Abstract
The Nedd8 activating enzyme (NAE) launches the transfer of the ubiquitin-like protein Nedd8 through an enzymatic cascade to covalently modify a diverse array of proteins, thus regulating their biological functions in the cell. The C-terminal peptide of Nedd8 extends deeply into the active site of NAE and plays an important role in specific recognition of Nedd8 by NAE. We used phage display to profile C-terminal sequences of Nedd8 that can be recognized by NAE for the activation reaction. We found that besides the native Nedd8 sequence ending with 71LALRGG76, NAE can accommodate diverse changes at the Nedd8 C-terminus including Arg and Ile replacing Leu71, Leu, Ser and Gln replacing Ala72, and substitutions by bulky aromatic residues at positions 73 and 74. We also observed that short peptides corresponding to the C-terminal sequences of the Nedd8 variants can be activated by NAE to form peptide~NAE thioester conjugates. Once NAE is covalently loaded with these Nedd8-mimicking peptides, they can no longer activate full length Nedd8 for its transfer to the neddylation targets such as the cullin subunits of cullin-RING E3 ubiquitin ligases (CRLs). We have thus developed a new method to inhibit protein neddylation via Nedd8-mimicking peptides.
Keywords: Nedd8, Nedd8 activating enzymes, phage display, peptides, enzyme inhibitors
Introduction
An increasing number of cellular proteins are being identified to be posttranslationally modified by Nedd8, a ubiquitin like protein (UBL), to regulate their biological activities in the cell. [1–5] Similar to UB, Nedd8 is transferred through an enzymatic cascade composed of E1, E2 and E3 enzymes to its modification targets. [6–11] One important class of cellular proteins modified by Nedd8 is represented by the cullin-RING E3 ligases (CRLs) that catalyze protein ubiquitination. [12–13] The attachment of Nedd8 to the cullin subunit induces a conformational change in CRL that facilitates UB transfer from the E2 enzyme to the substrate proteins bound to CRL. [14] It has been estimated that there are more than 300 CRLs expressed in human cells and they are responsible for designating 20% of cellular proteins to be degraded by the proteasome after their modification by UB. [15–16] Thus Nedd8 plays an important role in CRL regulation that underlies normal cell physiology. Besides CRL, Nedd8 has also been found to modify other key cellular targets including p53, [17] epidermal growth factor receptor (EGFR), [18] transforming growth factor β (TGF-β) type II receptor, [19] cell cycle regulating transcription factor E2F-1, [20] caspases, [21] ribosomal proteins, [22] histone H4, [23] and Parkin, [24] an E3 UB ligase associated with the early induction of Parkinson’s disease.
Protein modification by Nedd8 can be mediated by either the canonical Nedd8 transfer cascade or the UB transfer cascade. The Nedd8 transfer cascade is composed of the Nedd8 activating E1 enzyme (NAE), [8, 25] E2 enzymes specific for Nedd8 such as Ubc12 (Ube2M) and Ube2F, [7, 9] and E3 enzymes such as Dcn1 and Hrt1 for cullin modification. [10–11] Alternatively the NAE-Ubc12 cascade for Nedd8 transfer can relay with E3 enzymes of the UB transfer cascade such as mouse double minute 2 (Mdm2), Casitas B-lineage lymphoma (c-Cbl), and inhibitors of apoptosis (IAPs) so that Nedd8 can be conjugated to the cellular proteins targeted by E3 enzymes in protein ubiquitination pathways. [17–19, 21] Most recently Nedd8 was found to cross react with the UB activating E1 enzyme (UAE) to enable Nedd8 conjugation to cellular proteins through the UB transfer cascade. [26–28] Due to this atypical mode of Nedd8 transfer, hetero-conjugated Nedd8-UB chains are formed in the cell. [29]
Because of the critical role of protein neddylation in cell biology, the cascade enzymes for Nedd8 transfer have been an intense focus of drug discovery efforts. MLN4924, an adenosine sulfamate analog, was found to be a potent inhibitor of NAE due to the formation of a covalent adduct of the compound with the C-terminus of Nedd8. [16, 30] MLN4924 has been shown to block ubiquitination and degradation of CRL substrates and inhibit tumor growth. Currently MLN4924 is in clinical trials for the treatment of solid tumors and hematological malignancies. [31–32] Other NAE inhibitors have also been developed based on natural product structures and an inorganic rhodium complex. [33–35]
Here we identified short peptides as inhibitors of Nedd8 transfer through the NAE-Ubc12 cascade by carrying out phage selection of a Nedd8 library with randomized C-terminal residues based on their reactivity with NAE. We found that the C-terminal sequences of Nedd8 variants selected by phage display are preferentially recognized by NAE in the activation reaction. The short peptides corresponding to the C-termini of the Nedd8 variants from phage selection can function as Nedd8 mimics to form peptide~NAE conjugates (“~” designates a thioester bond) and block the loading of full length Nedd8 on NAE. These Nedd8-mimicking peptides can thus be used as road blocks to inhibit Nedd8 transfer through the cascade.
