Summary
Antiviral vaccines have been the most successful biomedical intervention for preventing epidemic viral disease. Vaccination for smallpox in humans and rinderpest in cattle was the basis for disease eradication, and recent progress in polio eradication is promising. While early vaccines were developed empirically by passage in live animals or eggs, more recent vaccines have been developed because of the advent of new technologies, particularly cell culture and molecular biology. Recent technological advances in gene delivery and expression, nanoparticles, protein manufacturing, and adjuvants have created the potential for new vaccine platforms that may provide solutions for vaccines against viral pathogens for which no interventions currently exist. In addition, the technological convergence of human monoclonal antibody isolation, structural biology, and high throughput sequencing is providing new opportunities for atomic-level immunogen design. Selection of human monoclonal antibodies can identify immunodominant antigenic sites associated with neutralization and provide reagents for stabilizing and solving the structure of viral surface proteins. Understanding the structural basis for neutralization can guide selection of vaccine targets. Deep sequencing of the antibody repertoire and defining the ontogeny of the desired antibody responses can reveal the junctional recombination and somatic mutation requirements for B-cell recognition and affinity maturation. Collectively, this information will provide new strategic approaches for selecting vaccine antigens, formulations, and regimens. Moreover, it creates the potential for rational vaccine design and establishing a catalogue of vaccine technology platforms that would be effective against any given family or class of viral pathogens and improve our readiness to address new emerging viral threats.
Keywords: viral immunity, vaccine development, monoclonal antibodies, neutralizing antibody, structure-based vaccine design, vaccines
Introduction
We are in a transitional time for antiviral vaccines. There are currently licensed vaccines that protect against 15 viral diseases (Table 1), but none have been licensed since the human papilloma virus (HPV) virus-like particle (VLP) in 2006. For the remaining emerging and re-emerging viruses for which vaccine development is a high public health priority, it may be necessary to exploit new technologies that identify critical antigenic sites, induce potent and targeted T-cell responses, and develop novel delivery systems. In addition, more creative options for manufacturing and distribution should be explored, particularly for viruses where the disease burden is greatest in resource-poor settings. In this review, I describe some examples of how new technologies are advancing vaccine design and production, guiding future development options, or opening avenues for rapid responses to emerging viruses. This is not intended to be an exhaustive list of recent achievements, but rather the presentation of selected stories and thoughts to generate curiosity and new ideas.
Table 1.
Licensed antiviral vaccines in the United States
| Virus | Number of serotypes covered by vaccine | Year of licensure | Platform | Technology required |
|---|---|---|---|---|
| Yellow Fever | 1 | 1938/1961 | Live-attenuated | Live animal passage/Egg culture |
| Smallpox | 1 | 1944/2007 | Live-attenuated | Live animal passage/Cell culture |
| Influenza A and B | 3 (H1N1, H3N2, and type B) | 1945/2003 | Inactivated and live-attenuated | Egg culture |
| Poliovirus | 3 | 1955/1963 | Inactivated and live-attenuated | Cell culture |
| Measles | 1 | 1963 | Live-attenuated | Cell culture |
| Mumps | 1 | 1967 | Live-attenuated | Cell culture |
| Rubella | 1 | 1969 | Live-attenuated | Cell culture |
| Rabies | 1 | 1980 | Inactivated | Cell culture |
| Adenovirus | 2 (types 4 and 7) | 1980 | Live attenuated | Cell culture |
| Hepatitis B | 1 | 1986 | Virus-like particle | Molecular biology |
| Japanese encephalitis | 1 | 1992/2009 | Inactivated | Live animal passage/Cell culture |
| Hepatitis A | 1 | 1994 | Inactivated | Cell culture |
| Varicella | 1 | 1995 | Live-attenuated | Cell culture |
| Rotavirus | 5 (G1, G2, G3, G4, P[8]) or 1 (G1P[8]) | 2006 | Live-attenuated | Molecular biology |
| Human papillomavirus | 4 (types 6, 11, 16, 18) or 2 (types 16, 18) | 2006 | Virus-like particle | Molecular biology |
Goals of vaccination
For an individual, the goal of vaccination is to prevent or modify disease. From a public health perspective, the goal is to control the spread of a pathogen within a population. Ideally, personal protection and epidemic control would lead to elimination or eradication of the pathogen. Vaccine effectiveness –central to achieving these goals—is determined by several critical factors: (i) biological challenges that must first be identified then overcome; (ii) technical advances that solve operational or logistical hurdles; and (iii) social and political will of communities, organizations, and governments. These latter issues are largely influenced by the environmental conditions and resources of the settings in which vaccination programs are implemented.
Disease prevention is the minimal goal for a vaccine. Most vaccines are licensed on the basis of clinical endpoints, meaning that vaccinated subjects experience a clear reduction in magnitude or frequency of disease manifestations. These could include incidence of hospitalizations, clinic visits, rash, fever, influenza-like respiratory symptoms, or laboratory abnormalities. Vaccine efficacy is secondarily determined by microbiological evidence of a reduction in the incidence or burden of infection by a particular pathogen. For example, the HPV vaccine was licensed based on its ability to prevent cervical neoplasia and not necessarily based on prevention of infection (1). For other viruses, such as human immunodeficiency virus (HIV) or herpes simplex virus (HSV), the primary objective is to prevent infection or completely clear virus after infection. For these particular viruses, achieving sterilizing immunity or abortive infection is of prime importance because of the pathogens’ abilities to establish latency, evade the immunologic clearance by the host, and cause relapsing, recurring, or persistent disease. Vaccine-mediated prevention of infection is relatively difficult to achieve, as most vaccines do not induce sterilizing immunity but facilitate rapid clearance of virus-infected cells to reduce the burden of disease and prevent persistent infection. For example, the varicella vaccine does not avert infection but prevents persistent infection with wildtype strains of varicella zoster virus (VZV) when given to children and reduces the incidence of herpes zoster and postherpetic neuralgia when given to adults (2).
A charitable vaccine is one that prevents pathogen transmission without direct benefit to the vaccinated individual. Currently there are no licensed antiviral vaccines that fit this definition. However, one could envision the possibility of immunizing pregnant women to provide passive immunity to newborn infants for diseases, such as respiratory syncytial virus (RSV), that may be otherwise unavoidable and lethal in neonates and occur before active immunization is possible. This would fit within current standards of prenatal care that ensures expectant mothers have immunity against rubella to protect infants from congenital rubella syndrome. This example and the principle of providing prenatal care to the mother-infant pair justifies a charitable immunization that may be of little direct benefit to the mother. A similar argument could be made in the future for maternal immunization with a cytomegalovirus vaccine, when one is successfully developed. Likewise, healthcare workers and family members are often compelled to receive vaccines in part to prevent severe disease from infections like influenza but largely to protect vulnerable patients who may not be protected by more direct treatment.
