generation, Two-photon excited fluorescence, NADH, FAD, Redox ratio
1. Introduction
Loss of adipose tissue due to trauma or disease is characterized not only by a loss of tissue volume, but also by a loss of tissue function. Over the last few decades, it has become clear that in addition to acting as a storage system for fat and energy, adipose tissue serves a key role in the endocrine system [1] and disruptions in signaling functionality can result in diabetes, cardiovascular disease and hypertension [2–4]. Therefore, the development of quantitative methods to assess adipose tissue function is of direct relevance not only for tissue engineering applications in the context of tissue replacement, but also for improved understanding of numerous diseases and the effects of corresponding treatments. Non-invasive, optical imaging modalities are well-suited for this purpose as they can be implemented in multi-modal imaging platforms that simultaneously provide functional, morphological and biochemical information about natural and synthetic biomaterial scaffolds, extracellular matrix components (such as collagen and elastin) and cells. Such images can be acquired based entirely on endogenous sources of contrast, thus, obviating the need for any processing steps that can in principle introduce artifacts or prevent dynamic monitoring of the sample.
In recent years, microscopic imaging modalities that rely on non-linear light-matter interactions have been increasingly employed to characterize the morphology and function of cells and other tissue components. Such methods include two-photon excited fluorescence (2PEF) [5–12], second-harmonic generation (SHG) [6, 12–15], third-harmonic generation (THG) [16, 17], coherent anti-Stokes Raman scattering (CARS) [18–20] and stimulated Raman scattering (SRS) [21] microscopic imaging. These approaches typically employ near-infrared wavelengths for sample illumination and achieve deeper penetration than confocal microscopy. In addition, the non-linear nature of the optical interactions dictates that they occur only at the focus of the beam, minimizing photodamage and photobleaching during scanning of 3D volumes, which is important both for tissue viability and for enabling optimal detection of endogenous signals that are generally weak.
2PEF is particularly well-suited to assess cell function and morphology, using natural sources of fluorescence such as nicotinamide adenine dinucleotide (phosphate) (NAD(P)H), flavin adenine dinucleotide (FAD), keratins, lipofuscin, retinol and porphyrins [22, 23]. A number of investigators have developed approaches to quantify the signal emanating from NADH and/or FAD as a measure of the metabolic status of the cell, but also to assess some morphological aspects, such as nuclear to cytoplasmic ratio and mitochondrial organization [24–27]. Both lifetime fluorescence [9, 28, 29, 30] and traditional intensity based [5, 6, 10, 24, 25] 2PEF measurements have shown promise as methods that can provide quantitative biomarkers of stem cell differentiation. For example, we recently demonstrated that dynamic changes in the redox fluorescence ratio, defined as the ratio of intensities corresponding to FAD/(NADH+FAD), can be correlated to specific adipogenic differentiation stages of adipose derived stem cells over the course of six months [10]. However, to assess differentiation in that case, it was necessary to sacrifice scaffolds at distinct time-points and perform invasive immunohistochemical and PCR-based assays. Thus, direct correlations between the metabolic status of a cell and its level of differentiation as assessed by lipid droplet formation were not possible.
Third-harmonic generation (THG) is an additional contrast mechanism [31] that is compatible with 2PEF imaging. THG imaging is usually performed using an excitation wavelength in the 1.0–1.7 µm excitation range, and signal is detected at one third of this wavelength. Differing from SHG, it does not require molecular asymmetry and can be produced by any medium. However, in biological THG imaging no signal is generally observed from homogeneous media, as a consequence of destructive interference resulting from the axial phase shift (the Gouy shift) experienced by the excitation beam near its focus [31]. When the beam is focused at the interface between two media (a) and (b), the signal approximately scales as |αa - αb|2, where α = χ(3)/ n3ω (n3ω - nω), χ(3) is the third-order nonlinear susceptibility, n3ω is the refractive index at the harmonic frequency, and (n3ω - nω) is the refractive index dispersion [16]. This mechanism produces contrasted images where optical inhomogeneities of size comparable to the beam focus are highlighted on a dark background. In particular, a strong signal is observed at the interface between the (aqueous) cytoplasm of cells and a lipidic or absorbing organelle a few 100s of nm in size [16]. Recent studies have shown that THG can be used to visualize the morphology of unstained tissues [32–36], and in particular to detect lipid structures [16, 37–39]. Importantly, THG imaging is best achieved with focused infrared femtosecond pulses, and is easily combined with 2PEF imaging [16, 35, 40, 41].
Therefore, one primary goal of this study was to develop a method for studying correlations between lipid droplet formation and cell metabolism in differentiating engineered tissues over extended time periods, based on combined THG and 2PEF measurements. We examined silk scaffolds embedded with mesenchymal stem cells that were exposed either to propagation or adipogenic differentiation media, and acquired THG and 2PEF images from the same scaffolds at 2, 5 and 9 weeks following the induction of differentiation. We developed an image processing workflow to identify cells and lipid droplets, and to extract quantitative metrics associated with the redox ratio, lipid content, lipid droplet number and lipid droplet size in each cell included in the analysis. Through this cell-by-cell quantification approach, we also assessed potential correlations between the metabolic and lipid droplet readouts within individual tissues. Collectively, the label-free biomarkers described in this study can be used to assess adipogenic differentiation in engineered tissue constructs, and more generally, could serve as measures of adipose tissue function for disease diagnostics and drug screening efficacy.
2. Materials and Methods
2.1 3D Adipose Tissue Engineering
Hexafluoro 2 propanol (HFIP) silk solution was used to generate silk scaffolds based on methods established in previous studies [10, 42, 43]. Scaffolds measured 8 mm in diameter and 0.6 mm thick; pores were created using 200 µm diameter salt crystals. Human mesenchymal stem cells (hMSCs) were seeded in the scaffolds and two experimental groups were cultured, each with different media reported previously to either induce adipogenic differentiation (DMEM plain, high glucose, with L-glutamine: 434 mL, 0.1 mM Non-essential amino-acids: 5 mL, 1% Pen-Strep: 5 mL, 10% FBS: 50 mL, 50 µM Indometacin, 1000x stock solution: 500 µL, 0.5 mM Isobutyl Xanthine 100x stock solution: 5 mL, 1 µM Dexamethasone: 500 µL, 5 µg/mL insulin: 250 µL) or allow the hMSCs to propagate undifferentiated (DMEM plain, high glucose with L-glutamine: 440 mL, 1% Non-essential amino-acids: 5 mL .1% Pen-Strep: 5 mL, 10% FBS: 50 mL, 2 ng/mL bFGF: 10 µL) [43]. The hMSC-seeded scaffolds were placed in standard 12-well plates and maintained in culture within a standard 37°C, 5% CO2 incubator.