Results
Phage selection of the Nedd8 library with a randomized C-terminus
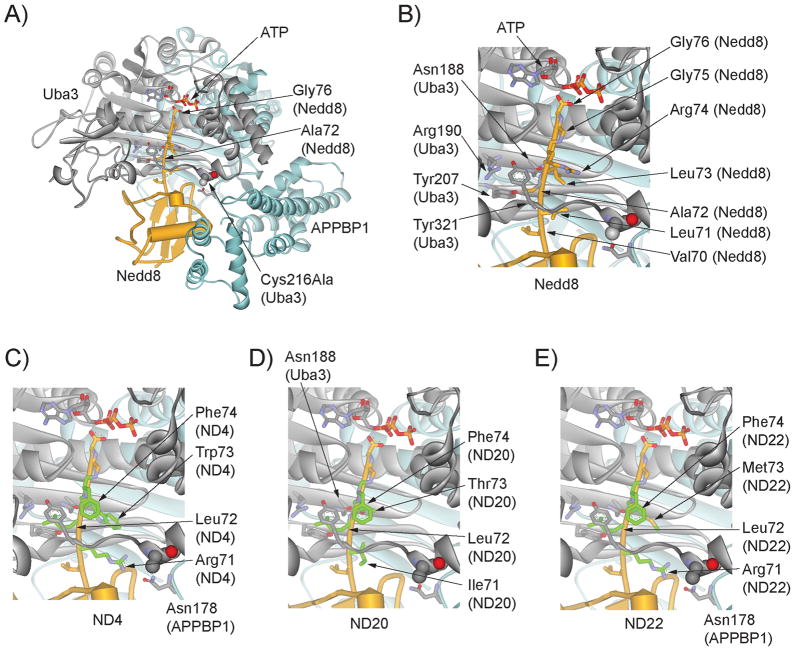
We previously used phage display to profile the recognition of UB C-terminal sequence by UAE and found that UB variants with alternative C-terminal sequences can be activated by UAE and transferred to downstream E2 enzymes. [36] We were thus interested in using phage display to profile the recognition of Nedd8 C-terminal sequence by NAE. Nedd8 is activated by NAE following the same mechanism of UB activation by UAE. [25] A crystal structure of the NAE-Nedd8-ATP triple complex shows that the C-terminal peptide of Nedd8 extends deeply into the ATP binding pocket of NAE so that the carboxyl end of Nedd8 can react with ATP (Figure 1A) to form a Nedd8-AMP conjugate. [37] The activated Nedd8 carboxylate is then captured by the catalytic Cys residue of NAE to form the Nedd8~NAE thioester conjugate.
Figure 1.
Crystal structure of the NAE-Nedd8-ATP complex and modelled structures of Nedd8 variants binding to NAE. A) Binding of wt Nedd8 to NAE as shown in the triple complex with ATP (PDB ID 1R4N). [37] Nedd8 is shown in yellow, the Uba3 subunit of NAE in grey and the APPBP1 subunit of NAE in cyan. The catalytic Cys residue of NAE in the crystal structure was mutated to Ala (Cys216Ala) and it is shown in a space-filling model. B) Interactions of the C-terminal peptide of wt Nedd8 (70VLALRGG76) with the NAE active site residues. The C-terminal residues of Nedd8 are shown in stick models and the NAE residues forming the binding pocket around the Ala72 residue of Nedd8 are also shown. C), D) and E), modelled structures of Nedd8 variants ND4 (70VRLWFGG76), ND20 (70VILTFGG76) and ND22 (70VRLMFGG76) in complex with NAE.
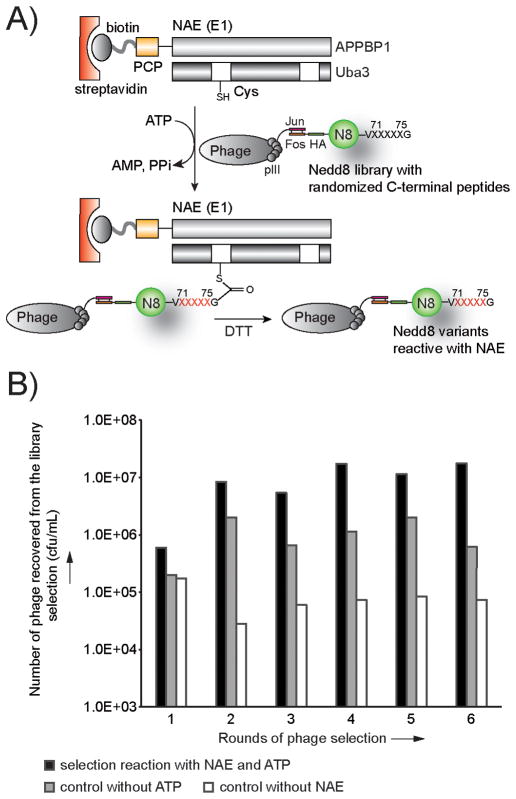
Nedd8 has a native sequence of 71LALRGG76 at its C-terminus. We randomized residues 71–75 of Nedd8 in the library but kept Gly76 intact because the C-terminal Gly residue is conserved across UB and various UBLs, and is vitally important for conjugation to their cellular targets. [38] We then displayed the Nedd8 library on the surface of M13 phage with the pJF3H phagemid. [39–40] To select for Nedd8 variants that were reactive with NAE, we constructed a fusion protein between the NAE subunit APPBP1 and the peptidyl carrier protein (PCP), and coexpressed the fusion protein with Uba3 to assemble a functional heterodimeric NAE (Figure 2A). [41] We labelled the PCP-NAE fusion with biotin by using the enzyme Sfp phospho-pantetheinyl transferase to attach a biotin-phosphopantetheinyl conjugate to PCP. [42] Subsequently we bound the biotinylated PCP-NAE to a streptavidin plate and added the phage library of Nedd8 and ATP to the plate. If the Nedd8 variants displayed on phage were recognized by NAE for the activation reaction, they would form thioester conjugates with NAE on the plate to enable the affinity selection of phage particles displaying the catalytically active Nedd8 variants. Finally the phage captured on the plate were eluted with dithiothreitol (DTT) that cleaved the Nedd8~NAE thioester conjugate. The eluted phage were amplified for iterative rounds of selection.