Eradication of a viral disease is the ultimate indicator of vaccine success. Thus, it is useful to understand some of the elements required for this to occur. It has been achieved only once in the history of human disease when the eradication of smallpox was declared in 1980 (3). More recently in 2011, the Food and Agriculture Organization announced the eradication of the veterinary disease rinderpest—caused by a morbillivirus—that had been responsible for outbreaks in cattle for centuries (4, 5). These successful eradication campaigns included the following common elements: (i) extensive organizational structure and strong management with contributions and advocacy from broad coalitions of governments, professional organizations, and local communities; (ii) support across religious, cultural, and economic boundaries that facilitated educational campaigns, political will, and cogent cost:benefit analyses to deal with the inevitable logistical setbacks, social instability, anti-vaccine sentiments, and funding requirements; (iii) technical innovations like the bifurcated needle for vaccinia inoculation or the thermostable vaccine formulation for rinderpest that simplified the process of vaccine delivery to resource-poor areas; (iv) strategic decisions, based on field work, detailed epidemiology, and mathematical modeling, that revised the vaccination approach at critical junctures like transitioning from mass vaccination campaigns to ring vaccination for smallpox or vaccination of targeted populations for rinderpest; (v) engagement of local indigenous people to implement the final stages of the vaccination campaign; and (vi) reliable diagnostic algorithms and surveillance to ensure the disease did not re-emerge.
Exciting progress is being made on polio eradication with autochthonous cases in the first quarter of 2013 found in only 3 countries (Afghanistan, Pakistan, and Nigeria), and just small numbers of type 1 and type 3 wildtype viruses remaining (6). Sadly, the effort that remains is large and dangerous because of the groups who use vaccine-related issues for their purposes of disrupting society and maintaining chaos in order to acquire power. The measles eradication effort has also made significant progress since 2000, although large outbreaks beginning in 2008 have caused setbacks in some regions (7).
It is only feasible to attempt global eradication of a viral pathogen when there is a single host for the virus and no other animal reservoir that sustains transmission. Because of the threat of bioterrorism, advanced sequencing and molecular technologies, and the increasing frequency of emerging diseases from disrupted ecologies, there will always be an actual or virtual reservoir and potential for re-emergence for most viruses, so the term ‘elimination’ or the use of qualifiers may be more appropriate than the term ‘eradication’. Nevertheless, the goal of eradication remains an important consideration even at the early stages of vaccine development, and should be kept in mind as the vaccine approach, diagnostic tools, and epidemiological information are developed for the next viral pathogen.
What’s old is new: how vaccines work and lessons from HIV vaccine development
The principle that vaccines induce adaptive immune responses defined by the properties of specificity and memory has been understood for many decades. It has also been known for more than 30 years that antibody-mediated effector mechanisms are designed for preventing infection and T-cell-mediated effector mechanisms are best for recognizing and clearing virus-infected cells. There are many nuances in how cellular and humoral responses collaborate to establish immunity, but awareness that antibody can prevent and determine the frequency of infection, while T cells control viral clearance, influence the patterns of response, and often determine the severity of illness, is a well-accepted paradigm.
Neutralizing antibody
Most antiviral vaccines work by inducing antibodies specific for the surface glycoproteins of enveloped viruses or the capsid proteins of non-enveloped viruses. Antibody is the primary element of adaptive immunity that is designed to pre-exist at protective levels and be present during re-exposure to a viral pathogen. Antibody is the only component of the adaptive immune response that can recognize a virus before it has infected a cell. Pre-existing antibody can act with the speed of innate immunity, but with more specificity, higher avidity, and targeted functionality. Therefore, a major immunological goal for immunization is the induction of durable antibody responses. Protective antibody responses work best when they are neutralizing and inhibit infection. Neutralization can occur by three major mechanisms. First, aggregation or immobilization of the virus reduces the infectious inoculum by preventing the virus from reaching the target cell. A second mechanism involves antibody directly blocking attachment of the virus to the target cell by covering the receptor binding domain. Third, neutralization can occur post-attachment by preventing entry or uncoating through fusion inhibition. Thus, neutralizing antibodies need to recognize oligomeric surface proteins in their native state or in intermediate forms that may be transiently present before completion of the fusion process. These epitopes are generally conformational and often quaternary epitopes, and not often present on monomeric forms of viral proteins.
Cytolytic CD8+ T cells
Although antibodies are recognized as the primary mechanism of vaccine-induced protective immunity, it is rare for deficiency of antibody to be associated with the severity of a viral illness. The exception is infection by picornaviruses that can be lethal in the setting of immunoglobulin deficiency. In contrast, illness severity from most viral diseases is much greater and sometimes lethal when T-cell deficiencies are present. Therefore, virus-specific T-cell responses are important for controlling virus infection and limiting the severity of disease. However, no vaccine has ever been licensed in the US based solely on the induction of T-cell-mediated immunity. A brief review of the history of HIV vaccine development and the evolving concepts of immunity can illustrate the importance of these basic principles.
HIV vaccine development
There have been four major vaccine concepts evaluated for efficacy against HIV infection (Fig. 1). The first involved a subunit monomeric gp120, one component of the major HIV-1 surface glycoprotein, adjuvanted with alum. The hypothesis was that antibody to viral surface glycoproteins should be the major determinant of protection against infection. Two studies were performed, one in men-who-have-sex-with-men (MSM) in the U.S. and Europe, and the other in injection drug users (IDUs) in Thailand. The trials ended in 2003 when no evidence of efficacy was found in either trial. This was expected by the scientific community, as the antibody induced by this product did not neutralize commonly transmitted CCR5-dependent primary isolates of HIV-1 in vitro. During these trials, it was found that laboratory-passaged isolates of HIV-1 were relatively easy to neutralize, while primary isolates were more difficult to neutralize (8), and broadly neutralizing antibodies were rare even in chronically infected individuals.
