SUMMARY
c-Myc oncogenic activity is thought to be mediated in part by its ability to generate DNA replication stress and subsequent genomic instability when deregulated. Previous studies have demonstrated a nontranscriptional role for c-Myc in regulating DNA replication. Here, we analyze the mechanisms by which c-Myc deregulation generates DNA replication stress. We find that overexpression of c-Myc alters the spatiotemporal program of replication initiation by increasing the density of early-replicating origins. We further show that c-Myc deregulation results in elevated replication-fork stalling or collapse and subsequent DNA damage. Notably, these phenotypes are independent of RNA transcription. Finally, we demonstrate that overexpression of Cdc45 recapitulates all c-Myc-induced replication and damage phenotypes and that Cdc45 and GINS function downstream of Myc.
INTRODUCTION
DNA replication involves the stepwise assembly of the prereplicative complex on chromatin (Machida et al., 2005; Méchali, 2010; Remus and Diffley, 2009). The origin recognition complex (ORC) is recruited first, followed by the Cdc6- and Cdt1-dependent loading of the mini chromosome maintenance (MCM)2–7 helicase. MCMs are present on DNA in excess, as inactive double hexamers (Blow et al., 2011; Evrin et al., 2009; Remus and Diffley, 2009). Replication initiation occurs upon the action of S phase CDKs and CDC7, which promote Cdc45 and GINS binding to and subsequent activation of the MCMs (Costa et al., 2011; Remus and Diffley, 2009). DNA is then unwound by the Cdc45-MCM2-7-GINS (CMG) complex (Fu et al., 2011; Gambus et al., 2006; Ilves et al., 2010; Pacek et al., 2006), which also facilitates the loading of DNA polymerase-α to initiate DNA synthesis (Heller et al., 2011; Labib, 2010; Mimura and Takisawa, 1998).
The mechanisms that determine the exact sites of replication initiation are not well understood in metazoans. Genome-wide and fine-mapping studies suggest that chromatin context, transcriptional interference, epigenetic regulation, and specific sequence bias play a role in positioning origins (Gilbert, 2010; Méchali, 2010). More replication origins are assembled than those actually used; these additional, dormant origins are critical to prevent genomic instability at times of replication fork failure and ensure the timely completion of DNA replication (Anglana et al., 2003; Ge et al., 2007; Lucas and Hyrien, 2000; Woodward et al., 2006). In addition, proper availability of dormant origins appears to protect against tumorigenesis (Kawabata et al., 2011).
The c-Myc proto-oncogene (Myc) is an essential regulator of cell growth, and its expression is frequently altered in cancer (Dang et al., 2006; Grandori et al., 2000). Myc regulates transcription of several genes that control key cellular functions including cell growth and cell-cycle progression (Fernandez et al., 2003; Patel et al., 2004). When deregulated, Myc promotes oncogenesis, as demonstrated in different cell types and in transgenic mice (Adams et al., 1985; Kovalchuk et al., 2000). However, the mechanism of Myc-induced oncogenesis remains unclear. Myc may contribute to tumorigenesis by over-stimulating cell growth and metabolism, and/or by causing genomic instability (Classon et al., 1987; Pelengaris et al., 2002). The latter effect has been attributed in part to the role of Myc in regulating DNA replication (Cole and Cowling, 2008; Dominguez-Sola et al., 2007; Sankar et al., 2009). Replication of DNA in the absence of Myc is impaired, whereas Myc overexpression triggers signs of origin hyperactivation. Deregulated Myc also activates a DNA damage response (Dominguez-Sola et al., 2007); however, the aberrant DNA structure that is responsible has not been identified.
Here, we used DNA combing to investigate the consequences of Myc overexpression on the spatiotemporal program of DNA replication, and on the stability of replication forks in Xenopus-cell-free extracts and mammalian cells. We show that Myc initiates premature origin firing, increases origin density, and leads to asymmetrical fork progression and DNA damage. We further demonstrate that Cdc45 overexpression phenocopies all Myc-dependent phenotypes and that the Myc-induced replication alterations are manifested only when Cdc45 or GINS are available, indicating that Myc functions upstream of the CMG complex in a pathway that leads to activation of replication origins. Finally, we provide insight into the mechanism by which Myc facilitates initiation of DNA replication.
RESULTS
Myc Influences the Spatiotemporal Pattern of Origin Activity
Myc positively regulates DNA replication (Herold et al., 2009; Pelengaris et al., 2002), and our previous studies helped identify the nontranscriptional role of Myc in replication. Myc directly binds to DNA and appears to influence the activity of replication origins (Dominguez-Sola et al., 2007). To get insights into the mechanism of Myc-induced replication, we used molecular DNA combing (Marheineke et al., 2009) and cell-free extracts derived from Xenopus eggs to monitor origin activity under normal or Myc-overexpressing conditions. The Xenopus system recapitulates semiconservative, chromosomal DNA replication but does not support RNA transcription (Srinivasan and Gautier, 2011).