2.2 Two-photon excited fluorescence and third harmonic generation image acquisition
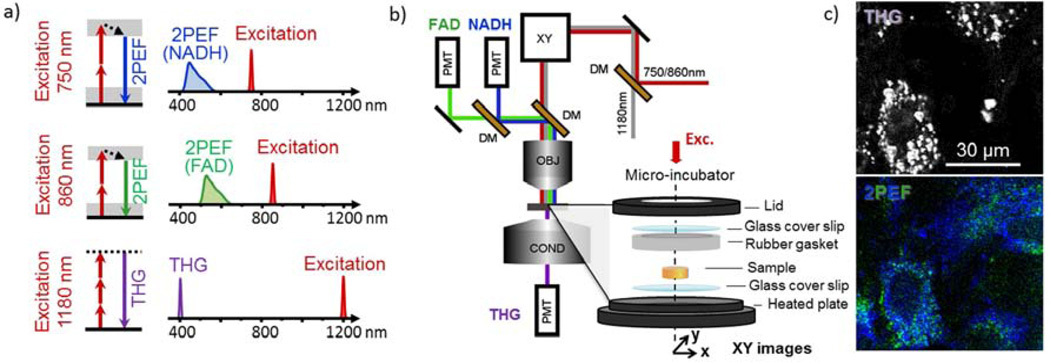
A schematic of the experimental arrangement is shown in Fig. 1. The principles of operation and relevant excitation/emission wavelengths are shown in Fig. 1a, while Fig. 1b illustrates the optical setup. Imaging was performed on a custom-built laser scanning microscope incorporating a femtosecond Titanium:Sapphire (TiS) laser (750–980 nm, 80 MHz, Chameleon, Coherent Inc., CA, USA), an optical parametric oscillator (1180 nm, 80 MHz, KTP-OPO, APE, Germany), galvanometer mounted mirrors (GSI Lumonics, USA) and a water-immersion objective (40× 1.1NA Zeiss, Germany). Ti:S and OPO powers were controlled with wave plates and polarizers, and beams were combined using a dichroic mirror (1000-DCXR Chroma). Signals were detected in the epi- and trans-directions by photomultiplier modules (SensTech, UK) and lab-designed counting electronics. Scanning and acquisition were synchronized using lab-written LabVIEW software and a multichannel I/O board (PCI-6115, National Instruments, USA). Fluorescence was collected in the backward (epi) direction using a dichroic mirror (695dcxru, Chroma, USA) and 2PEF signals were directed toward two independent detectors using a dichroic mirror (Semrock). THG was collected in transmission.
Figure 1.
Experimental Setup and Principles of 2PEF and THG. a) Two-photon excitation occurred at 750 and 860nm, with emission collected with bandpass filters centered at 450 and 582nm, which resulted primarily from NADH and FAD, respectively. THG excitation occurred at 1180nm, with emission collected with a bandpass filter centered at 377nm. b) Schematic of the custom experimental setup used for this work. Heated micro-incubator chamber consisting of a lid with an imaging window fitted with a cover glass, rubber gasket to protect the sample and a heated base with a coverslip fitted imaging window is shown at bottom left. c) Examples of 2D optical sections from hMSC embedded silk scaffolds using 2PEF and THG microscopy.
Two 2PEF and one THG image stack were acquired sequentially with 750, 860 and 1180nm excitation, respectively. Detection was done on separate channels equipped with 450/80, 582/75 and 377/50 nm filters, respectively (Semrock, USA). Fig. 1c shows typical 2PEF and THG images. 2PEF signal detected from hMSCs at 450 nm with 750 nm excitation emanates primarily from NADH, while the 2PEF emission at 582 nm using 860 nm excitation is attributed mostly to FAD [6, 10, 23]. Voxel size was typically 0.6×0.6×2 µm3. Voxel dwell time was approximately 5µs per image channel. Image volume thickness varied between typically 30 µm and 100 µm. Image stacks of AM- and PM-seeded scaffolds were recorded at three time points after seeding: weeks 2, 5, and 9.
2.3 Tissue mounting for longitudinal observations
To ensure sterility and cell viability for the duration of the experiment, scaffolds were placed prior to each imaging session in between two sterile coverslips of a heated confocal imaging chamber (model RC-30 WA, Warner Instruments LLC, Hamden CT), while immersed in media (Fig 1b). The temperature was maintained at 37°C throughout imaging using a temperature controller (model TC-344B, Warner Instruments LLC). Images from both experimental groups were then analyzed to assess tissue development.
2.4 Image processing: spatial isolation of cells and lipid droplets
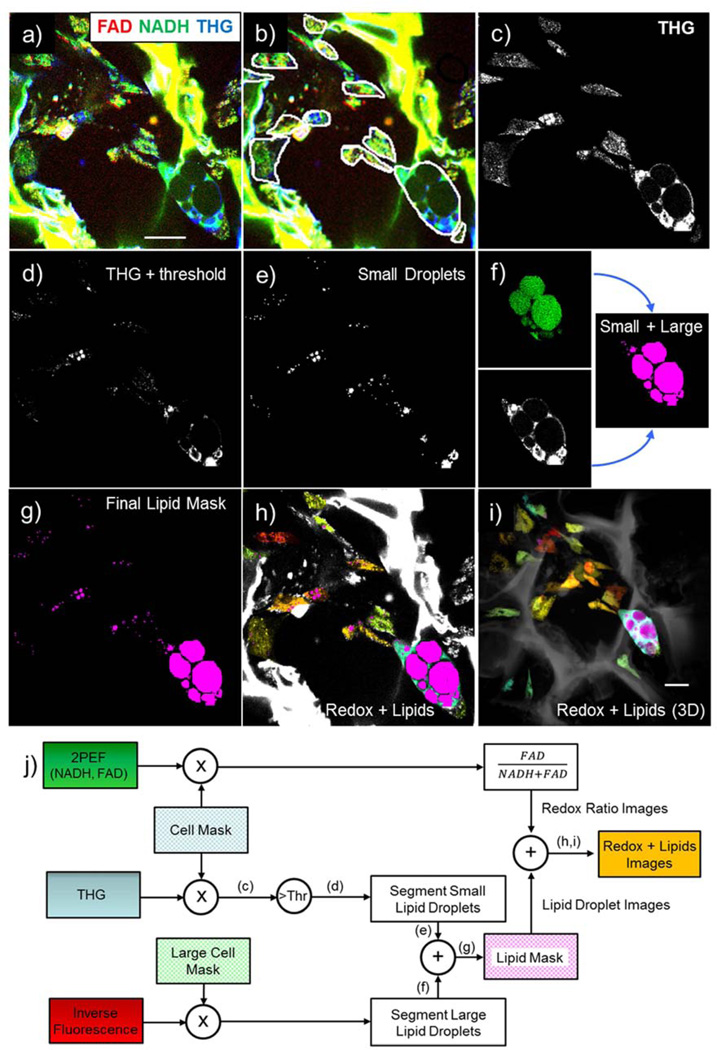
Cells and lipid droplets were identified and segmented using algorithms written in Matlab software (Mathworks, Natick, MA). Fig. 2 illustrates the image processing approach we developed to measure cell and lipid droplet characteristics. Fig. 2a shows representative initial images, where the THG image (blue) was overlaid with the NADH (green) and FAD (red) 2PEF images. For the initial part of the analysis consisting of cell border extraction, we found it difficult to develop a fully automated algorithm that yielded accurate results for all image stacks. The strong endogenous silk fluorescence compared to the weak cellular fluorescence was one of the reasons that prevented consistent outlining of cell borders in a manner that truly represented the shape of the cells. We therefore isolated individual cells from each other and nearby silk using an interactive manual-circling approach based on the overall morphology and relative signal intensity (Fig. 2b). Cells were first selected from all depths of the imaging volumes. In order to account for the fact that a single cell may be visible at different depths within the image volume, each cell was identified first within a single 2D field and then image slices above and below the initial image plane were examined and segmented. . Cell border identification was followed by lipid segmentation using an automated algorithm. This algorithm was designed to take advantage of two properties of lipid droplet signals: lipid droplets were expected to (i) yield higher THG intensity than other cellular structures and (ii) have a round shape. Within the cell contour mask illustrated in Fig. 2b, the THG signal was used for lipid droplet segmentation (Fig. 2c). First, an intensity threshold was computed using a multiple of the average THG signal within the cell mask. This threshold was determined for each AM stack in order to maximize lipid structure signal and minimize signal from non-lipid structures. The same threshold multiplier was then applied to the corresponding PM scaffold for each time point. Threshold multiplier values were 1.5, 1.8, and 1.0 for weeks 2, 5, and 9, respectively. The result of applying the threshold to the THG signal is illustrated in Fig. 2d. Each thresholded THG image (2D) was then assessed for pixel connectivity and organized into connected objects. The roundness of connected objects was evaluated based on eccentricity, where the eccentricity of a circle is 0, and the eccentricity of an ellipse is between 0 and 1. For the work presented here, 0.5 was selected as a threshold for eccentricity. Each object that did not meet the eccentricity requirement for roundness (eccentricity>0.5) was then subject to an incremental increase in intensity thresholding. This process was repeated until all objects were either deemed to be round enough to be lipid droplets or small enough to be considered debris. An object was determined to be too small if it was 4 or fewer connected pixels. A 4-pixel minimum limit was established because that was determined to be the smallest droplet size where eccentricity was still an accurate measure of roundness. The end result of this step in the lipid droplet segmentation is illustrated in Fig. 2e.