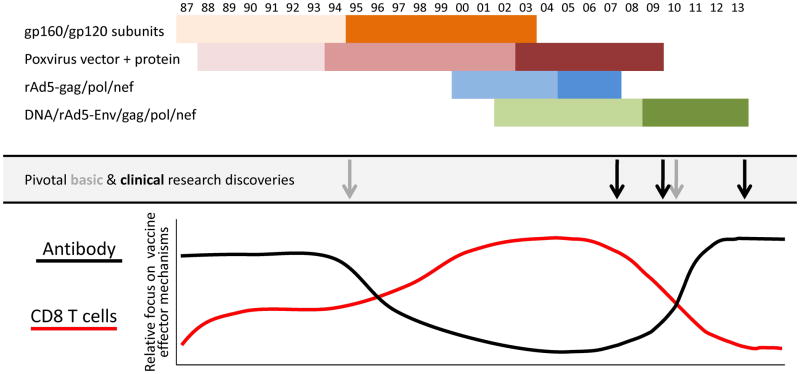
Fig. 1. History of HIV vaccine development and concepts of vaccine-mediated protection.
HIV vaccine development programs that advanced to efficacy evaluation are depicted as early Phase I and II studies lighter shades and efficacy studies in darker shades. The Poxvirus+protein includes Phase I trials of live recombinant vaccinia followed gp160 subunit boosting then Phase I/II and efficacy studies of replication-defective recombinant canarypox vectors, hence 3 separate phases are shown. The light grey arrows indicate important basic research findings and the black arrows indicate conclusions from efficacy trials that impacted the perceived value of antibody and CD8 T-cell responses in vaccine-induced immunity shown in graphical form. The light grey arrow in 1995 represents the realization that primary HIV isolates were difficult to neutralize and that available antigen designs could not elicit broadly neutralizing antibodies. The light grey arrow in ~2010 represents the discovery of multiple broadly neutralizing human mAbs using new technologies to isolate immunoglobulin genes from individual B cells.
The second concept involved recombinant poxvirus vectors expressing both HIV envelope glycoproteins and internal structural and regulatory proteins followed by boosting with a recombinant subunit envelope. The underlying hypothesis for this approach was that combining the induction of CD8+ T-cell responses to antigens delivered by the gene-based vector with the antibody responses boosted by the envelope subunit protein would provide an additional level of protection from breakthrough infections. These studies began with live replication-competent recombinant vaccinia vectors boosted with gp160 (9–11), but evolved to replication-defective canarypox vectors (ALVAC) boosted with the same gp120 protein used in the initial failed trials (12, 13). At the time, CD8+ T-cell responses were measured by Cr51-release assays, as there were promising immunogenicity studies suggesting HIV-specific CD8+ T cells were being induced (14), although at a relatively low frequency. There was a low expectation that this approach would be successful (15), especially considering the outcome of the gp120 studies and the relatively poor immunogenicity from U.S. studies (16). Nevertheless, this product concept advanced to efficacy testing in a general population cohort in Thailand in a trial called RV144.
In the meantime, there was a growing pessimism about the likelihood of inducing broadly neutralizing antibodies with currently available antigens, and at the same time, there were several major advances in the ability to detect and analyze T-cell responses including tetramers to measure epitope-specific CD8+ T cells by flow cytometry (17), a method for intracellular cytokine staining of peptide-stimulated T cells (18), and new instrumentation and analysis tools for flow cytometry (19). This coincided with the demonstration that CD8+ T cells were important for controlling HIV-1 replication and were a major determinant of the viral load setpoint (20, 21). The ability to measure CD8+ T-cell responses combined with the demonstration of their importance in HIV biology and the lack of available antigen designs that could elicit neutralizing antibodies resulted in the HIV vaccine field turning toward developing vaccines based on T-cell mediated protection. This was exemplified by a vaccine developed by Merck based on delivering internal and regulatory proteins, Gag, Pol, and Nef, with a gene-based replication-defective recombinant adenovirus serotype 5 (rAd5) vaccine vector that induced a biased CD8+ T-cell response. Without a surface glycoprotein antigen, there was no chance for this vaccine concept to induce neutralizing antibody responses, and even CD4+ T cells were a minor component of the response relative to CD8+ T cells. The candidate vaccine was evaluated first in Ad5 seronegative MSM then expanded to Ad5 seropositive MSM in the Americas in a study referred to as the STEP trial. The study was then extended to a general population cohort at multiple sites in South Africa in a trial called Phambili. In September 2007, it was announced that the STEP study had failed to prevent HIV infection, and in fact, the rate of infection was higher among uncircumcised subjects who were vaccinated than in placebo recipients and even higher in uncircumcised men who were Ad5-seropositive prior to vaccination. Therefore, vaccinations in the STEP and Phambili studies were immediately discontinued and subjects were unblinded. In the subsequent analysis, it was found that there was a minor influence by CD8+ T cells on the sequence of HIV genes within certain dominant CD+8 T-cell epitopes on which some immunological pressure was being exerted (22). However, the magnitude, breadth, and location of these CD8+ T-cell effectors were not sufficient to significantly influence HIV viral load or disease progression.
A fourth vaccine concept attempted to achieve a more balanced response. By priming with DNA prior to boosting with rAd5, more CD4+ T-cell responses were induced, and CD8+ T cells were amplified. In addition to expressing Gag, Pol, and Nef, Env proteins representing the three major HIV-1 subtypes were expressed with the intention of inducing antibodies that could neutralize HIV in vivo, although it was known that neutralization against the Tier 2 more difficult to neutralize primary isolates was not achieved in vitro. In nonhuman primate models (NHP) of SIV infection using analogous SIV vaccine constructs, there was evidence that acquisition of infection could be reduced against some challenge strains of SIV and viral load in animals with breakthrough infections could be reduced against other strains (23, 24). In addition, the rAd5 vector used in this product was designed to not contain the E4 genes, which distinguished it from the rAd5 vector used in the STEP and Phambili studies because E4 gene products are required for production of adenovirus structural proteins like fiber and hexon. Therefore, the E4-deleted rAd5 vector produced less antigenic competition between vector-specific proteins and the recombinant vaccine antigens, and made less Ad5 proteins to stimulate vector-specific immune responses. The phase IIb efficacy study for this product (HVTN 505) began enrollment in 2009 in North American MSM who were circumcised and Ad5-seronegative. Later that year, it was announced that the RV144 trial in Thailand with the ALVAC-gp120 prime-boost had shown partial efficacy with about 30% reduction in acquisition of HIV. This supported the continuance of the HVTN 505 study, because in prior immunogenicity assessments, it induced a similar level and quality of Env-specific antibody and a much higher frequency of CD8+ T cells than the RV144 regimen. Nevertheless, in March 2013 vaccinations in the HVTN 505 study were discontinued following the recommendation by an independent Data and Safety Monitoring Board (DSMB) based on reaching futility for achieving efficacy for either reduction of HIV acquisition or lowering of the viral load set point in subjects infected despite vaccination (http://www.niaid.nih.gov/news/newsreleases/2013/Pages/HVTN505April2013.aspx).