Figure 2.
Phage selection of the Nedd8 library based on its reactivity with NAE. A) The phage selection scheme. The Nedd8 protein was displayed on the surface of M13 phage with the pJF3H phagemid that expressed Nedd8 with an N-terminal Fos peptide so that Nedd8 could be anchored on the phage surface by high affinity binding of the Jun-Fos peptides. The pJF3H phagemid gave rise to monovalent display of Nedd8 variants on phage particles. C-terminal residues of Nedd8 covering the sequence 71LALRG75 were randomized (“X” in black). Phage selection specifically enriched C-terminal sequences of Nedd8 variants that can be recognized by NAE for activation (“X” in red). B) Titer of the eluted phage from each round of Nedd8 library selection with NAE. In the selection reaction, both NAE and ATP were provided for the formation of Nedd8~NAE thioester conjugates. In the control reactions, either ATP or NAE was excluded from the reaction mixture. The titer numbers of the phage were plotted on a logarithmic scale (cfu, colony forming unit).
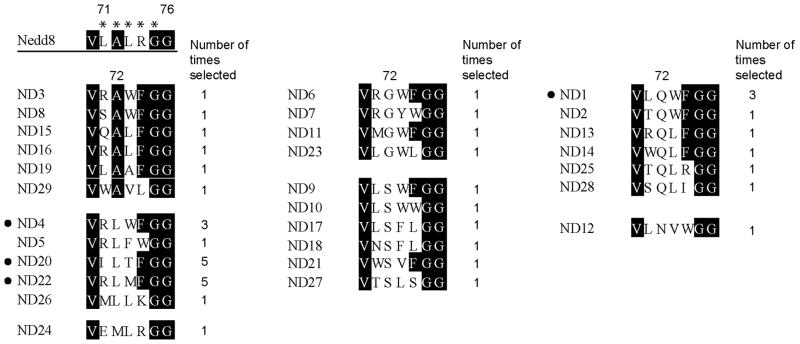
During the library selection, we found that the number of phage particles from the selection reaction were steadily increasing compared to the controls without the addition of ATP or the coating of NAE on the plate (Figure 2B). This suggested that Nedd8 variants reactive with NAE were enriched. Alignment of the C-terminal sequences of the phage clones after six rounds of selection showed that the C-terminal profiles of the selected Nedd8 variants are clearly different from the wild type (wt) Nedd8. As shown in Figure 3, Leu71 of the wt Nedd8 is preferentially replaced with Arg, and Ala72 of wt Nedd8, previously assigned as a key site for NAE recognition, [43] is most often replaced with Leu, Gln and Ser. The alignment also shows that positions 73 and 74 were typically occupied by residues with bulky aromatic side chains such as Phe and Trp in place of the wt Leu and Arg residues, respectively. Interestingly only the wt Gly residue was selected at position 75. This matches the previous observation that the double Gly motif at the end of Nedd8 is indispensable for protein neddylation. [44]
Figure 3.
Alignments of the C-terminal sequences of Nedd8 clones selected by phage display. Nedd8 C-terminal residues 71LALRG75 (designated by the stars) were randomized in the library. Nedd8 variants were grouped based on the identities of the residues selected at position 72 replacing Ala in wt Nedd8. The number of times each clone was identified among 41 sequenced clones after the 6th round of phage selection is also shown. Clones with their names next to bullet points were the most abundant after phage selection and they were further characterized for activation by NAE.
Kinetic characterization of the activation of Nedd8 variants and the corresponding C-terminal peptides by NAE
We expressed the Nedd8 variants ND1, ND4, ND20 and N22 from phage selection to further assay their activities with NAE since they were the most abundant clones in the selected phage pool (Figure 3). Nedd8 variants were expressed with their N-termini fused to glutathione transferase (GST) since it has been reported that the stand alone Nedd8 protein is prone to unfold. [45] The ATP-PPi exchange assay[46–47] of the Nedd8 variants showed that ND20 and ND22 have higher activity than wt Nedd8 (Figure 4A). Measurements of kcat and K1/2 of the Nedd8 variants with NAE under saturating concentrations of ATP found that ND20 displays a K1/2 of one third of wt Nedd8 (Table 1) while kcat values were unchanged taking into account the experimental errors. As a result, the kcat/K1/2 of ND20 (150 μM/min) is three fold higher than wt Nedd8 (43 μM/min). ND22 also has a slightly higher kcat/K1/2 (53 μM/min) due to the decrease in K1/2. These results suggest that phage selection increased the binding affinity of the selected Nedd8 variants with NAE due to variation of the C-terminal sequences and these changes provided Nedd8 variants with a higher reactivity with NAE. We also found that not all Nedd8 variants from the selection were more reactive with NAE than the wt Nedd8. Kinetic characterization of ND1 and ND4 showed that these mutants are less active than wt Nedd8 due to a significant increase in K1/2 (Table 1).
Figure 4.