Figure 2.
Image Processing Workflow. a) Unprocessed images (red= FAD, green= NADH, blue= THG). b) Manually identified cells. c) Intracellular THG signal used for lipid segmentation. d) THG image after intensity thresholding. e) Small droplet segmentation. f) 2-step large droplet segmentation- inverse fluorescence auto-segmentation with manual adjustment aided by THG, g) Merged (i.e. small and large) lipid mask. h) Redox color-coded image superimposed on THG-based lipid image. i) Redox color-coded and THG-based lipid 3D projection. j) Schematic representation of image processing workflow
For larger cells, an additional step in the lipid segmentation process was required. Due to the homogeneity of third-order susceptibility within lipid droplets, no THG signal was detected from the center of larger droplets. As a result, larger cells (volume greater than 80,000 pixels) were subject to an extra processing step (Fig. 2f). Specifically, we generated images that represented the inverse of the fluorescence signal within cell borders by inverting the signal intensity of 4 fluorescence excitation/emission wavelength pairs- 750 nm/450 nm, 750 nm/582 nm, 860 nm/450 nm and 860 nm/582 nm- and averaging their values on a per-pixel basis (Fig. 2f, top left subpanel). We then combined the average inverse fluorescence image with THG segmented images, resulting in the right subpanel of Fig. 2f. Lipid droplets segmented using inverse fluorescence signal were subjected to a larger minimum droplet size (20 pixels) in order to minimize detection of non-lipid structures within the inverse fluorescence images. The overall lipid droplet mask for the optical section is presented in Fig. 2g. The average NADH and FAD fluorescence for each cell was estimated based on the mean intensity of the 750/450nm and 860/582nm channels, respectively, along with the corresponding redox ratio (defined as FAD/(NADH+FAD)). Finally, cells were color-coded according to their redox ratios as shown in Fig. 2h for a single section and for the entire volume, shown as a depth-projection in Fig. 2i. Image stacks were also visualized as 3D projections using Image J Software (NIH, USA) and Osirix Imaging Software (Pixmeo, Geneva, Switzerland). Specifically, the output images from Matlab were assembled into TIFF stacks in Image J and then exported to Osirix for 3D rendering. Cells within silk scaffolds were color-coded with the jet color map based on redox ratio values within a fixed range of 0.005 to 0.2, so that image volumes from different time points and groups can be compared visually. A schematic representation of the image processing workflow is included in Fig. 2j.
2.5 Statistical analysis
To assess the significance of changes in redox ratio and lipid content between experimental groups and between time points of the study, statistical analysis was performed in JMP 10 software (SAS, Cary, NC, USA). Differences between culture groups within a time point (t-tests) and across time points were assessed using analysis of variance (ANOVA). Tukey HSD tests were performed for lipid content metrics to evaluate significance between each time point/culture group pairing. Significance was calculated at the α=0.05 level. Correlations were evaluated between redox ratio and several measured lipid content metrics and assessed for significance.
3. Results
3.1 Extraction of functional features of 3D engineered tissues from 2PEF and THG images
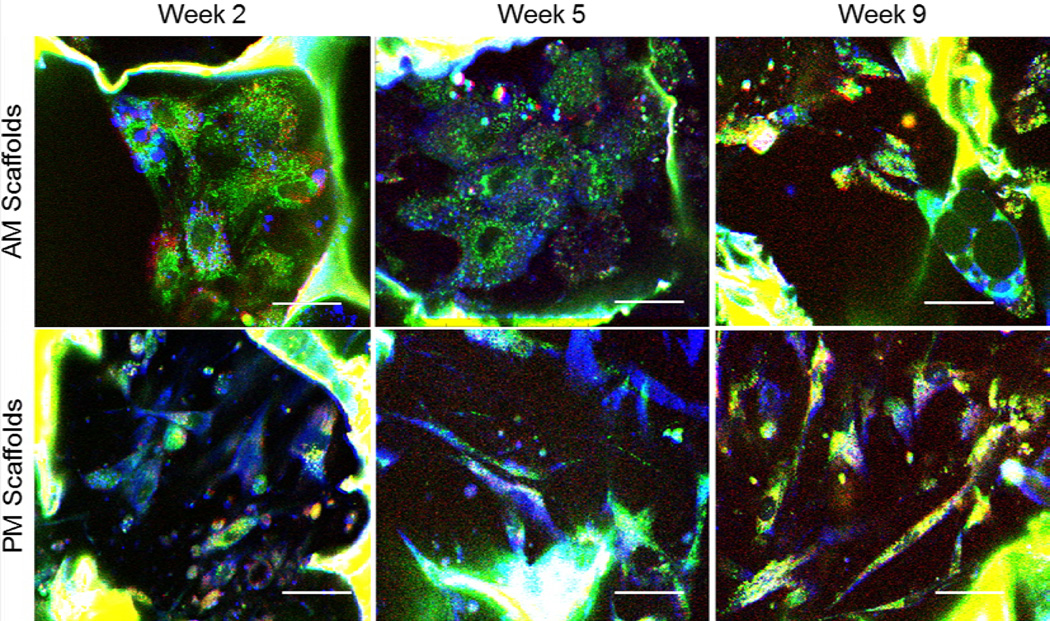
A total of 251 cells were manually identified from a total of 6 image volumes taken at weeks 2, 5, and 9 from the same stem cell embedded silk scaffolds exposed to either adipogenic (AM) (30, 19, and 30 cells, respectively) or propagation medium (PM) (24, 47, 101 cells, respectively). Representative sections of the detected 2PEF channels corresponding primarily to NADH (green) and FAD (red) fluorescence are shown along with the corresponding THG images (blue) from the AM and PM scaffold at each time point (Fig. 3 and Supplemental video). Silk scaffolds are readily visible in the 2PEF images due to silk strong fluorescence and in the THG images, where only pore edges are visible due to the contrast mechanism of THG microscopy. In addition, the THG images reveal the presence of small lipid droplets in the form of bright intracellular round structures with strong THG signal (particularly visible in the AM-treated scaffolds for the week 2 and 5 time-points), as well as larger droplets in the form of hollow round features (see Fig. 3), particularly in week 9 images of the AM scaffolds. Cells within the PM scaffold appear to have the characteristic elongated, undifferentiated morphology throughout the 9 weeks of culture.