An extensive effort on the correlates of infection risk in the RV144 study revealed several surprising findings. Antibody responses to the V2 region of HIV-1was correlated with reduced risk of HIV-1 infection (25). This is a region which happens to be bound by PG9, a broadly neutralizing monoclonal antibody (mAb), shown to bind V2 in a glycan dependent manner (26). A sieve analysis comparing virus sequences from infected placebo recipients to those of infected vaccinees showed there were significant sequence differences in the region of antibody binding consistent with selection pressure (27). This supported the possibility that antibody to V2 was mediating protection in vivo even though neutralizing antibody responses against Tier 2 primary isolates had not been detected in vitro in vaccinee sera. There is an integrin-binding motif in this part of V2 (LDI) that can interact with α4β7 on mucosal CD4+ T cells (28), providing a plausible mechanism by which V2 antibody might protect against HIV infection that would not be detected in neutralizing antibody assays. However, it seems equally plausible that there are other factors present in vivo at the site of infection that result in virus neutralization and are not captured by currently available in vitro neutralizing assays. The presence of mucus, complement, Fc receptor-bearing cells, and other elements of the mucosal environment could contribute to virus aggregation or sequestration and reduce the likelihood of virus reaching a susceptible target cell. Some data that support these speculations are that gp120 alone induced antibodies with similar V2 specificity, but did not protect. While the study populations experienced different exposure routes for HIV-1 transmission and the magnitude of risk was also different with incidence rates >3% in the gp120 studies and <1% in the Thailand cohort, it was also found that the patterns of IgG isotype response were also different with the ALVAC/gp120 vaccine inducing a more IgG1/IgG3 biased response and the gp120 alone inducing more IgG2/IgG4 responses (29). These observations have opened new areas of inquiry into mechanisms of in vivo virus neutralization (30) that may inform not only vaccine development for HIV but for other mucosal pathogens as well.
The sobering lack of ability to predict the outcomes of HIV vaccine efficacy trials has at times been a polarizing force in the field but has also led to a series of observations that have redirected the scientific emphasis back to the importance of antibodies for vaccine-induced protection. In addition, the failures have stimulated the use of new technologies to better understand the requirements for inducing broadly neutralizing antibodies that are reviewed next. The process has also helped reframe some basic principles of vaccine development that are listed in Table 2.
Table 2.
General principles for achieving vaccine-induced immunity
|
New technologies for old problems
In recent decades, major advances in viral vaccine development have coincided with the emergence of new technologies. Therefore, selected technologies that are likely to influence future vaccine design are discussed briefly. In particular, the confluence of new techniques to isolate human monoclonal antibodies, solve atomic structures of viral surface proteins, and obtain and analyze large volumes of sequence information to describe both virus and antibody evolution, in combination with classical virological and serological analyses have opened a new era in vaccine development that hopefully will lead to more effective vaccine antigens to prevent current and future viral threats. Much of this technology has been driven by the quest to understand HIV immunity and develop a vaccine to prevent HIV. However, the lessons learned from the work on HIV are now translating to potential solutions for old problems like pandemic influenza and respiratory syncytial virus.
Isolation and synthetic production of human monoclonal antibodies
The original technology for producing murine monoclonal antibodies by immortalizing B cells as hybridomas (31) has been a cornerstone of modern biology and resulted in the Nobel Prize for Milstein and Kohler in 1984. Since then, human monoclonal antibodies have been produced by phage display (32), in genetically modified mice (33, 34), or using EBV transformation (35), stimulation with TLR agonists (36), B-cell factors associated with germinal center survival (37), or producing human hybridomas (38) for immortalization of antibody-producing B cells. However, in 2008, a transformative technology was introduced, in part made possible by the advances in high-throughput sequencing technology. Using RT-PCR, both the heavy and light chain immunoglobulin (Ig) genes were amplified from single B cells and cloned into expression vectors (39). This allowed large scale isolation and synthetic production of human monoclonal antibodies by transfection of producer cells in vitro. This technology was rapidly adapted for sorting plasmablasts and cloning Ig genes to identify high affinity influenza-specific antibodies (40) and used to isolate broadly neutralizing antibodies against HIV (41). Identifying new human monoclonal antibodies has now become routine, limited only by the imagination for how to select a particular antibody specificity.
High-throughput sequencing
Advances in sequencing technology have influenced many aspects of biology and are now providing new insights into the maturation of antibody responses and characterization of antigenic content of vaccine products. The cost and time required for sequencing has declined precipitously and the quantity and accuracy has increased as bioinformatic analysis tools have improved. This has allowed rapid sequencing of antibody genes isolated from individual B cells and large amounts of data to be collected on the V genes contained within the B-cell repertoire. Similar data can be generated for the α and β components of T-cell receptors (TCRs) to describe the repertoire of T cells (42, 43). Once a CDR3 sequence motif is identified for an antibody with the desired functional properties, that information can be used to interrogate deep sequencing data from the entire B-cell repertoire and select particular heavy or light chain alleles for further analysis. This type of analysis over time allows one to track the ontogeny of an antibody response as it undergoes somatic mutations and affinity maturation. Alternatively, deep sequencing can be used to selectively evaluate plasmablasts that are present in blood 5–7 days after an antigenic stimulus and that are highly enriched for antibodies specific for the original antigen. Until recently, only single-cell sequencing was able to reliably pair the heavy and light chain genes and provide the necessary information to produce a functional antibody. While deep sequencing has been able to evaluate the repertoire and track ontogeny of selected variable regions within the IgH and IgL genes, there was not a solution for pairing the genes. New sequencing technologies that operate within emulsion droplets may allow bar-coding of sequences derived from individual T cell or B cells and solve this problem (44, 45), so within the next few years thousands of paired antibody or TCR genes could be derived from a single sample. The field of bioinformatics and analysis software will have to keep up with the rapid pace of change to accommodate this exponential increase in data.