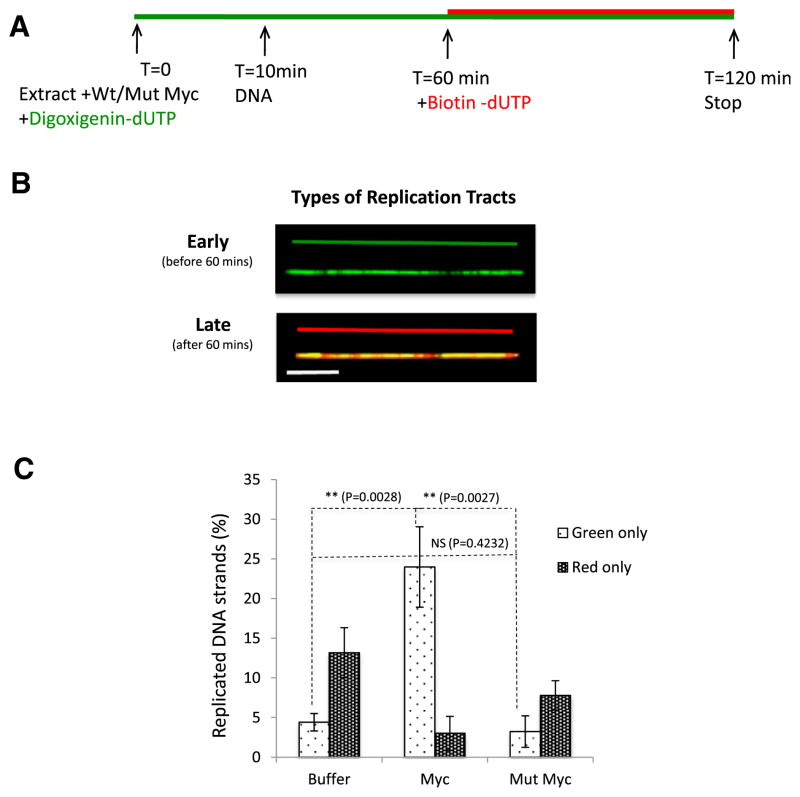
The experimental strategy is depicted in Figure 1A. Replicating chromosomal DNA is sequentially labeled with two nucleotide analogs that can be differentially detected with specific antibodies. Addition of the nucleotide analogs at different times in the experiment allowed us to differentiate between stretches of early- (before 60 min) and late- (after 60 min) replicating DNA (Figure 1B), monitor fork movement, fork symmetry, and interorigin distance.
Figure 1. Myc Triggers Premature Activation of Replication Origins.
(A) Schematic representation of the experiments.
(B) Types of DNA fibers scored in (C). Since digoxygenin-dUTP is not removed prior to addition of biotin-dUTP, DNA incorporating green and red nucleotides results in orange tracks. Scale bar is 10 kb.
(C) Percentage of fibers that replicate early (green, before 60 min) or late (red, after 60 min) in control, Myc-, and mutant-Myc-supplemented extracts. Number of DNA molecules analyzed: NBuffer = 120; NMyc = 106.
Error bars represent SD of the mean. See also Figure S1.
First, we monitored the impact of adding recombinant Myc protein on the timing of origin firing. To this end, we quantified DNA fibers in which replication was initiated and completed within 1 hr (green only) and fibers in which replication was initiated after 1 hr and progressed until termination of the reaction at 2 hr (red only). The average length of DNA molecules was calculated using YO-YO-iodide staining, which stains both replicated and unreplicated DNA. We observed that DNA supplemented with either buffer (73.42 kb) or recombinant Myc (74.89 kb) did not significantly differ in length (p = 0.2407; Figure S1A). However, the distribution of replicated DNA (Figure S1B) showed that DNA replicated upon Myc overexpression was shorter than controls (83.18 kb for buffer versus 69.34 kb for Myc).
The addition of full-length Myc protein (75 nM) resulted in 25% of DNA fully replicating before 1 hr when compared to the 5% of early replicated fibers in the buffer control (Figure 1C). In contrast, addition of recombinant Myc protein with a C-terminal truncation removing the bHLH domain and thus abrogating DNA binding (amino acid residues 337–420; Mut Myc) had no significant impact on the timing of origin firing (Figure 1C). Myc therefore triggers early origin firing consistent with our previous data showing an early burst in radiolabeled nucleotide incorporation in Myc-supplemented extracts (Dominguez-Sola et al., 2007).