Figure 3.
Endogenous TPEF and THG images can be overlaid to co-localize lipid droplets within cells. The same scaffold is imaged after 2, 5 or 9 weeks in either adipogenic (top row) or propagation (bottom row) media. Green=NADH fluorescence, Red= FAD fluorescence, Blue= third harmonic generation. Scale bar= 30µm
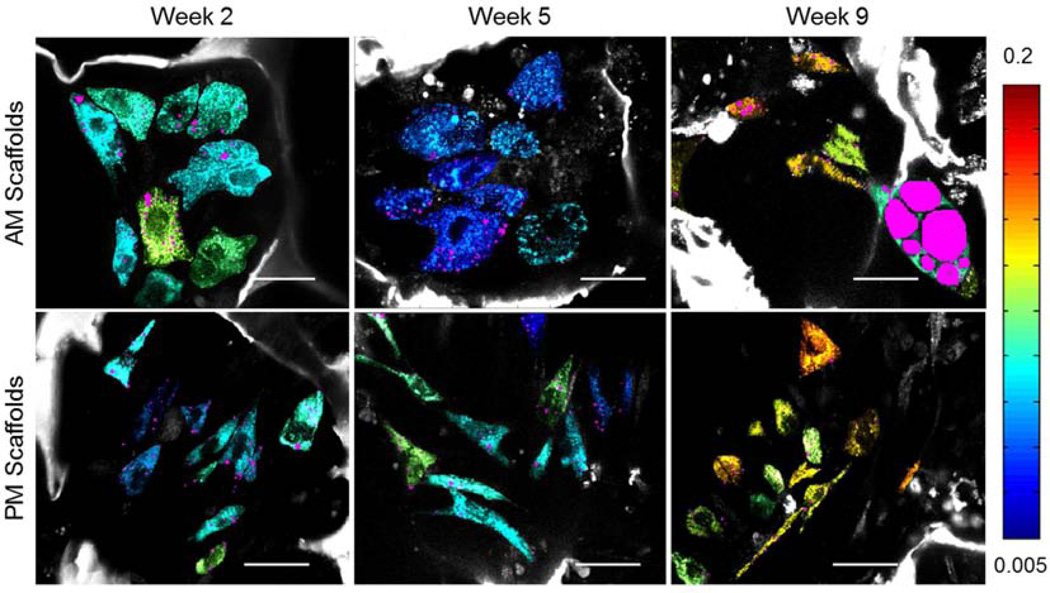
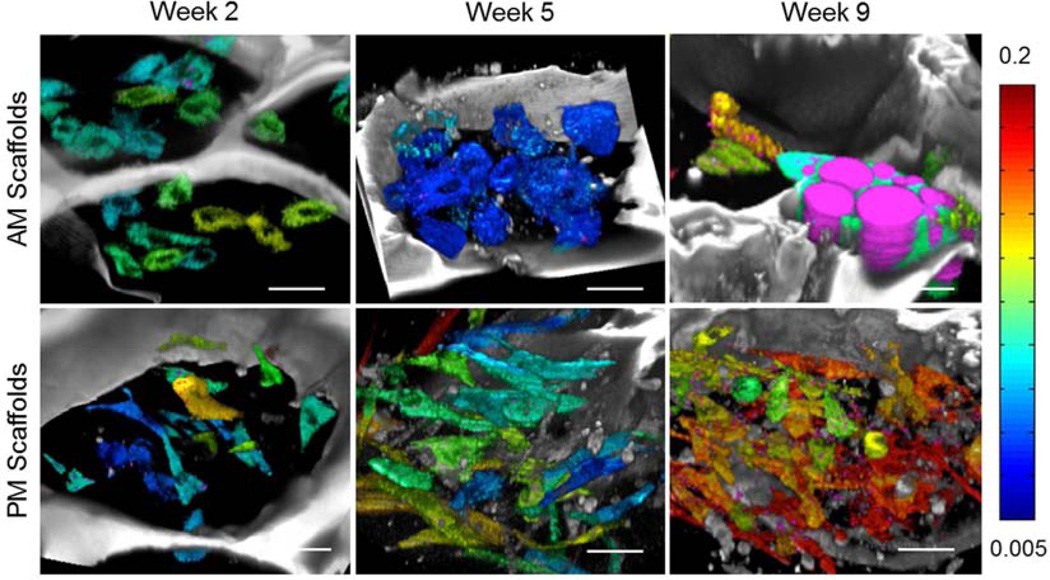
The average value of NADH and FAD fluorescence was extracted for each individual cell and the redox ratio was calculated as FAD/(NADH+FAD). Corresponding redox colored cells are shown in Fig. 4 (jet colormap) along with objects identified as lipid droplets (magenta) based on the approach outlined in Fig. 2 and described in the Methods section (see also Supplemental videos). 3D reconstructions of the examined volumes are included in Fig. 5 and Supplemental videos. Significant changes in redox as a function of time are clearly apparent for the cells within the scaffold undergoing adipogenic differentiation, with little change in cell number per scaffold pore. In contrast, cell proliferation is evident within the PM scaffold, where it is associated with an increased redox ratio at week 9.
Figure 4.
Redox-based color-coded images extracted based on endogenous TPEF images are superimposed on THG-based lipid content (magenta) images. The same scaffold is imaged after 2, 5 or 9 weeks in either adipogenic (top row) or propagation (bottom row) media. Scale Bar= 35µm. Redox scale color bar is shown on the right. Endogenous silk fluorescence is shown in grayscale.
Figure 5.
3-D reconstructions of silk scaffolds (grayscale) embedded with hMSCs (jet color map) and exposed to either adipogenic (top row) or propagation (bottom row) media for 2, 5 or 9 weeks. Different hues within the jet color map (shown on right) represent different cellular redox ratios estimated based on endogenous TPEF images. Lipid droplets identified based on analysis of corresponding THG images are shown in magenta.
3.2 Quantitative assessment of metabolic status during differentiation using the optical redox ratio
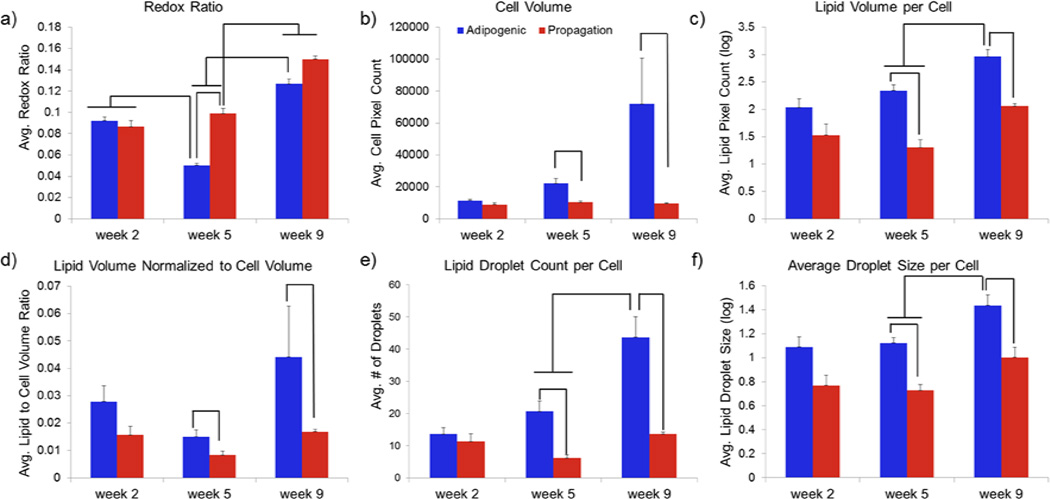
The dynamically changing redox trends illustrated in Figs. 4 and 5 are summarized in Fig. 6a. The overall redox ratio of the two scaffolds was similar during the initial time point. However, the redox ratio of the AM scaffold was significantly lower than that of the PM one for the 5 (p<0.0001) and 9 (p<0.05) week time-points. Additionally, we observed a significant decrease in redox ratio in the AM group between weeks 2 and 5, followed by a significant increase between weeks 5 and 9 (p<0.0001). A significant increase in redox was detected also for the PM scaffold during week 9 (p<0.0001).