Deep sequencing technology has also changed the way viruses and, consequently, virus vaccines can be evaluated. HIV exists as a swarm or quasispecies as do most RNA viruses. Deep sequencing technology has allowed the analysis of the swarm instead of the analysis of individual viruses and has provided the basis for understanding the impact of immune pressure on the evolution of virus strains in ways that were not possible 10 years ago. For example, SIV infection can be evaluated by isolating individual variants by single genome amplification (46). In the case of these viruses, multiple isolates have to be individually sequenced to describe the breadth of existing quasispecies. Using deep sequencing, the entire swarm can be analyzed for its diversity, even though the connection between sequences from different regions of the genome may not be discernable. Applications of this technology have also contributed to the evaluation of vaccine safety. For example, when applied to licensed viral vaccine preparations in a vial, it was revealed that sequences from porcine circovirus-1 (PCV1) were in the rotavirus vaccine (Rotarix®) (47). This is a highly prevalent nonpathogenic virus from pigs that sometimes contaminates commercially available trypsin but does not infect humans, so distribution of the vaccine was reinstituted. In addition, for vaccines produced in avian or primate cells there were some remnants of endogenous avian leucosis or simian retrovirus sequences detected which were incomplete and not unexpected as a component of cellular DNA. Therefore, the findings did not affect safety or availability of the vaccines but has established a new standard that may need to be met for future vaccines. If manufacturers do not complete these analyses themselves, it is now relatively simple now for others to do it, which can lead to complex regulatory matters for the developer. These examples where sequencing has been used to evaluate market preparations of vaccines have also revealed minor genetic variation within vaccine viruses and suggests that the extent of variation may need to be acknowledged for future live-attenuated vaccines.
Structural biology and antigen design
Antigen design has always been an empirical process in the past and has generally been based on information derived from linear sequences. This one dimensional view of protein antigens is relevant for T-cell epitopes which are linear peptide sequences ranging in length between 8 and 11 amino acids and for some linear antibody epitopes. However, as noted above, most antibodies with neutralizing activity bind conformational epitopes that must be considered in 3 dimensions and are often quaternary, binding multiple protomers within the oligomeric protein complex. Structural biology and the ability to more rapidly identify and solve atomic-level structures of viral surface proteins has changed our ideas of how to define the antigenic target for a vaccine-induced response, and has raised the possibility of structure-based vaccine design (48). An immunoglobulin molecule was first crystallized in 1965 (49), and the first viral surface glycoprotein to be crystallized was influenza hemagglutinin in 1977 (50). The first structure of a glycoprotein-antibody complex was solved in 1990 (51). Since then, many structures have been solved and the evaluation of antigen-antibody complexes at the atomic level has provided the basis for specific antigen designs. Using tools developed by computational biologists, the concept of epitope scaffolds has been explored in which individual antibody epitopes are mounted on another protein and constrained in ways that maintain the chemistry and structure of the domain that interacts with the antibody of interest. This has been done with partial success for the HIV-1 gp41 epitopes for mAb 2F5 and mAb 4E10, and for the RSV F motavizumab epitope (52–54). These synthetic constructs have elicited antibodies that recognize the intended epitope but lack significant neutralizing activity, suggesting that epitopes may need to be recognized in a larger context to achieve the desired functional properties. Therefore, more work is needed to define the immunogenicity rules for individual epitopes as epitope scaffold designs are refined. In the meantime, the design of whole protein antigens is advancing based on the structural information gained from studying the interaction of antibodies with viral proteins. In some cases, antigenic sites need to be revised to be more accessible to antibodies in the naive repertoire, and in other cases, metastable antigenic sites need to be stabilized to preserve antibody recognition.
New paradigms emerging from technological convergence
The confluence of isolating broadly neutralizing human monoclonal antibodies, sequence analysis of antibody ontogeny, and structural analysis of antibody-antigen interactions is having a profound effect on the fields of HIV and influenza vaccine development. These two pathogens share the property of extreme genetic and antigenic variation, making the induction of broadly neutralizing antibodies (BNAbs) a shared objective. The moderate success of the RV144 trial showing a correlation with antibodies to the V2 region and the failure of T-cell-based vaccine approaches has led to a concentrated effort to understand the mechanisms for inducing broadly neutralizing antibody as the basis for the next generation of candidate HIV vaccines. Human monoclonal antibodies have now been obtained for the four antigenic sites on HIV gp160 associated with broad neutralization (41, 55–57), and the structures of the antibody interacting with the antigenic sites have been solved (26, 55, 57, 58). Not surprisingly, all the BNmAbs have usual features. In particular, they are characterized by either long CDR3 loops (meaning the parental B cells will come from rare recombination events and have low prevalence in the repertoire) or the variable domains have extremely high proportion of somatic mutations (meaning the induction of these antibodies may require prolonged or repeated antigen exposure).
Work is now focused on how antibody responses develop to these sites and how these antibodies can be elicited by vaccines. One line of inquiry explores the finding from RV144 that V2-specific antibodies were associated with protection. Human monoclonal antibodies were isolated from recipients of the RV144 vaccine, and V2-specific monoclonal antibodies were obtained. The crystal structures of mAbs CH58 and CH59 interacting with the V2 loop are consistent with the findings from the sieve analysis and the PG9 epitope structure mentioned above and provide the basis for designing better V2 structures and for isolating more quaternary antibodies to this region that may better stabilize the gp160 trimer for refined structural analysis (59). Another series of studies has been done with antibodies to the CD4 binding site. The original structure of the gp120 monomer (60, 61) and the definition of the CD4 binding site (CD4BS) on gp120 (62) served as the basis for designing a ‘resurfaced stabilized core’ molecule (RSC3) that was used as a flow cytometry probe to isolate VRC01, a broad and potent CD4BS mAb (56, 58). With the structure-designed probe and the sequence motif available from VRC01 and other similar molecules, a large number of CD4BS BNmAbs have now been isolated or identified from related VH gene sequences. They have allowed a detailed analysis of how these antibodies neutralize HIV based on atomic level structure that show how the angle of approach, rotation, and surrounding structures affect access to the epitope. In addition, deep sequencing of the B-cell repertoire over the timecourse of infection to define the sequential somatic mutations required to achieve high affinity binding has been possible (63). Thus, understanding the ontogeny of the VRC01-like antibody maturation has provided a roadmap for how gp160 can first engage the germline antibody and how the antigen and antibody co-evolve by reciprocally escaping and establishing the binding interactions that affect neutralizing activity. Direct evaluation of the developing CD4BS antibody response and diversification of gp160 over time in a chronically HIV-infected person has suggested that immunizing sequentially with a series of gp160-derived antigens could push the somatic mutation process far enough to attain antibodies with broad neutralizing activity (64).