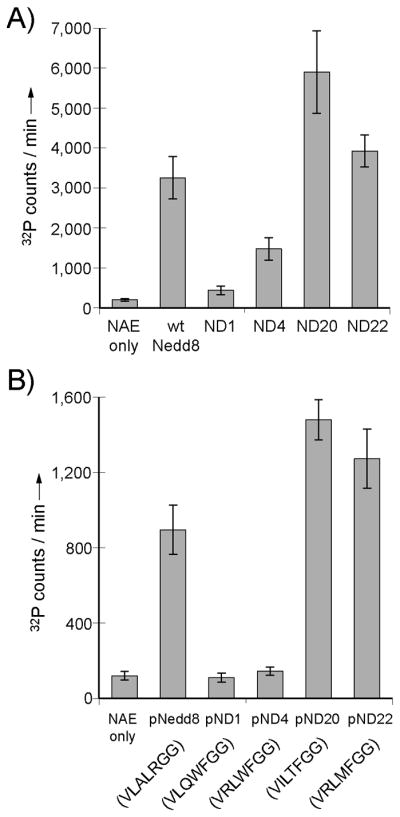
Results of ATP-PPi exchange assay of Nedd8 variants and the corresponding C-terminal peptides catalyzed by NAE. A) Activities of phage selected Nedd8 variants from phage selection with NAE. B) Activities of the short peptides corresponding to the C-terminal sequences of the Nedd8 variants.
Table 1.
Kinetic characterization of the Nedd8 variants enriched by phage selection and the Nedd8-mimicking peptides derived from the C-terminal sequences of the Nedd8 variants.
| K1/2 (μM) | kcat (min™1) | kcat/K1/2 (μMmin™1) | |
|---|---|---|---|
| Full-length proteins | |||
| wt Nedd8 (71LALRGG76) | 1.3 ± 0.2 | 55 ± 12 | 43 |
| Nedd8 variants from phage selection | |||
| ND1 (71LQWFGG76) | 150 ± 20 | 22 ± 9 | 0.15 |
| ND4 (71RLWFGG76) | 4.5 ± 1.1 | 23 ± 7 | 5.1 |
| ND20 (71ILTFGG76) | 0.45 ± 0.03 | 67 ± 10 | 150 |
| ND22 (71RLMFGG76) | 1.0 ± 0.3 | 52 ± 18 | 52 |
| Nedd8-mimicking peptides | |||
| pNedd8 (70VLALRGG76) | 88 ± 25 | 26 ± 7 | 0.29 |
| pND20 (70VILTFGG76) | 42 ± 11 | 28 ± 3 | 0.66 |
| pND22 (70VRLMFGG76) | 51 ± 16 | 29 ± 8 | 0.57 |
Since ND20 and ND22 display an increased affinity with NAE due to their C-terminal variations, we assayed if the short peptides corresponding to their C-terminal sequences can be better recognized by NAE for the activation reaction than the peptide with the native C-terminal sequence of Nedd8. We synthesized peptides pND20 and pND22 with the sequence of 70VILTFGG76 and 70VRLMFGG76. The numbering of the peptide residue follows that of the full length Nedd8 protein and “p” stands for peptide. We assayed the ATP-PPi exchange activity of the peptide catalyzed by NAE and found that based on the kcat/K1/2 values, pND20 and pND22 are twice as active as pNedd8, the peptide with the native C-terminal sequence of Nedd8 (Figure 4B and Table 1). At the same time, the activities (kcat/K1/2) of pND20 and pND22 are 65- and 75-fold lower than the wt Nedd8 protein (Table 1). The significant decreases in the activities of the peptides from the full length protein are largely due to the increases in K1/2 of the peptides (Table 1). This suggests that other interfaces of the globular fold of Nedd8 make important contributions in binding to NAE. Nevertheless the pND20 and pND24 peptide have a K1/2 about half of the pNedd8 peptide in the ATP-PPi exchange reaction, suggesting that phage selection indeed identified new C-terminal sequences of Nedd8 that are preferentially recognized by NAE over the wt Nedd8.
Assaying the transfer of Nedd8 variants and the corresponding peptides through the NAE-Ubc12 cascade for cullin modification
Once Nedd8 is activated by NAE to form the Nedd8~NAE conjugate, a catalytic Cys residue of the E2 bound to NAE can react with the C-terminal carboxylate of Nedd8 through a thioester exchange reaction that leads to the formation of a Nedd8~E2 conjugate. Subsequently Nedd8~E2 can bind to the Rbx1 subunit of CRL to deliver Nedd8 to cullin by forming an amide bond between the C-terminal carboxylate of Nedd8 and a Lys residue of the cullin subunit. Since the Nedd8 variants from phage selection can be activated by NAE as demonstrated by the ATP-PPi exchange assay (Figure 4A and Table 1), we further tested if the Nedd8 variants can be covalently charged to NAE and be further transferred to Ubc12 (E2) and cullin. We incubated the GST fusions of the Nedd8 variants with NAE, Ubc12 and the Rbx1-cullin 3 complex, and assayed the formation of Nedd8 conjugates via Western blot of the reaction mixtures with an anti-GST antibody. As shown in Figure 5, wt Nedd8 could form covalent conjugates with NAE, Ubc12 and cullin 3. The Nedd8 variant ND20 was most reactive with the Nedd8 transfer cascade; it could not only be efficiently transferred to NAE and Ubc12, but also further to cullin 3. In comparison, ND4 and ND22 were less active. They could form covalent conjugates with NAE and Ubc12, yet no significant formation of a Nedd8-cullin conjugate was observed. ND1 failed to form a covalent conjugate with NAE and as expected could not be transferred to Ubc12 and cullin 3. These results suggest that the phage selected Nedd8 variants with non-native C-terminal sequences may vary significantly in their activities with the cascade enzymes at different stages of Nedd8 transfer.
Figure 5.