Figure 6.
Dynamic changes in redox ratio and lipid content in 3D silk scaffolds embedded with hMSCs and treated with either adipogenic (blue) or propagation (red) media. Mean a) redox ratio per cell b) cell volume (pixels) c) Lipid volume per cell d) lipid to cell volume ratio e) lipid droplet number per cell and f) droplet size per cell. Error bars represent standard errors of the mean. Significant differences noted in graphs are evaluated at p<0.05.
3.3 Quantitative assessment of lipid biosynthesis during stem cell differentiation
We extracted a number of morphological metrics, based on the identification of the cell and lipid droplet borders, as shown in Fig. 6b-f. Consistent with the visual representations in Figs. 4 and 5, the average cell volume, as assessed by the number of pixels within each identified cell, did not change as a function of time within the volume examined in the PM scaffold (Fig. 6b). In contrast, while the AM scaffold cells had similar volume to that of the PM scaffold cells at week 2, they became significantly larger at weeks 5 and 9 (p<0.05) (Table 1). Interestingly, the standard deviation in observed cell volumes also increased as a function of time in the AM scaffolds, indicating the presence of cells at various stages of differentiation (Table 1). Non-invasive imaging modalities are uniquely capable to provide this type of information.
Table 1.
Means and standard deviations of the cell volumes (in pixels) in the adipogenic medium (AM) and propagation medium (PM) groups.
| AM | PM | |
|---|---|---|
| Week 2 | 11555±3755 | 8905±5275 |
| Week 5 | 22290±12603 | 10379±5490 |
| Week 9 | 71926±156830 | 9574±4014 |
The average lipid content per cell changed only moderately over time in the PM scaffold (Fig. 6c). In contrast, a significant increase in lipid content was detected at weeks 5 and 9 for the AM scaffolds relative to the corresponding PM scaffold time points (p< 0.0001). A significant increase in lipid content was also detected between the week 5 and week 9 AM scaffold time points (p<0.05). Furthermore, the lipid-to-cell volume ratio was higher in the AM than the PM scaffolds during week 5 and 9 (p< 0.05) (Fig. 6d). In addition, we observed a small decrease in the lipid-to-cell volume ratio in both PM and AM scaffolds between week 2 and week 5. This observation suggests that cell volume initially increases at a larger rate than the volume occupied by lipids. However, this effect is limited and the observation requires further validation. Finally, the lipid-to-cell volume score identified the most prominent differences between AM- and PM-treated scaffolds at week 9.
To further characterize the lipid accumulation within each cell of the scaffolds examined, we quantified the average number (Fig. 6e) and volume (Fig. 6f) of the identified lipid droplets. We found that both the number and average size of lipid droplets in each cell increased significantly over time in the AM-treated scaffold. Droplet numbers did not change significantly within the PM group and differences in average droplet size were observed only when comparing week 2 and 9 PM image data. In addition, both metrics were higher for the AM than the PM scaffolds with differences between the two groups becoming significant (p<0.05) between weeks 5 and 9.
3.4 Correlations between lipid content and redox ratio
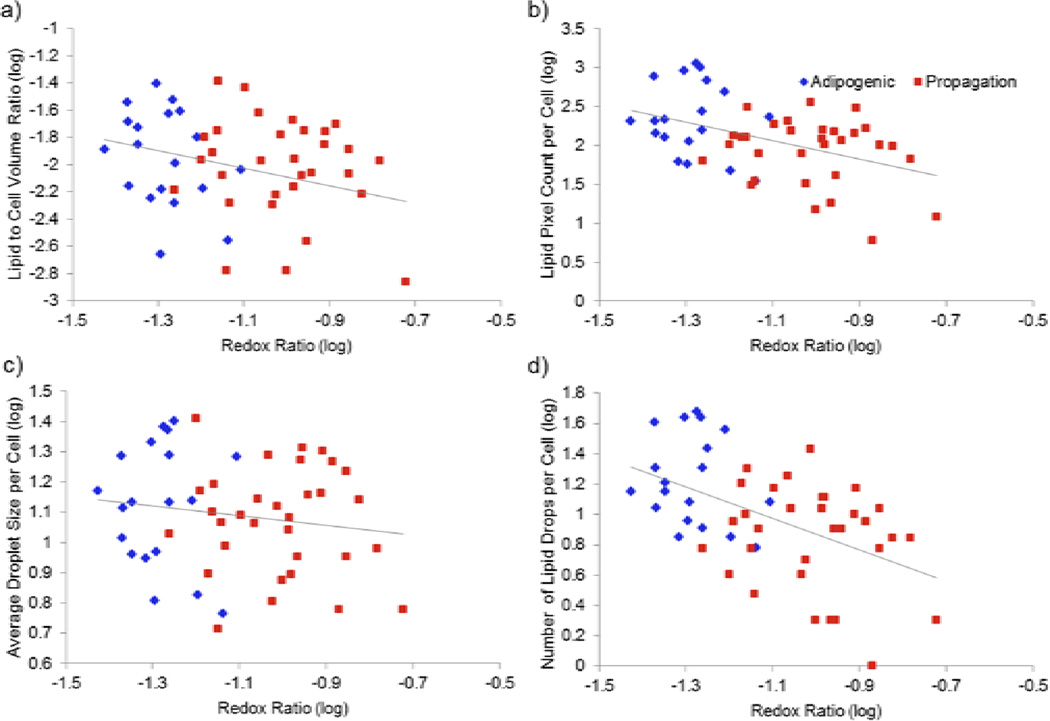
To evaluate possible links between cell metabolism and lipid biosynthesis, we examined correlations between the extracted redox ratio and the various lipid content and droplet metrics. Significant inverse correlations were identified between the redox ratio and the lipid to cell volume ratio, the overall lipid content and the number of droplets per cell, for the data collected during week 5 and 9 (Table 2). There was not a significant correlation between redox ratio and the average droplet size per cell for any of the time points other than week 9. Correlations were generally stronger during week 5 than during week 9 with the lipid content and number of droplets exhibiting more negative correlation coefficient values than the average droplet size.
Table 2.