The need for a more universal influenza vaccine that would reduce the need for yearly immunization and provide broader protection against pandemic threats has similarly led to intense efforts to identify and induce broadly neutralizing antibodies that are effective across multiple strains of influenza. Most neutralizing activity is directed against the globular head domain of the influenza hemagglutinin (HA). However, in 1993 a mononuclear antibody directed against the stem region of HA was reported to have cross-neutralizing activity, and in 2008, these antibodies were rediscovered and their interaction with epitopes in the HA stem was defined structurally (68). Neutralizing HA head-specific antibodies also have hemagglutination-inhibition (HAI) activity, which is an assay typically used as a surrogate marker of neutralization. Stem-specific antibodies do not have HAI activity; thus, their activity must be directly measured by neutralization assays. As a consequence, they have often not been accounted for in clinical trials that most commonly have an HAI endpoint. Therefore, prior studies of candidate influenza vaccines may not have fully explored the utility of stem-specific antibodies as a mechanism of protection across multiple subtypes. The stem-specific monoclonal antibodies tend to recognize HA subtypes within one of the two groups of HA molecules. The initial stem-specific monoclonal antibodies recognized HA from group 1 viruses (including H1, H2, H5, and H9 that have been associated with human infection). More recently group 2 (H3 and H7) HA stem-specific monoclonal antibodies have also been identified (69), and one reportedly neutralizes subtypes in both HA groups (70). Influenza stem-specific monoclonal antibodies may be analogous to the BNmAbs for HIV that interact with the membrane proximal domain of gp41, the influenza stem equivalent, that are known to be induced infrequently even during chronic infection. However, stem-specific monoclonal antibodies are commonly induced by influenza infection or vaccination (71–75), so it is not well understood why there is not substantial cross-protection in humans who are repeatedly exposed to influenza virus and vaccines. Nevertheless, it may be possible to exploit these epitopes on the HA stem to develop vaccine antigens that elicit neutralizing antibodies that cross-protect against seasonal strains and potentially provide some protection against pandemic strains of influenza. Therefore, extensive efforts are ongoing using human monoclonal antibodies and structural biology to guide new antigen designs to elicit these antibodies. Cross-neutralizing monoclonal antibodies directed to the head domain have also been reported, although they appear to be rarer than cross-reactive stem antibodies. One possible explanation for this comes from the analysis of the mAb C05, which is an HA head-specific monoclonal antibody with broad neutralization activity (76). It is characterized by a long CDR3 loop that reaches into the recessed receptor binding domain rather than interacting primarily with the rim which is the more common binding site for subtype-specific monoclonal antibodies. This is reminiscent of BNmAbs to HIV that often have long CDR3 loops that are uncommon in the normal immunoglobulin repertoire. Thus, development of a universal influenza vaccine or a vaccine for HIV, have some shared challenges and hopefully also some shared solutions.
Unlike HIV and influenza, respiratory syncytial virus (RSV) is a virus with relatively little genetic variation, and despite its ubiquity has only two major antigenic subtypes. RSV infects everyone by 3 years of age (77) and continues to reinfect every 3–10 years throughout life. Mortality is greatest in those under 1 year of age (78), but the frail elderly (79) and immunocompromised individuals (80) also experience severe and often fatal infections. RSV entry is mediated by the F glycoprotein, which is a class I fusion protein analogous to HIV gp160 and influenza HA. Neutralizing antibody is known to provide protection against hospitalization based on the use of licensed palivizumab in infants at high risk of severe disease (81). However, induction of potent neutralizing activity in naïve infants or boosting neutralizing antibody titers in adults by investigational vaccines or natural infection has been difficult.
The understanding of fusion proteins has been advanced by defining the atomic structure of the proteins in their pre- and post-fusion state. For influenza HA, the pre-fusion structure was solved first and was facilitated by its stability at neutral pH. For paramyxoviruses, the structure of the post-fusion 6-helix bundle was reported in 2005 (82). Pre-fusion structures have been more difficult because of their instability when expressed as soluble proteins, but the PIV5 pre-fusion F structure was reported in 2006 (83) and had been used to model the potential structures for other paramyxovirus pre-fusion molecules. The RSV F post-fusion structure was reported by two groups in 2011 (84, 85), and recently the structure of RSV pre-fusion F was solved (86). This was the first example of paired pre- and post-fusion structures for a paramyxovirus fusion protein from the same virus and revealed a number of important findings. First, the modeling of one pre-fusion molecule can predict the general features of another, but the structural details and chemistry are not sufficient for the purposes of optimal antigen design. Second, there are significant rearrangements that occur between pre- and post-fusion molecules to accomplish membrane fusion, but there are also structurally stable domains between to the two conformations. Therefore, some epitopes may only be present on the pre-fusion structure, although some important antigenic sites may be preserved in both conformations. Third, the pre-fusion structure is the native functional form of the protein required for membrane fusion and entry, so antibodies specific to the pre-fusion structure may have greater neutralization potency, and the post-fusion structure may expose new antigenic sites that have no functional relevance for neutralization.
For RSV, screening for antibodies with neutralizing activity that did not bind post-fusion F yielded antibodies that were pre-fusion-specific. The Fab portions of these antibodies were used to stabilize the pre-fusion conformation and allowed the atomic level structure to be solved. In addition, a new antigenic site was revealed at the apex of the pre-fusion F trimer that was quaternary in nature and involved sequences from both the F1 and F2 cleavage products (antigenic site Ø). Antibodies to antigenic site Ø turn out to have much higher potency than antibodies to other recognized epitopes and are on the order of 50-fold more potent than palivizumab. Once the structural details were available, it was possible to design stabilizing mutations, and produce a pre-fusion F protein that was antigenically and physically stable. This representation of the native F trimer could then be used as a probe to select other pre-fusion specific antibodies and as a vaccine antigen to elicit potent neutralizing antibodies. Without the ability to screen, sequence, and synthesize monoclonal antibodies, examine the structure of the antibody-antigen interaction, and utilize the tools of computational biology, this new candidate vaccine antigen could not have been realized. The preceding work in HIV and influenza made the discovery of a new RSV vaccine target possible and has established a new paradigm using multiple new technologies to identify vulnerable sites on viral surface proteins that can be applied to vaccine development for other virus pathogens (Fig. 2).
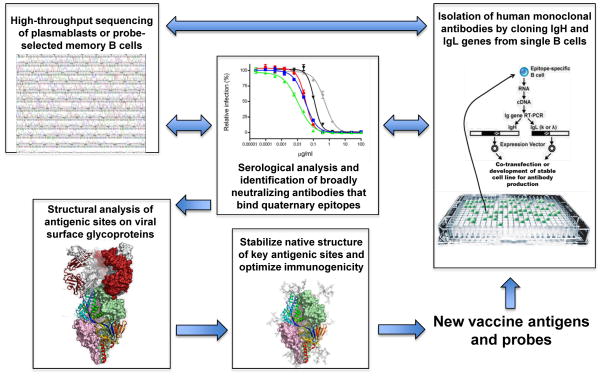
Fig. 2. Technological convergence on antigen design.