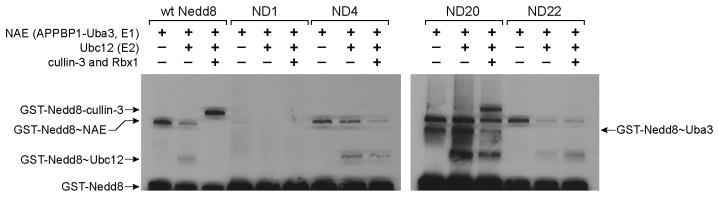
Western blot analysis of the transfer of Nedd8 variants from phage selection to NAE, Ubc12 and cullin 3. Nedd8 variants were expressed with an N-terminal GST fusion. The formation of Nedd8 conjugates was detected by Western blot with an anti-GST antibody.
To assay the transfer of the peptides derived from the C-terminal sequences of the Nedd8 variants through the cascade, we conjugated biotin to the N-terminal amino groups of peptides pNedd8, pND20 and pN22, and probed the Western blot of the transfer reaction with a streptavidin-horseradish peroxidase (HRP) conjugate. As shown in Figure 6A, pNedd8 with the C-terminal sequence of wt Nedd8 could form conjugates with NAE, Ubc12 and cullin 3. In contrast pND20 and pND22 could efficiently form peptide~NAE conjugates, yet they were defective in being transferred to Ubc12 and cullin 3. These results suggest that the wt sequence of the Nedd8 C-terminus could be important for Nedd8 transfer through the cascade enzymes and eventually to its modification targets at the end of the cascade. The peptide~NAE conjugates can be cleaved with 100 mM DTT confirming the formation of a thioester linkage between the peptides and NAE (Figure S1 in the Supporting Information). Since pND20 and pND22 can be activated and form covalent conjugate with NAE just as the Nedd8 protein, we refer to these peptides as “Nedd8-mimicking peptides”.
Figure 6.
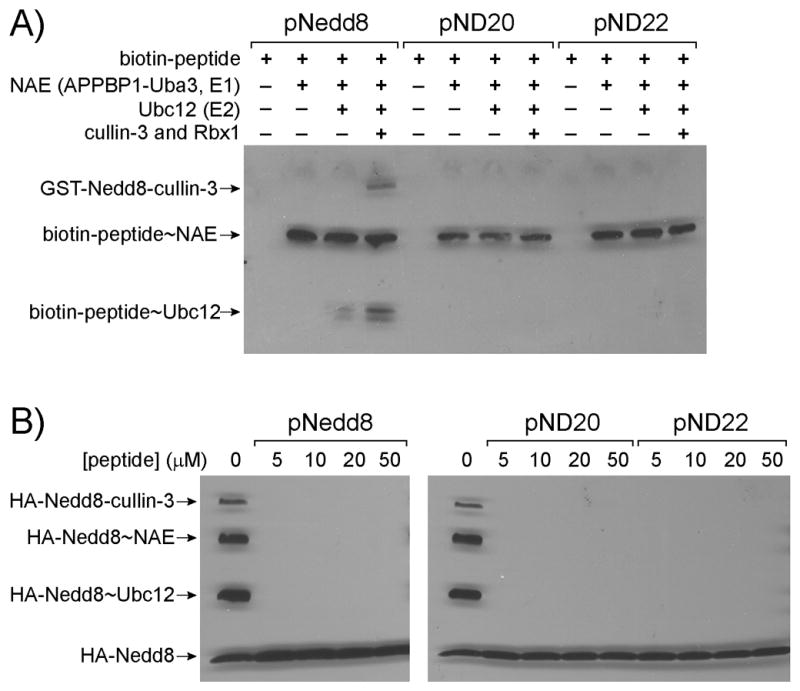
Peptide transfer through the Nedd8 enzymatic cascade and inhibition of Nedd8 transfer through the cascade by selected peptides. A) Loading of biotin-labelled peptides on NAE, Ubc12 and cullin 3. The Western blot was probed with a streptavidin-HRP conjugate. B) Inhibiting Nedd8 loading on NAE by Nedd8-mimicking peptides. NAE, Ubc12 and cullin 3-Rbx1 were first incubated with the Nedd8-mimicking peptides followed by the addition of HA-Nedd8. Attachment of HA-Nedd8 to cascade enzymes and cullin 3 was probed with an anti-HA antibody.
Inhibiting Nedd8 transfer through the NAE-Ubc12 cascade for cullin modification by Nedd8-mimicking peptides
We previously used phage selection to identify UB variants with alternative C-terminal sequences that can be activated by UAE. [36] We showed that short peptides derived from the C-terminal sequence of the UB variants are reactive with UAE to form thioester conjugates with UAE and E2s. [48] We referred to these peptides as “UB-mimicking peptides”. We found that they can effectively block the transfer of UB through the cascade enzymes due to their covalent attachment to UAE and E2s. We were thus interested in testing if the Nedd8-mimicking peptides from this study could block Nedd8 modification of cullin by inhibiting Nedd8 transfer through the NAE-Ubc12 cascade.
To test the activities of the peptides on blocking Nedd8 transfer, we preincubated peptides pNedd8, pND20 and pND22 with NAE, Ubc12 and cullin for 1 hour in the presence of ATP to allow for the loading of the peptides on NAE and further transfer of peptides to Ubc12 and cullin. We next added full length Nedd8 with an N-terminal HA tag to allow for the formation of HA-Nedd8 conjugates with the cascade enzymes and cullin. Western blot analysis of the reaction probed with an anti-HA antibody showed that preincubation of the peptides at concentrations as low as 5 μM with the cascade enzymes allowed complete blocking of Nedd8 modification of cullin 3 due to the inhibition of Nedd8 to form thioester conjugates with NAE and Ubc12 (Figure 6B). We also noted that peptide pNedd8 with the wt C-terminal sequence of Nedd8, and peptides pND20 and pND22 with sequences identified by phage selection, inhibited the formation of Nedd8 conjugates with NAE and Ubc12 to similar extents.