Correlation coefficients (R) and their significance (p values) between cell redox ratio and various lipid metrics at each time point.
| Lipid metric | Week 2 | Week 5 | Week 9 | |||
|---|---|---|---|---|---|---|
| R | p | R | p | R | p | |
| Lipid to Cell Volume Ratio | −0.28 | 0.053 | −0.17 | 0.042* | −0.39 | <0.0001* |
| Lipid Pixel Count per Cell | −0.19 | 0.21 | −0.44 | 0.001* | −0.37 | <0.0001* |
| Average Droplet Size per Cell | −0.14 | 0.35 | −0.16 | 0.28 | −0.40 | 0.005* |
| Number of Droplets per Cell | −0.18 | 0.23 | −0.50 | 0.0002* | −0.40 | <0.0001* |
4. Discussion
The ability to non-invasively assess key cell functional characteristics, such as metabolism and lipid biosynthesis, could have broad and significant implications not only in the context of tissue engineering and regenerative medicine, but also for a number of afflictions, such as diabetes, cancer and cardiovascular disease. In this study, we presented initial investigations that demonstrate the potential of such measurements in three dimensional engineered tissues consisting of silk biomaterial scaffolds embedded with stem cells undergoing adipogenic differentiation or exposed to propagation media. Specifically, we relied on endogenous NADH and FAD signals acquired through 2PEF to detect and quantify the cells’ redox ratio, and on the intrinsic THG signal of lipids to assess the progression of lipid droplet formation. We detected significant levels of heterogeneity among individual stem cells, especially ones progressing towards adipocytes, illustrating the highly dynamic and variable process of differentiation. We reported dynamic changes in the redox ratio as lipid droplets are forming and growing larger and we identified how these changes are correlated over time, which provides unique insights into these complex metabolic pathways within tissues without disturbing development.
Previous work has demonstrated that stem cells undergoing adipogenic differentiation initially exhibit a decrease in the 2PEF-based redox ratio, while propagating MSCs exhibit a measurable increase in redox ratio over time [6, 10]. Consistently, the significant increase in redox ratio measured between weeks 5 and 9 in PM scaffold cells may be related to growth arrest upon cells reaching confluence in the silk scaffolds (Fig. 5, bottom right panel). Furthermore, a significant decrease in redox ratio was observed in the current study between weeks 2 and 5 (p<0.0001) in the AM scaffold, and a significantly lower redox ratio compared to the PM scaffold was detected in the adipogenic group at weeks 5 and 9 (Fig. 6a). In previous work it was suggested that this decrease in redox ratio was associated with increased flux through mitochondrial pathways associated with de novo fatty acid synthesis [10]. Specifically, NADH accumulation by increased pyruvate dehydrogenase activity was proposed to outpace ATP demands and NADH oxidation by the electron transport chain. However, without the ability to non-destructively measure lipid droplet accumulation in previous work, Oil Red O staining was employed at specific time points to understand the relationships among cell redox ratio, fatty acid synthesis, and lipid droplet development [6, 10]. This study offers the first non-invasive 3D assessments of lipid droplet composition within engineered human adipose tissue using label-free non-linear microscopy, which facilitates comparisons among cell redox state and lipid metabolism on a cell-by-cell basis. The significant negative correlation among lipid metrics and redox ratio among cells within individual time points (Fig. 7) supports our previous hypothesis linking redox ratio decreases with fatty acid synthesis and lipid droplet accumulation.
Figure 7.
The redox ratio of individual hMSCs growing in a silk scaffold and treated with either adipogenic (blue) or propagation (red) media for 5 weeks is shown relative to the same cell’s a) lipid to cell volume ratio, b) lipid pixel count, c) average droplet size, and d) number of lipid droplets.
Previous multi-modal imaging studies have demonstrated the ability of THG, CARS or SRS to effectively isolate lipid droplets within cells or tissues while being compatible with 2PEF [16, 44]. THG requires a less complex optical setup compared to CARS/SRS since signal is obtained from a single excitation beam. However, since THG can detect objects other than intracellular lipid droplets (albeit usually with lower signals) and because it highlights only the boundaries of objects larger than the excitation wavelength, the development of image processing techniques to isolate specific organelles like lipid structures is all the more critical for the use of THG in quantitative imaging approaches. Due to the challenges in isolating lipid droplet signals from other interfaces, and the lack of THG signal in the center of larger droplets, no THG studies, up to this point, have quantified lipid droplet accumulation. Using CARS, Brackmann et al. explored the relationship between lipid droplet size and frequency in yeast cultures under different metabolic conditions [18]. While only small differences in lipid size were observed between the induction of fermentative and respiratory metabolism, large increases in the average lipid droplet size, frequency, and heterogeneity within cultures were measured after adaptation to long-term starvation [18]. While that study relied on specific media conditions and cell types to understand the relationship between droplet organization and metabolism, the current study can simultaneously provide data on both metabolic pathway utilization and its effects on lipid droplet accumulation through 2PEF-based redox ratios of metabolic cofactors and THG-based lipid metrics (Fig. 6).
Through correlations among redox ratio and lipid metrics at individual time points, it is evident that increased lipid droplet frequency and size are associated with lower redox ratios and AM scaffolds (Figs. 6 and 7), but additional insight into lipid metabolism can be gained through comparisons between time points. Specifically, the AM redox ratio reaches a minimum at week 5, which corresponds to a period of the lowest lipid-to-cell volume. This effect may be related to the previously reported occurrence of mitochondrial biogenesis [10, 45]. The increased cell volume at week 5 in the current study suggests that mitochondrial biogenesis may occur during the initial weeks following differentiation in order to facilitate fatty acid synthesis and increased oxidative phosphorylation. This increase in mitochondrial biogenesis relative to lipid droplets and minimum redox ratio at week 5 may correspond to peak de novo fatty acid synthesis within the AM scaffold. The eventual storage of these fatty acids into large lipid droplets is detected in week 9, by the increase in lipid volume per cell, increased lipid droplet frequency, and increased average droplet size (Fig. 6). The increase in average droplet size and reduced flux through lipid synthesis pathways, as measured by the increase in redox ratio, between weeks 5 and 9 in the AM scaffold (Fig. 6a) suggests that the maturing adipocytes are reducing fatty acid production and lipid droplet fusion is initiating. Indeed, previous work has established that new droplet formation decreases and droplet fusion increases as differentiating adipocytes mature [46, 47]. Furthermore previous long-term studies of the redox ratio suggested an increase in redox ratio and decrease in new droplet formation after week 8 [10]. Tracking individual cells over time in future work with a THG/2PEF system may help to elucidate the dynamic changes in mitochondrial structure and function and their relationship to lipid droplet formation and fusion.
The quantitative lipid metrics developed in this study, when coupled with the redox ratio, serve as quantitative biomarkers to measure lipogenesis and help characterize engineered adipose tissue development, but these methods have broader applicability. Assessments of adipocyte metabolism in vivo will benefit from these lipid droplet measures when de novo fatty acid synthesis is not as necessary for lipid droplet formation. In the case of reorganization of existing fat in the body, measuring lipid content has the potential to provide a more sensitive biomarker than perhaps redox ratio due to lack of de novo lipogenesis. Furthermore, while this study focuses on lipogenesis upon adipogenic differentiation, these techniques are also sensitive to the increases in droplet fission associated with lipolysis [48]. The inclusion of these THG metrics is particularly important given the production of NADH during both de novo fatty acid synthesis during lipogenesis and beta oxidation of fatty acids during lipolysis. Additionally, this automated method to measure lipid droplet accumulation may have applicability for the diagnosis of alcoholic liver disease, diabetes, or cancer [3, 49, 50]. In summary, by combining label-free non-invasive, quantitative assessments of both cell metabolism and lipid droplet characteristics using endogenous sources of contrast through multi-modal 2PEF/THG imaging system, we provide a powerful set of tools to monitor functional development over time within individual engineered 3D tissues.