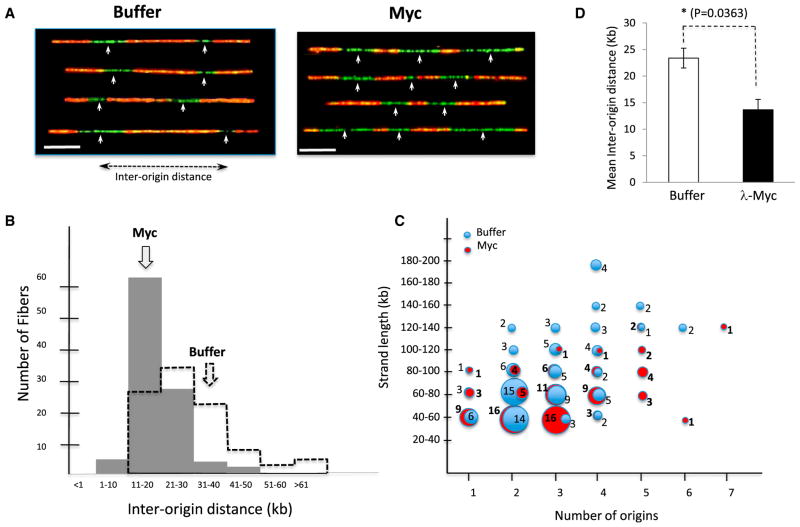
To evaluate the impact of Myc on the spatial distribution, i.e., density of origins, we next measured the distance between consecutive origins of replication. Origins that fire within the initial 60 min and continue to elongate past 60 min yield green tracks (digoxigenin-dUTP) flanked by orange tracks (digoxigenin-dUTP + biotin-dUTP). In DNA molecules where multiple origins fire, the fibers showed alternate stretches of green and red. The midpoint of each green track was scored as a replication origin. The distance from one origin to another is the “inter-origin distance” (Figure 2A) measured in kilobases. We scored DNA fibers that contain two origins or more, which represented about 60% of the total DNA molecules. Comparison of DNA from buffer and Myc-supplemented extracts shows that origins are more closely spaced following addition of Myc and thereby caused a shift in the distribution of interorigin distances (Figure 2B). The mean distance between origins was 29.83 kb (SD = 4.2 kb) in control and 19.2 kb (SD = 2.3 kb) upon Myc overexpression (Figure 2B). The number of origins per DNA fiber was also greater upon Myc overexpression (Figure 2C) despite the fact that the average length of replicated DNA was slightly shorter in Myc-treated extracts than in control (Figure S1B).
Figure 2. Myc Overexpression Decreases Interorigin Distance in Xenopus Extracts and Mammalian Cells.
(A) Representative images of DNA molecules scored in (B) and (C). White arrows indicate the putative position of replication origins. Scale bar is 10 kb.
(B) The interorigin distance is plotted against the percentage of DNA molecules in buffer and Myc-supplemented extracts. Number of origins analyzed: NBuffer = 117 NMyc = 118.
(C) Number of origins was plotted against DNA length (kb). The numbers on/beside the circle represent the number of DNA fibers with a given number of origins.
(D) B cells isolated from wild-type or λ-Myc mice were sequentially labeled with CldU and IdU. The combed DNA from cells was stained for early- (green) and late-(red) replicating tracts, and interorigin distances were plotted as described in (B).
Error bars represent SD of the mean. See also Figure S1.
Next, we investigated the effect of Myc overexpression on DNA replication in mammalian cells. We used B cells from mice expressing a Myc transgene (λ-Myc mice), a mouse model in which Myc deregulation promotes the development of immature B cell neoplasias (Kovalchuk et al., 2000). B cells were isolated from splenic mononuclear cell suspensions, stimulated to proliferate in vitro, and sequentially labeled with nucleotide analogs (5-chloro-2-deoxyuridine- green followed by 5-iodo-2-deoxyuridine- red). Interorigin distance was measured following the isolation and staining of combed DNA molecules. The DNA of B cells isolated from transgenic λ-Myc mice had shorter interorigin distances than their controls (15.04 kb versus 22.07 kb) (Figure 2D). Thus, constitutive overexpression of Myc in murine B cells resulted in a significant decrease in inter-origin distance, as did the addition of recombinant Myc protein to Xenopus extracts.