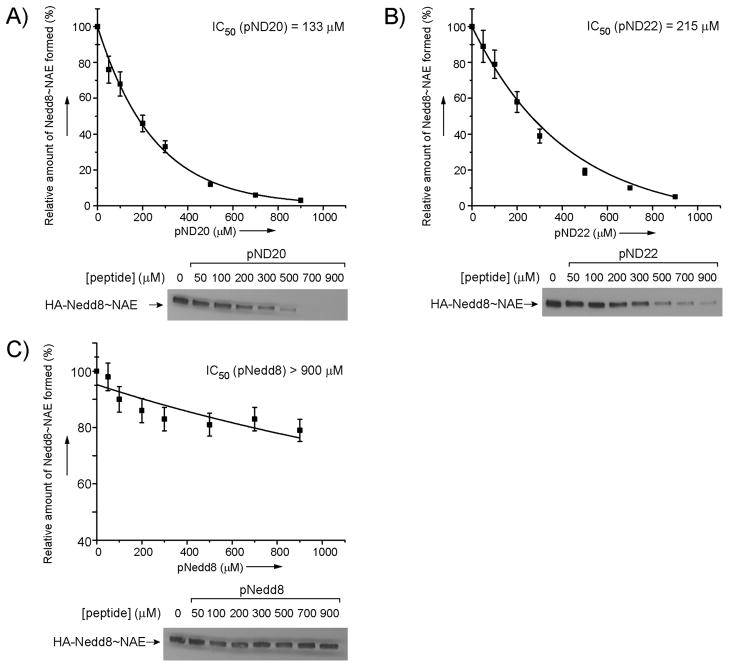
Next we measured the IC50 values of the peptides in inhibiting the formation of Nedd8~NAE conjugates. We incubated NAE with increasing concentrations of the peptides in the presence of 1 μM of HA-Nedd8 and ATP to allow competition between the peptides and Nedd8 for reacting with NAE. After a 15-minute incubation, the Nedd8-NAE conjugate was separated from the rest of the components by running the reaction on an SDS-polyacrylamide gel. The Western blot of the gel was probed with an anti-HA antibody to detect Nedd8 conjugates. The intensities of the band corresponding to the Nedd8~NAE conjugate were quantified by densitometry (Figure 7). [49] We found that pND20 and pND22 have observed IC50 values of 133 μM and 215 μM, respectively, in inhibiting the loading of Nedd8 on NAE. In contrast, the pNedd8 peptide with the C-terminal sequence of wt Nedd8 could not efficiently compete with full length Nedd8 in NAE loading at a concentration as high as 900 μM. These results suggest that some of the short peptides identified by phage selection are better recognized by NAE than the C-terminal peptide of wt Nedd8. As the Nedd8-mimicking peptides pN20 and pN22 function as competitive inhibitors of Nedd8 and form thioester conjugates with NAE, they can block Nedd8 loading on NAE and its subsequent transfer through the cascade enzymes. In this sense, the Nedd8 mimicking peptides are mechanism-based inhibitors of the Nedd8 transfer cascade.
Figure 7.
Inhibition of Nedd8 loading on NAE by the Nedd8-mimicking peptides pN20 (A), pND22 (B) and pNedd8 (C). Increasing concentrations of peptides were incubated with Nedd8 and NAE. The amount of the Nedd8~NAE conjugate formed in each reaction was quantified by Western blots probed with an anti-HA antibody. IC50 values of the NAE inhibition by the peptides were calculated by plotting the densities of the bands corresponding to the Nedd8~NAE conjugate against the concentrations of the peptides. The Western blots of Nedd8~NAE formed in the inhibition reactions are shown underneath the plots.
Discussion
In this study we used phage display to profile the recognition of the C-terminal sequence of Nedd8 by NAE. We found that NAE can accommodate diverse changes in residues 71 to 74 of Nedd8 and still activate the Nedd8 variants and initiate their transfer through the cascade for protein modification. We also observed that Nedd8 variants from phage selection may be stalled at different stages of the cascade, apparently due to the differences in their C-terminal sequences. ND4 and ND22 can be transferred to NAE and Ubc12 to form thioester conjugates, but they are incapable of being transferred from Ubc12 to cullin. In contrast ND20 can be transferred all the way through the cascade to modify cullin proteins (Figure 5). Nedd8 conjugation to the cullin protein induces a drastic rearrangement in CRL so that the UB~E2 conjugate can approach the substrate protein bound to CRL to facilitate UB transfer to the substrate protein. [14, 50] It would be of interest to assay if the Nedd8 variants with the non-native C-terminal sequences can activate CRL in a similar manner as wt Nedd8.