5. Conclusion
This study demonstrates a semi-automated method that integrates 2PEF-based metabolic readouts with THG-based lipid droplet metrics, to provide quantitative cell-by-cell statistics for characterizing 3D engineered adipose tissue development. Sensitivity to adipogenesis is demonstrated by monitoring individual silk scaffolds seeded with MSCs that were cultured in either adipogenic differentiation or MSC propagation media over a period of 9 weeks. We observed a decrease in redox ratio of the AM scaffold between weeks 2 and 5, followed by an increase between weeks 5 and 9, indicative of maturing engineered adipose tissue. These metabolic changes corresponded to increases in lipid pixel count per cell, average lipid droplet size per cell, and number of lipid droplets per cell at weeks 5 and 9 in the AM group. These findings support our working hypothesis that the decrease in redox ratio during adipogenic differentiation is associated with fatty acid synthesis and lipid droplet accumulation. By establishing metrics to quantify both metabolism and the organization of lipid droplets, we can measure both the rate of lipogenesis and its cumulative effects to more fully characterize the development of 3D engineered tissues. These quantitative, high-resolution non-linear optical microscopy methods can enable dynamic assessments of tissues without the need for sample destruction, which hold promise for improving the rate of in vitro protocol optimization and assessing the status of engineered implants in vivo.
Supplementary Material
V1: Sequential 2D optical sections within an image volume showing the 2PEF and THG intensities from a silk scaffold embedded with hMSCs treated with adipogenic differentiation media for 9 weeks.
V2: Sequence of 2D sections demonstrating individual cell redox ratios and lipid droplets (magenta) that were extracted from recorded 2PEF and THG images, respectively. Silk scaffold and other non-segmented regions are displayed in grayscale. See Figure 4 for the “jet” colormap used to define cell redox ratios.
V3: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with adipogenic media for two weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 360x290x42 µm.
V4: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with propagation media for two weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 260x240x60 µm.
V5: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with adipogenic media for five weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 180x180x32 µm.
V6: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with propagation media for five weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 250x235x78 µm.
V7: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with adipogenic media for nine weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 480x450x100 µm.
V8: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with propagation media for nine weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 320x320x60 µm.
Acknowledgements
This research was supported by NIBIB/NIH under grant numbers R01EB007542 and P41EB002520, by NIAMS/NIH under grant number F32AR061933, and by Agence Nationale de la Recherche under contracts ANR-09-PIRI-0013 and ANR-10-INBS-04. We thank Marie-Claire Schanne-Klein, Willy Supatto, Delphine Débarre, and Pierre Mahou for scientific discussions on nonlinear microscopy of tissues. We also thank Aneta Czajkowska and Evangelia Bellas for assistance with the tissue engineering protocols.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kershaw E, Flier J. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab. 2004;80(6):2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- 2.Ren J, Pulakat L, Whaley-Connell A, Sowers J. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J Mol Med. 2010;88:993–1001. doi: 10.1007/s00109-010-0663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, et al. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121(6):2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice WL, Kaplan DL, Georgakoudi I. Quantitative biomarkers of stem cell differentiation based on intrinsic two-photon excited fluorescence. J Biomed Opt. 2007;12(6):060504. doi: 10.1117/1.2823019. [DOI] [PubMed] [Google Scholar]
- 6.Rice WL, Kaplan DL, Georgakoudi I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One. 2010;5(4):e10075. doi: 10.1371/journal.pone.0010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgakoudi I, Rice WL, Hronik-Tupaj M, Kaplan DL. Optical spectroscopy and imaging for the noninvasive evaluation of engineered tissues. Tissue Eng Part B Rev. 2008;14(4):321–340. doi: 10.1089/ten.teb.2008.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgakoudi I, Quinn KP. Optical imaging using endogenous contrast to assess metabolic state. Annu Rev Biomed Eng. 2012;14:351–367. doi: 10.1146/annurev-bioeng-071811-150108. [DOI] [PubMed] [Google Scholar]
- 9.Stringari C, Sierra R, Donovan PJ, Gratton E. Label-free separation of human embryonic stem cells and their differentiating progenies by phasor fluorescence lifetime microscopy. J Biomed Opt. 2012;17(4):046012. doi: 10.1117/1.JBO.17.4.046012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn KP, Bellas E, Fourligas N, Lee K, Kaplan DL, Georgakoudi I. Characterization of metabolic changes associated with the functional development of 3D engineered tissues by non-invasive, dynamic measurement of individual cell redox ratios. Biomaterials. 2012;33:5341–5348. doi: 10.1016/j.biomaterials.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinegoni C, Ralston T, Tan W, Luo W, Marks DL, Boppart SA. Integrated structural and functional optical imaging combining spectral-domain optical coherence and multiphoton microscopy. Appl Phys Lett. 2006;88(5) [Google Scholar]
- 12.König K, Uchugonova A. Two-photon autofluorescence and second-harmonic imaging of adult stem cells. J Biomed Opt. 2008;13(5):054068. doi: 10.1117/1.3002370. [DOI] [PubMed] [Google Scholar]
- 13.Yannas IV, Tzeranis DS, Harley BA. So PTC. Biologically active collagen-based scaffolds: advances in processing and characterization. Philos Trans A Math Phys Eng Sci. 2010;368(1917):2123–2139. doi: 10.1098/rsta.2010.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JK, Kim UJ, Leisk GG, Bayan C, Georgakoudi I, Kaplan DL. Bone regeneration on macroporous aqueous-derived silk 3-D scaffolds. Macromol Biosci. 2007;7:643–655. doi: 10.1002/mabi.200700030. [DOI] [PubMed] [Google Scholar]
- 15.Wilson BC, Vitkin IA, Matthews DL. The potential of biophotonic techniques in stem cell tracking and monitoring of tissue regeneration applied to cardiac stem cell therapy. J Biophotonics. 2009;11:669–681. doi: 10.1002/jbio.200910079. [DOI] [PubMed] [Google Scholar]
- 16.Débarre D, Supatto W, Pena AM, Fabre A, Tordjmann T, Combettes L, et al. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat Methods. 2006;3(1):47–53. doi: 10.1038/nmeth813. [DOI] [PubMed] [Google Scholar]
- 17.Chu SW, Chen SY, Tsai TH, Liu TM, Lin CY, Tsai HJ, et al. In vivo developmental biology study using noninvasive multi-harmonic generation microscopy. Opt Express. 2003;11(23):3093–3098. doi: 10.1364/oe.11.003093. [DOI] [PubMed] [Google Scholar]
- 18.Brackmann C, Norbeck J, Akson M, Bosch D, Larsson C, Gustafsson L, et al. CARS microscopy of lipid stores in yeast: the impact of nutritional state and genetic background. J Raman Spectrosc. 2009;40:748–756. [Google Scholar]
- 19.Brackmann C, Bodin A, Akeson M, Gatenholm P, Enejder A. Visualization of the cellulose biosynthesis and cell integration into cellulose scaffolds. Biomacromolecules. 2010;11:542–548. doi: 10.1021/bm901153t. [DOI] [PubMed] [Google Scholar]