Myc Causes Aberrant Fork Progression
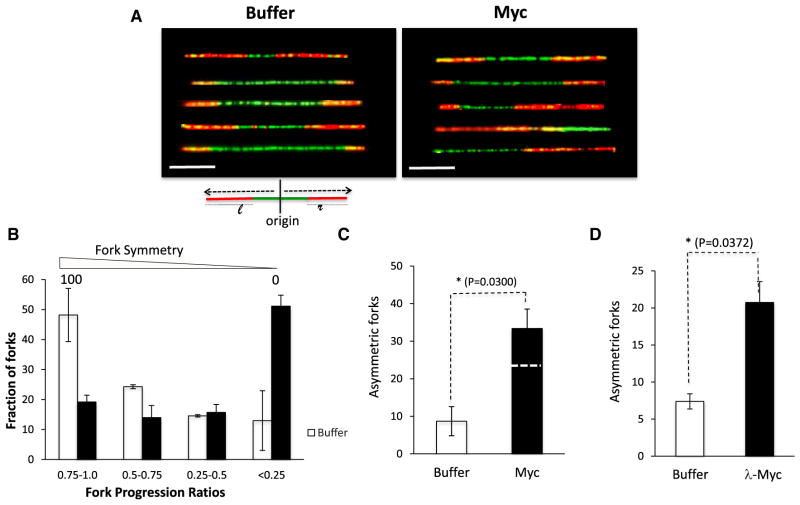
Myc overexpression triggers a robust DNA damage response (Dominguez-Sola et al., 2007; Karlsson et al., 2003). The aberrant DNA structure that elicits this response is not known. We hypothesized that stalled or collapsed replication forks could be the structures responsible for Myc-dependent DNA damage and therefore sought to assess the impact of Myc on fork progression. Following origin firing, replication elongation typically proceeds symmetrically on either side of the origin. To assess fork symmetry, we scored DNA molecules that were at least 40 kb in length and contained a single origin that fired before 60 min (green), flanked by tracks indicating fork progression (red) on one or both sides (Figure 3A). The DNA molecules with a single origin represented about 10%–15% of the total population of DNA fibers analyzed. To quantify the symmetry of fork progression, the extent of biotin-dUTP incorporation to the right and left of the origin was measured (length of red tracks that flank the origin, l: left, r: right) and the ratio between them (fork progression ratio [FPR]) was computed (Figure 3A). Perfectly symmetric forks have an FPR of 1, whereas unidirectional forks (with a single red track) have an FPR of 0. In the absence of exogenous Myc, 50% of forks were largely symmetrical, with an FPR value between 0.75 and 1.0. In contrast, the addition of Myc resulted in 50% of forks being highly asymmetrical with FPR values between 0 and 0.25 (Figure 3B). Notably, Myc addition did not yield many forks with partial asymmetry (FPR values between 0.25 and 0.75). This observation suggests that Myc overexpression does not slow down fork progression but rather triggers fork-stalling events that occur in close proximity to the origin.
Figure 3. Myc Causes Replication Fork Stalling in Xenopus Extracts and Mammalian Cells.
(A–D) The DNA fibers that had only one origin and contained forks that progressed either uni- or bidirectionally were analyzed.
(A) Representative images of DNA molecules that incorporated digoxigenin (green) or biotin (red) in control (buffer) or upon Myc overexpression (Myc). Scale bar is 10 kb.
(B) Fork progression ratio (FPR) of DNA fibers in control and Myc-supplemented extracts. FPR is calculated by measuring the lengths of red tracts and dividing the smaller value by the larger one.
(C) Fraction of DNA fibers that had unidirectional fork progression in control and Myc-supplemented extracts. The dashed line for Myc represents the corrected value. n ≥ 100; the p value reflects the adjusted value.
(D) B cells isolated from wild-type or λ-Myc mice were isolated and processed as in Figure 2D. Asymmetric forks were graphed as in Figure 3C. Error bars represent SD of the mean.
We also analyzed the number of “acutely” asymmetric forks (unidirectional fork progression) and observed that Myc-supplemented extracts had a significantly higher percentage of completely asymmetric forks (Figure 3C). These DNA molecules, in which replication did not progress bidirectionally following addition of biotin-dUTP, could result from DNA breakage or fork stalling or correspond to DNA molecules with an origin positioned close to the end of the DNA fiber. Since the YOYO-1 DNA dye is no longer detectable after the anti-biotin and anti-digoxygenin staining, we could not selectively count only those fibers that had unlabeled DNA at the end. However, since Myc expression results in increased origin density, the probability of an origin firing close to the end of a DNA fiber is higher in Myc-overexpressing extracts than in controls. Therefore, the contribution to fork asymmetry of origins positioned close to the end of a DNA strand is higher in Myc extracts than controls. Since the interorigin distance in Myc-supplemented extracts is one-third shorter than in controls (29.8 versus 19.2 kb), we applied a correction factor of 0.66 to the Myc value, which is shown as the dotted line in Figure 3C. Comparison of fork asymmetry in B cells isolated from λ-Myc mice or control mice revealed that the percentage of asymmetric forks was higher in λ-Myc B cells (Figure 3D) than in controls (20.74% versus 7.39%). The presence of significantly more asymmetric forks upon Myc expression suggests that Myc increases the stalling or collapse of replication forks.