NAE is the gate-keeping enzyme of the Nedd8 transfer cascade. It is responsible for the specific activation of Nedd8 and fencing off UB and other UBL proteins from entering the Nedd8 transfer cascade. A key element on Nedd8 for NAE recognition is the C-terminal peptide of Nedd8 that adopts an extended conformation reaching all the way to the ATP moiety that is bound to the Uba3 subunit of NAE (Figure 1A). [37] Uba3 residues Asn188, Arg190, Tyr207 and Tyr321 assemble a well-defined pocket for the binding of the Ala72 side chain of Nedd8 within its C-terminal sequence (71LALRGG76) (Figure 1B). The NAE binding pocket for the Ala72 residue of Nedd8 plays a key role in differentiating Nedd8 from UB. [43] UB shares a high sequence and structural homology with Nedd8 and its C-terminus has the sequence 71LRLRGG76 with just a single residue difference from Nedd8 at position 72. However, UB is not recognized by NAE for the activation reaction. As shown by the crystal structure of the Nedd8-NAE complex, [37] the Arg72 side chain of UB would be repelled by Arg190 of NAE (Figure 1B). Mutagenesis studies further showed that the Arg72Leu mutant of UB can be efficiently activated by NAE, confirming the role Arg190 plays in NAE differentiation of Nedd8 and UB based on electrostatic interactions. [37, 43, 51]
Our results obtained by phage selection of the Nedd8 library showed the bias of NAE against positively charged residues at position 72 of Nedd8 (Figure 3). None of the Nedd8 variants from the selection has a positively charged Arg, Lys or His side chain at position 72. We also found that Ala72 of Nedd8 can be replaced by both polar or hydrophobic side chains such as Ser, Gln or Leu. Matching with our selection results, it has been found that the Ala72Gln mutation in Nedd8 did not affect Nedd8 activation by NAE and the crystal structure of the Nedd8 mutant bound to NAE showed that the Gln72 side chain is engaged in hydrogen bonding interactions with the Arg190 side chain of Uba3. [43] Indeed we found that multiple Nedd8 variants were selected with an Ala72Gln mutation (Figure 3).
Besides the tolerance of residues other than Ala at position 72 of Nedd8, NAE can also accommodate diverse residues at positions 71, 73 and 74 of Nedd8 as suggested by the phage selection results (Figure 3). Based on the sequence alignment of the selected Nedd8 clones, Position 71 is mainly occupied by the wt Leu or a positively charged Arg residue, while positions 72 and 73 have a clear preference for bulky aromatic side chains. We modelled the binding of Nedd8 variants ND4, ND20 and ND22 with NAE based on the crystal structure of the NAE-Nedd8-ATP complex (Figure 1C, 1D and 1E). [37] The modelled structures suggest that the larger space between the C-terminal peptide backbone of the Nedd8 and the NAE active site residues, compared to the UAE in complex with UB, [52] makes it possible for NAE to accommodate multiple aromatic side chains at positions 71, 73 and 74 to replace Leu or Arg in wt Nedd8. The structural models show that space is somewhat limited for the Leu at position 72, however, slight rearrangements of the NEDD8 C-terminus should easily overcome these restrictions while the existing binding pocket is favourable for an aliphatic side chain reflecting its ability to interact with Ala in wt NEDD8. An Arg at position 71 as present in the variants ND4 and ND22 has the potential to engage in polar interactions with Asn178 in the crossover loop of the APPBP1 subunit of NAE (Figure 1C and 1E). Likewise a Thr at position 73 (ND20) can form hydrogen bonds with Asn188 of the Uba3 subunit of NAE (Figure 1D). Possibly these residues contribute to the decreased IC50 values of the pND20 and pND22 peptides while the Trp at position 73 of the pND4 peptide could simply be too large compared to Leu in wt pNedd8, and Thr and Met in the pND20 and pND22 peptides, respectively. As a result the pND4 peptide is much less active in the ATP-PPi exchange assay catalyzed by NAE (Figure 4B).
We also found that the Nedd8 variants ND20 and N22 selected by phage display have a higher affinity with NAE than the wt Nedd8 (Table 1). Correspondingly the 7-mer peptides pND20 and pND22 derived from the C-terminal sequences of the Nedd8 variants display a higher affinity towards Nedd8 than the peptide with the native C-terminal sequence of Nedd8 (Table 1). Peptides pND20 and pND22 can mimic the action of Nedd8 in forming Nedd8-AMP conjugates as shown by the ATP-PPi exchange assay and in forming thioester conjugates with NAE (Figure 4B and 6A). Once the peptides are conjugated to NAE, they block the catalytic Cys of NAE from being loaded with full length Nedd8. Furthermore the activity of NAE with Nedd8 cannot be regenerated by delivering the peptides to the downstream E2s since the peptides are defective in transferring from NAE to E2 (Figure 6A). In contrast, peptide pNedd8 with the wt C-terminal sequence of Nedd8 can form thioester conjugates with NAE and E2 in tandem and eventually be transferred to cullins.
Due to the optimization of Nedd8 C-terminal peptide-NAE interaction by phage selection, peptides pND20 and pND22 gained higher affinity with NAE than pNedd8; their IC50 values are at least 4–7-fold lower than pNedd8 in inhibiting Nedd8 loading on NAE (Figure 7). The Nedd8-mimicking peptides from phage selection therefore provide a new mechanism to inhibit NAE and the downstream Nedd8 transfer pathways. Further study on the effects of these peptides on regulating protein neddylation in the cell is worth pursuing.
Experimental Section
See the supporting information for experimental procedures on cloning, protein expression, construction of the Nedd8 library, phage selection of the library, and characterization of the Nedd8 variants and the Nedd8-mimicking peptides.