- 20.Le TT, Cheng JX. Single-cell profiling reveals the origin of phenotypic variability in adipogenesis. PloS One. 2009;4:e5189. doi: 10.1371/journal.pone.0005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slipchenko MN, Le TT, Chen H, Cheng JX. High-speed vibrational imaging and spectral analysis of lipid bodies by compound Raman microscopy. J Phys Chem B. 2009;113:7681–7686. doi: 10.1021/jp902231y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palero JA, de Bruijn HS, van der Ploeg van den Heuvel A, Sterenborg HJ, Gerritsen HC. Spectrally resolved multiphoton imaging of in vivo and excised mouse skin tissues. Biophys J. 2007;93:992–1007. doi: 10.1529/biophysj.106.099457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21(11):1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 24.Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri K, White JG, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A. 2007;104(49):19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xylas J, Quinn KP, Hunter M, Georgakoudi I. Improved Fourier-based characterization of intracellular fractal features. Opt Express. 2012;20(21):23442–23455. doi: 10.1364/OE.20.023442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitt J, Hunter M, Mujat C, McLaughlin-Drubin M, Münger K, Georgakoudi I. Diagnostic cellular organization features extracted from autofluorescence images. Opt Lett. 2007;32(22):3305–3307. doi: 10.1364/ol.32.003305. [DOI] [PubMed] [Google Scholar]
- 27.Levitt JM, McLaughlin-Drubin ME, Münger K, Georgakoudi I. Automated biochemical, morphological, and organizational assessment of precancerous changes from endogenous two-photon fluorescence images. PloS One. 2011;6(9):e24765. doi: 10.1371/journal.pone.0024765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright BK, Andrews LM, Markham J, Jones MR, Stringari C, Digman MA, et al. NADH distribution in live progenitor stem cells by phasor-fluorescence lifetime image microscopy. Biophys J. 2012;103:L7–L9. doi: 10.1016/j.bpj.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gratton E, Breusegem S, Sutin J, Ruan Q, Barry N. Fluorescence lifetime imaging for the two-photon microscope: time-domain and frequency-domain methods. J Biomed Opt. 2003;8(3):381–390. doi: 10.1117/1.1586704. [DOI] [PubMed] [Google Scholar]
- 30.Konig K, Uchugonova A, Gorjup E. Multiphoton fluorescence lifetime imaging of 3D-stem cell spheroids during differentiation. Microsc Res Tech. 2011;74:9–17. doi: 10.1002/jemt.20866. [DOI] [PubMed] [Google Scholar]
- 31.Barad Y, Eisenberg H, Horowitz M, Silberberg Y. Nonlinear scanning laser microscopy by third harmonic generation. Appl Phys Lett. 1997;70(8):922–924. [Google Scholar]
- 32.Oron D, Yelin D, Tal E, Raz S, Fachima R, Silberberg Y. Depth-resolved structural imaging by third-harmonic generation microscopy. J Struct Biol. 2004;147:3–11. doi: 10.1016/S1047-8477(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 33.Débarre D, Supatto W, Farge E, Moulia B, Schanne-Klein MC, Beaurepaire E. Velocimetric third-harmonic generation microscopy: micrometer-scale quantification of morphogenetic movements in unstained embryos. Opt Lett. 2004;29(24):2881–2883. doi: 10.1364/ol.29.002881. [DOI] [PubMed] [Google Scholar]
- 34.Tai SP, Lee WJ, Shieh DB, Wu PC, Huang HY, Yu CH, Sun CK. In vivo optical biopsy of hamster oral cavity with epi-third-harmonic-generation microscopy. Opt Express. 2006;14(13):6178–6187. doi: 10.1364/oe.14.006178. [DOI] [PubMed] [Google Scholar]
- 35.Olivier N, Luengo-Oroz MA, Duloquin L, Faure E, Savy T, Veilleux I, et al. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science. 2010;329(5994):967–971. doi: 10.1126/science.1189428. [DOI] [PubMed] [Google Scholar]
- 36.Witte Stefan, Negrean Adrian, Lodder Johannes C, De Kock Christiaan P J, Silva Guilherme Testa, Mansvelder Huibert D, Groot Marie Louise. Label-free live brain imaging and targeted patching with third-harmonic generation microscopy. Proc Natl Acad Sci U S A. 2011;108(15):5970–5975. doi: 10.1073/pnas.1018743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Débarre D, Beaurepaire E. Quantitative characterization of biological liquids for third-harmonic generation microscopy. Biophys J. 2007;92(2):603–612. doi: 10.1529/biophysj.106.094946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrar MJ, Wise FW, Fetcho JR, Schaffer C. In vivo imaging of myelin in the vertebrate central nervous system using third-harmonic generation microscopy. Biophys J. 2011;100(5):1362–1371. doi: 10.1016/j.bpj.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmerley M, Mahou P, Débarre D, Schanne-Klein MC, Beaurepaire E. Probing ordered lipid assemblies with polarized third-harmonic generation microscopy. Phys Rev X. 2013;3:011002. [Google Scholar]
- 40.Aptel F, Olivier N, Deniset-Besseau A, Legeais JM, Plamann K, Schanne-Klein MC, et al. Multimodal nonlinear imaging of the human cornea. Invest Opthalmol Vis Sci. 2010;51(5):2459–2465. doi: 10.1167/iovs.09-4586. [DOI] [PubMed] [Google Scholar]
- 41.Mahou P, Zimmerley M, Loulier K, Matho KS, Labroille G, Morin X, et al. Multicolor two-photon tissue imaging by wavelength mixing. Nat Methods. 2012;9:815–818. doi: 10.1038/nmeth.2098. [DOI] [PubMed] [Google Scholar]
- 42.Bellas E, Panilaitis BJB, Glettig DL, Kirker-Head CA, Yoo JJ, Marra KG, et al. Sustained volume retention in vivo with adipocyte and lipoaspirate seeded silk scaffolds. Biomaterials. 2013;34:2960–2968. doi: 10.1016/j.biomaterials.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mauney JR, Nguyen T, Gillen K, Kirker-Head C, Gimble JM, Kaplan DL. Engineering adipose-like tissue in vitro and in vivo utilizing human bone marrow and adipose-derived mesenchymal stem cells with silk fibroin 3D scaffolds. Biomaterials. 2007;28:5280–5290. doi: 10.1016/j.biomaterials.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Slipchenko MN, Zhu J, Buhman KK, Cheng JX. Coupling CARS with multiphoton fluorescence and high harmonic generation imaging modalities using a femtosecond laser source. Proc SPIE. 2009;7183:718315. [Google Scholar]
- 45.Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114(9):1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24(3):109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- 47.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 48.Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791(6):459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bozza PT, Viola JPB. Lipid droplets in inflammation and cancer. Prostaglandins Leukot Essent Fatty Acids. 2010;82(4–6):243–250. doi: 10.1016/j.plefa.2010.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
V1: Sequential 2D optical sections within an image volume showing the 2PEF and THG intensities from a silk scaffold embedded with hMSCs treated with adipogenic differentiation media for 9 weeks.
V2: Sequence of 2D sections demonstrating individual cell redox ratios and lipid droplets (magenta) that were extracted from recorded 2PEF and THG images, respectively. Silk scaffold and other non-segmented regions are displayed in grayscale. See Figure 4 for the “jet” colormap used to define cell redox ratios.
V3: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with adipogenic media for two weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 360x290x42 µm.
V4: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with propagation media for two weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 260x240x60 µm.
V5: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with adipogenic media for five weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 180x180x32 µm.
V6: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with propagation media for five weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 250x235x78 µm.
V7: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with adipogenic media for nine weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 480x450x100 µm.
V8: 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with propagation media for nine weeks. The jet color map is used to represent different redox ratios calculated based on recorded endogenous 2PEF images. Lipid droplets detected in corresponding THG images are shown in magenta. Stack size: 320x320x60 µm.