Cdc45 Recapitulates Myc-Dependent DNA Replication Stress
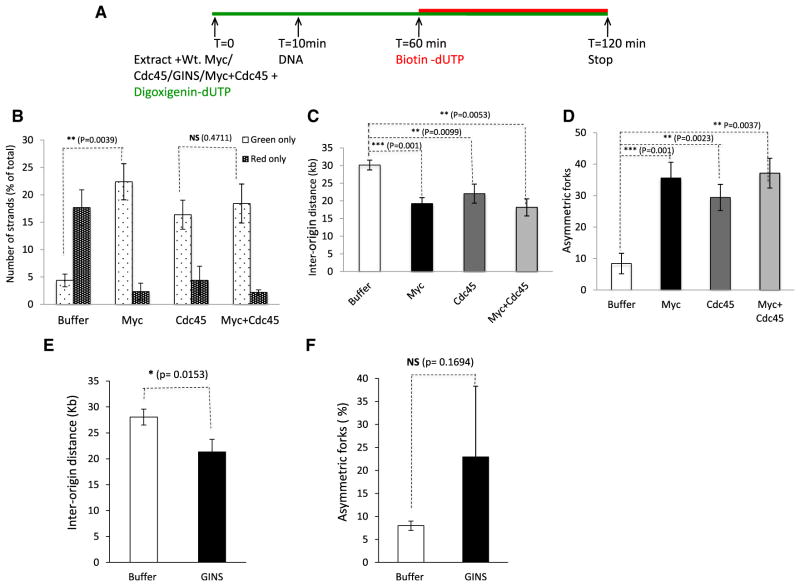
Since Myc overexpression increases the association of Cdc45 on chromatin (Dominguez-Sola et al., 2007), we decided to evaluate the role of Cdc45 in Myc-dependent replication stress. We initially assessed the consequences of Cdc45 overexpression on DNA replication. To do this, we used Cdc45-depleted extracts (Figure S2A) and first tested the activity of Cdc45 recombinant protein (Figure S2A); we found that recombinant Cdc45 was functional and rescued P32 incorporation in Cdc45-depleted extracts (Figure S2B). We then supplemented nondepleted extracts with Cdc45 (Figure 4A) and observed that overexpression of Cdc45 resulted in precocious origin firing (Figure 4B), increased origin density as reflected by decreased interorigin distance (Figure 4C), and increased asymmetry of replication forks (Figure 4D). Increased expression of Cdc45 thus recapitulated the phenotypes observed upon Myc overexpression. Next, we asked whether coexpression of Myc and Cdc45 has an additive or synergistic effect. We observed that cooverexpression of Myc and Cdc45 did not result in a higher fraction of early-replicating DNA fibers when compared with the effect of Myc or Cdc45 alone (Figure 4B). Similarly, coexpression of Myc and Cdc45 did not result in a further decrease of interorigin distance (Figure 4C) or any additional increase in fork asymmetry (Figure 4D). Since coexpression of both proteins did not augment the overexpression phenotype, these data strongly suggest that Myc and Cdc45 might function in the same pathway.
Figure 4. Cdc45 Recapitulates the Myc-Induced Replication Phenotype.
(A) Schematic representation of the experiment.
(B) Early- versus late-replicating DNA (green only versus red only) were plotted for control, Myc, Cdc45, and coexpression of Myc and Cdc45.
(C) The interorigin distances for buffer, Myc, Cdc45, and coexpression of Myc and Cdc45 were measured and plotted as in Figures 1 and 2.
(D) Graph showing the fraction of DNA that had unidirectional fork progression (FPR < 0.25) in control, Myc-, Cdc45-, and Cdc45- and Myc-supplemented extracts; n ≥ 100.
(E) Interorigin distance was measured after addition of recombinant GINS protein as described in Figure 2.
(F) Asymmetric forks were measured after addition of recombinant GINS as described in Figure 3.
Error bars represent SD of the mean. See also Figure S2.
Cdc45 protein associates with MCM2-7 and GINS to form the CMG complex, which controls origin firing and DNA unwinding. Thus, we tested whether GINS overexpression triggers DNA replication stress. We supplemented extracts with purified, recombinant GINS complex (Boskovic et al., 2007) and monitored the impact on DNA replication. We found that GINS overexpression increased origin density as reflected by decreased interorigin distance (Figure 4E). We also observed more asymmetry of replication forks in presence of added GINS, although the difference was not statistically significant (p = 0.1694) possibly due to sequence divergence between Xenopus and human GINS used in these experiments (Figure 4F).