The emergence of antibody gene sequencing, rapid isolation of human monoclonal antibodies, and structural analysis of antigenic sites on native viral surface proteins has provided a new paradigm for the development of vaccine antigens. In addition to being useful as candidate vaccine antigens, the stabilized viral proteins can be used as probes to select B cells producing better antibodies or to probe B-cell populations by deep sequencing to understand the ontogeny of antibody maturation. Therefore, the process can be cyclical. These reagents also provide new options for understanding viral pathogenesis. For example, the native surface protein can be used to probe for cellular receptors or ligands to study viral attachment and entry or intracellular signaling pathways that may be induced during virus infection. Furthermore, they provide tools for improving the serological evaluation of virus or vaccine-induced immune responses.
Prospects for difficult and emerging viruses
There are still many viral diseases that have significant impacts on public health for which vaccines are not available. Many of these remaining viral targets could be classified as ‘difficult’ based on the following characteristics: (i) infection is not self-limited, has a high frequency of severe disease, and often leads to persistence; (ii) the virus has evolved multiple mechanisms to alter and evade host immune responses, (iii) the host can be reinfected; (iv) T cells play a critical role in immunity; (v) there is significant genetic variation; (vi) the site of infection is the same as the major target organ for disease; (vii) there is integration of the viral genome or sequestration of the virus making it less accessible to immune effectors; (viii) animal models fail to recapitulate pathogenesis of human disease; and (ix) there is a long delay between initiation of infection and onset of adaptive cellular immunity (87). We also inevitably will be faced with emerging viral diseases for which vaccines would benefit the public health. Addressing these challenges will require new paradigms for discovery, development, manufacturing, and distribution (88). It may also require a different level of investment to fully understand viral pathogenesis and immunity.
Many prior successful vaccine development efforts have been achieved through empirical and incremental improvements in clinical efficacy using products based on whole virions, without understanding the underlying mechanisms of protection. To develop vaccines for the difficult viruses and to be prepared for emerging infections, we need to move from the age of empiricism to the age of rational design based on understanding fundamental immunology, structure, and function. In addition, we need to be creative in applying new technologies to vaccine science. The first vaccines for smallpox, rabies, and yellow fever were motivated by specific diseases and were generally achieved by empirical thinking and the focused intent of a small number of individuals. However, most vaccines successfully developed in the modern era have been more opportunistic. New technologies based on cell culture and molecular biology provided novel vaccine solutions for those prepared to see the potential utility.
We are now in a scientific era with an embarrassment of technological riches and have the opportunity to use these new tools to solve some of our most vexing vaccine design challenges. In particular, the ability to isolate human monoclonal antibodies, to analyze the ontogeny of antibody affinity maturation and evolution of virus quasispecies with new sequencing technology, and to characterize the interaction of antibodies and antigenic sites on virus surface proteins with atomic level detail, in combination with advanced methods for assessing antibody specificity and function, has provided a new paradigm for the design of vaccine antigens that may address existing and future viral challenges. Combining atomic level antigen design with new methods for gene delivery and protein manufacturing may allow us to identify vaccine platform technologies that would be likely to work against a particular class or family of viral pathogens. High throughput sequencing technology also provides new opportunities to discover, track, and perhaps predict future emerging viral diseases (89). Therefore, it is within our current technological capabilities to produce a ‘periodic table’ of existing viruses and produce a catalogue of vaccine approaches demonstrated to work for the finite number of virus families that exist. In the event of a new emerging virus pathogen, this pre-existing information would allow the rapid construction of vaccine candidates to develop if needed for outbreak control.
Notwithstanding the significant opportunities that currently exist, there are still some important gaps in our understanding of how to induce immune responses. One of the key issues for HIV and influenza vaccine development is the finding that the antigenic sites of greatest interest with the potential for broad cross-neutralization are not as immunodominant as antigenic site Ø appears to be for RSV. So far, the scientific understanding of how to make an antigenic site an immunogenic site is not sufficient to accomplish this on a routine basis. This is a critical challenge to overcome for rational structure-based antigen design. Other looming biological questions include the following: (i) What are the biological properties that make an immune response boostable? (ii) What is required to achieve durable immunity and sustained antibody production? (iii) What are the factors that would require organ- or site-specific localization of immune effectors for immunity, and how can those responses be induced? Finally, to fully realize the benefits that vaccines have to offer society, scientists need to better educate the public and policy makers about how vaccines work and communicate with more transparency about the complicated process of manufacturing biologics. If public trust and political will can be maintained to support continued scientific advances and new partnerships between governments or with nonprofit organizations and industry are established to manage the regulatory process and costs, new antiviral vaccines will contribute and add to the extraordinary track record of vaccines improving public health.
Acknowledgments
I thank Dr. Julie E. Ledgerwood and Dr. Kayvon Modjarrad for thoughtful advice and suggestions. This work was supported by the intramural funding from the National Institute of Allergy and Infectious Diseases.