Supplementary Material
Acknowledgments
This work was supported by a lab startup grant from the University of Chicago, a National Science Foundation CAREER award (1057092) to J.Y., a National Institute of Health grant 1R01GM104498 to J.Y. and H.K., a NIH grant R01-CA112282 to H.K., and the Deutsche Forschungsgemeinschaft (SCHI 425/6-1 and FZ 82) to H.S. This work was also funded in part by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (to J.Y. and H.K.). We thank Dr. Brenda A. Schulman of the Howard Hughes Medical Institute and St Jude Children’s Research Hospital (USA) for providing the expression plasmids of Nedd8 and NAE. We also thank Dr. Ning Zheng of the Howard Hughes Medical Institute and University of Washington for providing the expression plasmids of Ubc12 and the Rbx1-cullin3 protein.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.del Pozo JC, Estelle M. Proc Natl Acad Sci U S A. 1999;96:15342–15347. doi: 10.1073/pnas.96.26.15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Proc Natl Acad Sci U S A. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabut G, Peter M. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitby FG, Xia G, Pickart CM, Hill CP. J Biol Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 6.Gong L, Yeh ET. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 7.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. Mol Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Joazeiro CA. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 13.Hotton SK, Callis J. Annu Rev Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- 14.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarikas A, Hartmann T, Pan ZQ. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 17.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, Yarden Y. J Biol Chem. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 19.Zuo W, Huang F, Chiang YJ, Li M, Du J, Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, Deng H, Feng XH, Luo S, Gao C, Chen YG. Mol Cell. 2013;49:499–510. doi: 10.1016/j.molcel.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Loftus SJ, Liu G, Carr SM, Munro S, La Thangue NB. EMBO Rep. 2012;13:811–818. doi: 10.1038/embor.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J, Ditzel M, Meier P. Mol Cell. 2010;40:810–822. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma T, Chen Y, Zhang F, Yang CY, Wang S, Yu X. Mol Cell. 2013;49:897–907. doi: 10.1016/j.molcel.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Um JW, Han KA, Im E, Oh Y, Lee K, Chung KC. J Neurosci Res. 2012;90:1030–1042. doi: 10.1002/jnr.22828. [DOI] [PubMed] [Google Scholar]
- 25.Schulman BA, Harper JW. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjerpe R, Thomas Y, Chen J, Zemla A, Curran S, Shpiro N, Dick LR, Kurz T. Biochem J. 2012;441:927–936. doi: 10.1042/BJ20111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hjerpe R, Thomas Y, Kurz T. J Mol Biol. 2012;421:27–29. doi: 10.1016/j.jmb.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Leidecker O, Matic I, Mahata B, Pion E, Xirodimas DP. Cell Cycle. 2012;11:1142–1150. doi: 10.4161/cc.11.6.19559. [DOI] [PubMed] [Google Scholar]
- 29.Singh RK, Zerath S, Kleifeld O, Scheffner M, Glickman MH, Fushman D. Mol Cell Proteomics. 2012;11:1595–1611. doi: 10.1074/mcp.M112.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 31.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. Genes & cancer. 2010;1:708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soucy TA, Smith PG, Rolfe M. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 33.Leung CH, Chan DS, Yang H, Abagyan R, Lee SM, Zhu GY, Fong WF, Ma DL. Chem Commun. 2011;47:2511–2513. doi: 10.1039/c0cc04927a. [DOI] [PubMed] [Google Scholar]
- 34.Zhong HJ, Ma VP, Cheng Z, Chan DS, He HZ, Leung KH, Ma DL, Leung CH. Biochimie. 2012;94:2457–2460. doi: 10.1016/j.biochi.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Zhong HJ, Yang H, Chan DS, Leung CH, Wang HM, Ma DL. PLoS One. 2012;7:e49574. doi: 10.1371/journal.pone.0049574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Bhuripanyo K, Schneider J, Zhang K, Schindelin H, Boone D, Yin J. ACS Chemical Biology. 2012;7:2027–2035. doi: 10.1021/cb300339p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr, Holton JM, Schulman BA. Mol Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 38.Hochstrasser M. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbas CF, 3rd, Burton DR, Scott JK, Silverman GJ. Phage Display, A laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2000. [Google Scholar]
- 40.Crameri R, Suter M. Gene. 1993;137:69–75. doi: 10.1016/0378-1119(93)90253-y. [DOI] [PubMed] [Google Scholar]
- 41.Huang DT, Schulman BA. Methods Enzymol. 2005;398:9–20. doi: 10.1016/S0076-6879(05)98002-6. [DOI] [PubMed] [Google Scholar]
- 42.Yin J, Lin AJ, Golan DE, Walsh CT. Nature Protocols. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 43.Souphron J, Waddell MB, Paydar A, Tokgoz-Gromley Z, Roussel MF, Schulman BA. Biochemistry. 2008;47:8961–8969. doi: 10.1021/bi800604c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girdwood D, Xirodimas DP, Gordon C. PLoS One. 2011;6:e20089. doi: 10.1371/journal.pone.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitahara R, Yamaguchi Y, Sakata E, Kasuya T, Tanaka K, Kato K, Yokoyama S, Akasaka K. J Mol Biol. 2006;363:395–404. doi: 10.1016/j.jmb.2006.07.074. [DOI] [PubMed] [Google Scholar]
- 46.Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Proc Natl Acad Sci U S A. 1981;78:761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haas AL, Rose IA. J Biol Chem. 1982;257:10329–10337. [PubMed] [Google Scholar]
- 48.Zhao B, Choi CHJ, Bhuripanyo K, Villhauer EB, Zhang K, Schindelin H, Yin J. Org Lett. 2012;14:5760–5763. doi: 10.1021/ol3027736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yunus AA, Lima CD. Methods Mol Biol. 2009;497:167–186. doi: 10.1007/978-1-59745-566-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saha A, Deshaies RJ. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bohnsack RN, Haas AL. J Biol Chem. 2003;278:26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 52.Lee I, Schindelin H. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.