Myc-Induced DNA Replication Stress Requires Cdc45 and GINS
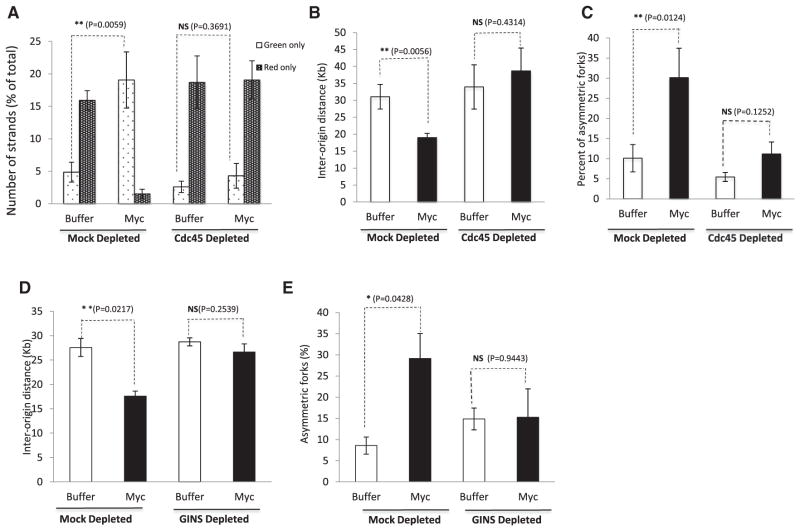
To further test the idea that Myc and the CMG complex work in the same pathway, we next investigated whether Myc-induced replication stress depended on Cdc45. We hypothesized that Myc increases origin firing through increased recruitment of Cdc45 to chromatin and that manifestation of the Myc phenotype requires Cdc45. To test this, we experimentally created conditions where Cdc45 becomes rate limiting. We first established partial Cdc45-depletion conditions (one round of depletion for 15 min) in which Cdc45 levels are reduced to approximately 20% of control levels (Figure S2A) but are sufficient to support chromosomal DNA replication to levels comparable to that observed in control extracts (Figure S2C). Reducing Cdc45 levels to about 20% of controls did not hinder normal replication (Figure S2C), but reduced the overall pool of available Cdc45 in the extracts (Figure S2A). We then overexpressed Myc in both mock-depleted extracts and in extracts partially depleted of Cdc45. As anticipated, overexpressing Myc in mock-depleted extracts resulted in the characteristic Myc phenotypes of early origin firing (Figure 5A), decreased interorigin distance (Figure 5B), and increased fork stalling (Figure 5C). In contrast, all these phenotypes were abrogated when Cdc45 was rate limiting (Figures 5A–5C and S3).
Figure 5. Cdc45 and GINS Are Required for Myc-Dependent Phenotypes.
(A) Early- versus late-replicating DNA fibers (green only versus red only) for buffer or Myc-supplemented extracts in mock- or Cdc45-depleted extracts.
(B) Interorigin distances for buffer (control) or Myc extracts in mock- or Cdc45-depleted conditions.
(C) Asymmetric fork progression for buffer or Myc extracts in mock- or Cdc45-depleted conditions were plotted as described in Figure 2; n ≥ 100.
(D and E) The GINS complex was partially depleted using Psf2 antibody. The effect of overexpressing Myc in mock- or GINS-depleted extracts was evaluated. The experimental setup and measurement of interorigin distances (D) and asymmetric forks (E) are described earlier (Figures 2 and 3).
Error bars represent SD of the mean. See also Figure S3.
Next, we evaluated the potential requirement of GINS for Myc-dependent DNA replication stress. Since GINS are essential for DNA replication (MacNeill, 2010), we established conditions for partial depletion of GINS using specific antibodies against Psf2 (Boskovic et al., 2007). This strategy, although reducing the amount of available GINS (Figure S2D), successfully preserved genomic DNA replication. Under these conditions, we found that overexpression of Myc in GINS-depleted extracts did not significantly alter interorigin distance (Figure 5D) and did not significantly affect the degree of fork asymmetry (Figure 5E). We conclude that Myc-dependent DNA replication stress requires the downstream activity of the CMG complex.
Mechanisms of Myc-Dependent DNA Damage
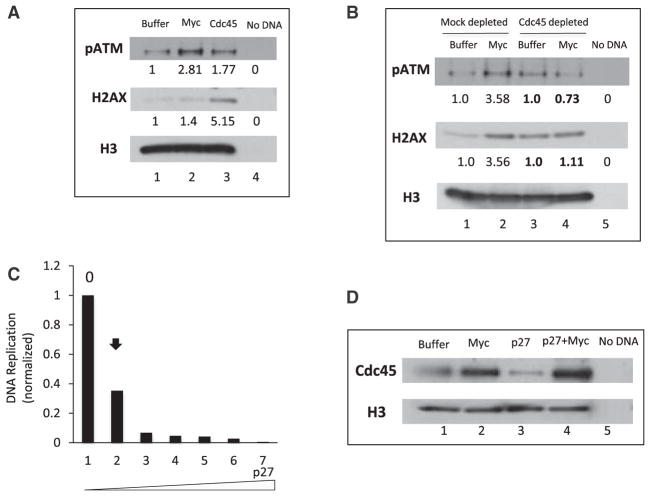
Our lab and others have previously described how Myc-dependent DNA replication stress results in DNA damage, a known consequence of Myc deregulation in different cell types (Dominguez-Sola et al., 2007; Karlsson et al., 2003). Given that Cdc45 is required for Myc-dependent replication stress, we next compared the ability of Myc and Cdc45 to trigger DNA damage. First, we monitored phosphorylation of the H2AX variant, which is a marker of double-stranded breaks (DSBs) and aberrant DNA structures following phosphorylation by ATM, DNA-PKcs, or ATR. As shown in Figure 6A, we confirmed that Myc expression resulted in elevated γ-H2AX. Notably, addition of recombinant Cdc45 protein to extracts also resulted in increased levels of γ-H2AX on chromatin (Figure 6A). The increase in γ-H2AX correlated with increased levels of the auto-phosphorylated form of ATM (p-ATM) in extracts overexpressing Myc or Cdc45 (Figure 6A), thus indicating the concomitant activation of ATM (Bakkenist and Kastan, 2003) upon Myc or Cdc45 overexpression. We conclude that Cdc45 recapitulates all aspects of Myc-dependent replication phenotypes: altered DNA replication patterns and DNA damage. To assess whether Myc was required for Cdc45-dependent DNA damage, we first established Myc-depletion conditions that allowed DNA replication to proceed