Footnotes
The author has no conflicts of interest to declare
References
- 1.Quadrivalent vaccine against human papillomavirus to prevent high-grade cervical lesions. N Engl J Med. 2007;356:1915–1927. doi: 10.1056/NEJMoa061741. [DOI] [PubMed] [Google Scholar]
- 2.Oxman MN, et al. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N Engl J Med. 2005;352:2271–2284. doi: 10.1056/NEJMoa051016. [DOI] [PubMed] [Google Scholar]
- 3.Henderson DA. Smallpox eradication. Public Health Rep. 1980;95:422–426. [PMC free article] [PubMed] [Google Scholar]
- 4.Roeder PL. Rinderpest: the end of cattle plague. Prev Vet Med. 2011;102:98–106. doi: 10.1016/j.prevetmed.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 5.de Swart RL, Duprex WP, Osterhaus AD. Rinderpest eradication: lessons for measles eradication? Curr Opin Virol. 2012;2:330–334. doi: 10.1016/j.coviro.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Progress toward eradication of polio - worldwide, january 2011-march 2013. MMWR. 2013;62:335–338. [PMC free article] [PubMed] [Google Scholar]
- 7.Global control and regional elimination of measles, 2000–2011. MMWR. 2013;62:27–31. [PMC free article] [PubMed] [Google Scholar]
- 8.Mascola JR, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 9.Graham BS, et al. Determinants of antibody response after recombinant gp160 boosting in vaccinia-naive volunteers primed with gp160-recombinant vaccinia virus. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Clinical Trials Network. J Infect Dis. 1994;170:782–786. doi: 10.1093/infdis/170.4.782. [DOI] [PubMed] [Google Scholar]
- 10.Graham BS, et al. Augmentation of human immunodeficiency virus type 1 neutralizing antibody by priming with gp160 recombinant vaccinia and boosting with rgp160 in vaccinia-naive adults. The NIAID AIDS Vaccine Clinical Trials Network. J Infect Dis. 1993;167:533–537. doi: 10.1093/infdis/167.3.533. [DOI] [PubMed] [Google Scholar]
- 11.Cooney EL, et al. Enhanced immunity to human immunodeficiency virus (HIV) envelope elicited by a combined vaccine regimen consisting of priming with a vaccinia recombinant expressing HIV envelope and boosting with gp160 protein. Proc Natl Acad Sci USA. 1993;90:1882–1886. doi: 10.1073/pnas.90.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitisuttithum P, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 13.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari G, et al. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton DR, et al. Public health. A sound rationale needed for phase III HIV-1 vaccine trials. Science. 2004;303:316. doi: 10.1126/science.1094620. [DOI] [PubMed] [Google Scholar]
- 16.Russell ND, et al. Phase 2 study of an HIV-1 canarypox vaccine (vCP1452) alone and in combination with rgp120: negative results fail to trigger a phase 3 correlates trial. J Acquir Immune Defic Syndr. 2007;44:203–212. doi: 10.1097/01.qai.0000248356.48501.ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 18.Openshaw P, et al. Heterogeneity of intracellular cytokine synthesis at the single-cell level in polarized T helper 1 and T helper 2 populations. J Exp Med. 1995;182:1357–1367. doi: 10.1084/jem.182.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roederer M, et al. 8 color, 10-parameter flow cytometry to elucidate complex leukocyte heterogeneity. Cytometry. 1997;29:328–339. doi: 10.1002/(sici)1097-0320(19971201)29:4<328::aid-cyto10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 20.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koup RA, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janes H, et al. MRKAd5 HIV-1 Gag/Pol/Nef vaccine-induced T-cell responses inadequately predict distance of breakthrough HIV-1 sequences to the vaccine or viral load. PloS One. 2012;7:e43396. doi: 10.1371/journal.pone.0043396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letvin NL, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letvin NL, et al. Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys. Sci Trans Med. 2011;3:81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolland M, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arthos J, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 29.Chung A, Robinson H, Ackerman ME, Michael NL, Kim J, Alter G. RV144 vaccination induces a different antibody effector function profile in comparison to VAX003. HIV Vaccines Keystone Symposium; Keystone, CO. 2012. p. Abstract #125. [Google Scholar]
- 30.Hope TJ. Moving ahead an HIV vaccine: to neutralize or not, a key HIV vaccine question. Nat Med. 2011;17:1195–1197. doi: 10.1038/nm.2528. [DOI] [PubMed] [Google Scholar]
- 31.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 32.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 33.Green LL, et al. Antigen-specific human monoclonal antibodies from mice engineered with human Ig heavy and light chain YACs. Nat Genet. 1994;7:13–21. doi: 10.1038/ng0594-13. [DOI] [PubMed] [Google Scholar]
- 34.Lonberg N, et al. Antigen-specific human antibodies from mice comprising four distinct genetic modifications. Nature. 1994;368:856–859. doi: 10.1038/368856a0. [DOI] [PubMed] [Google Scholar]
- 35.Steinitz M, Klein G, Koskimies S, Makel O. EB virus-induced B lymphocyte cell lines producing specific antibody. Nature. 1977;269:420–422. doi: 10.1038/269420a0. [DOI] [PubMed] [Google Scholar]
- 36.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 37.Kwakkenbos MJ, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Trans Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glanville J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci USA. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turchaninova MA, et al. Pairing of T-cell receptor chains via emulsion PCR. Eur J Immunol. 2013 doi: 10.1002/eji.201343453. [DOI] [PubMed] [Google Scholar]
- 45.DeKosky BJ, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keele BF, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Victoria JG, et al. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol. 2010;84:6033–6040. doi: 10.1128/JVI.02690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwong PD, Wilson IA. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat Immunol. 2009;10:573–578. doi: 10.1038/ni.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terry WD, Matthews BW, Davies DR. Crystallographic studies of a human immunoglobulin. Nature. 1968;220:239–241. doi: 10.1038/220239a0. [DOI] [PubMed] [Google Scholar]
- 50.Wiley DC, Skehel JJ. Crystallization and x-ray diffraction studies on the haemagglutinin glycoprotein from the membrane of influenza virus. J Mol Biol. 1977;112:343–347. doi: 10.1016/s0022-2836(77)80149-6. [DOI] [PubMed] [Google Scholar]
- 51.Bizebard T, Mauguen Y, Petek F, Rigolet P, Skehel JJ, Knossow M. Crystallization and preliminary X-ray diffraction studies of a monoclonal antibody Fab fragment specific for an influenza virus haemagglutinin and of an escape mutant of that haemagglutinin. J Mol Biol. 1990;216:513–514. doi: 10.1016/0022-2836(90)90378-Y. [DOI] [PubMed] [Google Scholar]
- 52.Correia BE, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 53.McLellan JS, et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J Mol Biol. 2011;409:853–866. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ofek G, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci USA. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao HX, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyatt R, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 61.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao HX, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 71.Kanekiyo M, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013 doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei CJ, et al. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci Trans Med. 2012;4:147ra114. doi: 10.1126/scitranslmed.3004273. [DOI] [PubMed] [Google Scholar]
- 74.Ledgerwood JE, et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 76.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 78.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 80.Hertz MI, Englund JA, Snover D, Bitterman PB, McGlave PB. Respiratory syncytial virus-induced acute lung injury in adult patients with bone marrow transplants: a clinical approach and review of the literature. Medicine. 1989;68:269–281. doi: 10.1097/00005792-198909000-00002. [DOI] [PubMed] [Google Scholar]
- 81.Palivizumab a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-RSV Study Group. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- 82.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc NAtl Acad Sci USA. 2011;108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Graham BS, Walker C. Meeting the Challenge of Vaccine Design to Control HIV and Other Difficult Viruses. In: Kaufmann SHE, Rouse BT, Sacks DL, editors. The Immune Response to Infection. Washington, DC: ASM Press; 2011. pp. 559–570. [Google Scholar]
- 88.Graham BS, Ledgerwood JE, Nabel GJ. Vaccine development in the twenty-first century: changing paradigms for elusive viruses. Clin Pharmacol Therapeut. 2009;86:234–236. doi: 10.1038/clpt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morse SS, et al. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]