with similar kinetics as control, undepleted extracts (data not shown). We then monitored DNA replication (interorigin distance) in these Myc-depleted extracts that were supplemented with either buffer or recombinant Cdc45. We found that addition of Cdc45 protein to Myc-depleted extracts shortened the interorigin distance when compared with buffer-supplemented extracts (Figure S4). This strongly suggests that Cdc45 exerts its function downstream of Myc. Next, we assessed whether Myc-dependent DNA damage, i.e., γ-H2AX and p-ATM, required Cdc45. We monitored chromatin-bound γ-H2AX and p-ATM following addition of Myc to control extracts or extracts partially depleted of Cdc45. Myc-dependent DNA damage was abrogated in extracts that were deficient in Cdc45 (Figure 6B). Therefore, both Myc-dependent replication stress and subsequent DNA damage require the activity of Cdc45 downstream of Myc.
Figure 6. Mechanisms of Myc-Dependent DNA Damage.
(A) Chromatin was incubated with extracts that were supplemented with buffer, Myc, or Cdc45 protein for 45 min. Chromatin-bound proteins were isolated and immunoblotted with phospho-ATM (pATM) and γH2AX antibodies (H2AX). Histone H3 was used as loading control. The numerical values indicate densitometric analysis of band intensity using ImageJ.
(B) Mock- or Cdc45-depleted extracts were supplemented with sperm chromatin and incubated for 1 hr. Chromatin-bound pATM and γH2AX were assessed as in (A). Band intensities were quantified using ImageJ; control/buffer was assigned a value of 1. Depleted extracts were compared with their corresponding mock-depleted controls.
(C) Sperm chromatin was replicated in extracts supplemented with P32 dCTP and increasing amounts of recombinant p27 protein. The replication assay was performed as described in Experimental Procedures. Radioactivity incorporated into DNA was quantified and plotted. The arrow indicates the concentration of p27 that inhibited DNA replication by 66%.
(D) Chromatin was isolated from extracts supplemented with buffer (lane 1), Myc alone (lane 2), p27 alone (lane 3), or Myc and p27 (lane 4). The concentration of p27 protein that inhibits DNA replication by 66% (as determined from Figure 6C) was used. Chromatin-bound proteins were blotted with Cdc45 antibodies. Histone H3 was used as a loading control.
Next, we attempted to gain insights into the mechanism by which Myc recruits Cdc45 to chromatin. Myc is known to antagonize p27, the CDK2/cyclin E inhibitor (Vlach et al., 1996), and CDK2/cyclin E complexes, in turn, modulate p27 (Xic1) activity at Xenopus replication origins (Furstenthal et al., 2001). Moreover, origin activation and firing require the local activity of CDK2/cyclin E complexes, which results in the recruitment of Cdc45 and GINS to primed origins (Mimura and Takisawa, 1998). Therefore, we hypothesized that Myc overexpression could lower the threshold of CDK activity required for origin activation and Cdc45 recruitment. To test our hypothesis experimentally, we added recombinant p27 protein at a concentration that significantly inhibited both DNA replication (Figure 6C), and Cdc45 loading onto chromatin (Figure 6D, lane 3). Under these inhibitory conditions, we overexpressed Myc and found that Cdc45 loading was restored (Figure 6D, compare lanes 3 and 4). This strongly suggests that Myc facilitates Cdc45 recruitment to chromatin and to replication origins by antagonizing the inhibitory activity of p27.
DISCUSSION
Aberrant DNA replication initiation is a critical source of genomic instability leading to chromosomal alterations (Davidson et al., 2006; Kawabata et al., 2011; Lebofsky and Walter, 2007; Shima et al., 2007). Our previous study demonstrating a nontranscriptional role for Myc in regulating the initiation of DNA replication prompted us to explore the mechanisms by which this commonly deregulated oncogene influences DNA replication. Notably, while several studies have documented the transcriptional functions of Myc in regulating S phase entry (Alexandrow and Moses, 1998; Classon et al., 1987; Heikkila et al., 1987; Pelengaris et al., 2002), our approach using Xenopus cell-free extracts allows us to specifically address the nontranscriptional functions of Myc in regulating replication and triggering oncogenic DNA damage (Dominguez-Sola et al., 2007; Lebofsky and Walter, 2007).
Myc Alters Overall DNA Replication Kinetics
Using molecular DNA combing, we observe that Myc overexpression triggers premature and more synchronous firing of replication origins than in control, untreated conditions. This is supported by the significant increase in DNA fibers that are fully replicated early. Consistent with the analysis of replication kinetics by radionucleotide incorporation, Myc accelerates S phase entry. Since Myc does not alter fork progression rate (Dominguez-Sola et al., 2007), our results indicate that Myc alters the intrinsic temporal program of DNA replication (Czajkowsky et al., 2008; Gilbert et al., 2010). Notably, Myc also influences the spatial regulation of DNA replication initiation, as seen by the decrease in interorigin distance and the increased incidence of fibers harboring clusters of origins in Xenopus and mammalian cells upon Myc overexpression. Of note, interorigin distance was shorter in B cells than the average values previously reported for mammalian cells. We believe this could be due in part to the average size of the DNA fibers analyzed (80 kb), which would skew the analysis toward short interorigin distance. In addition, we note that B cells stimulated to proliferate in vitro have a rapid doubling time.
Origin Firing and Myc
The spatiotemporal program of origin firing is thought to be regulated by several mechanisms including origin interference, activation of dormant origins, and localized activity of protein kinases. Interference is a mechanism by which newly fired origins inhibit firing of adjacent potential origins. Interference decreases with increased distances from the fired origins (Lebofsky et al., 2006). It is thought that origin interference helps in achieving complete replication of the genome by regulating origin spacing. The activation of dormant origins within unreplicated DNA then allows completion of S phase and is critical to achieve full replication during normal S phase (Lucas et al., 2000) or under stress conditions (Anglana et al., 2003; Courbet et al., 2008; Ge et al., 2007; Woodward et al., 2006). Myc could alter origin interference early by allowing clusters of origins to fire, thus disrupting the normal spacing and timing program (Berezney et al., 2000). Alternatively, since Myc triggers DNA damage (see below), it is conceivable that Myc-dependent increase in origin density could be due to firing of dormant origins as a consequence of replication stress. However, our findings do not favor this theory. In replication assays using radionucleotide incorporation, Myc-induced replication burst occurs very early. In fact, as replication proceeds, overall replication actually slows down due to checkpoint activation (Dominguez-Sola et al., 2007). Also, in DNA-combing experiments, Myc expression is accompanied by an increase of early firing origins (“green-only” fibers). The data suggest that the “green-only” tracks result from excessive early firing and not from the increased firing of dormant origins activated later in response to fork stalling. By triggering premature firing of normally silent origins, Myc expression could accelerate the precocious depletion of origins such that no dormant origins would be left to fire in case of replication problems. It is also possible that Myc can render origins more sensitive to origin-triggering factors whose concentration may otherwise limit firing early in S phase. Based on our results, we propose that one mechanism by which Myc stimulates origin firing is by lowering the threshold of CDK2/cyclin E activity required to antagonize the CDK inhibitor p27. Some studies suggest that Myc could affect CDK2 activity through multiple mechanisms. It has been proposed that Myc directly activates cyclin D2 transcription resulting in the sequestration of p27 and the subsequent increase in CDK2/cyclin E activity (Bouchard et al., 1999). Other reports indicate that Myc can regulate cyclin E/CDK2 activity in ways that are independent of cyclin D2 (Campaner et al., 2010; Deb-Basu et al., 2006; Hydbring et al., 2010; Vlach et al., 1996). We suggest that the direct (nontranscriptional) modulation of CDK2 activity, which we observe in transcriptionally inactive extracts, could be most relevant to the role of Myc in origin firing when cyclin/cdk complexes are readily available, which is the case in Xenopus extracts or cycling cells. In contrast, regulation of CDK2 activity by Myc via transcriptional activation of cyclin D2 could be more relevant to the role of Myc in facilitating the G0 to S transition in quiescent cells.
Myc, Fork Symmetry, and Subsequent DNA Damage
We have shown that a significant number of forks initiated in presence of excess Myc do not progress bidirectionally. Since Xenopus extracts are devoid of transcription, fork arrest cannot be the result of collisions between the replication and transcription machinery. Additionally, we find that Myc expression does not impact the fraction of forks with moderate asymmetry; the presence of excess Myc results in replication forks that are either symmetric or profoundly asymmetric, with only about 25% of fibers exhibiting intermediate phenotypes. The absence of a Gaussian distribution of fork progression ratios strongly suggests that fork stalling is an early event.
When stalled forks fail to restart, the replisome might disassemble and cause DNA breaks (Labib and Hodgson, 2007). Furthermore, the inaccurate restart of stalled replication forks can cause genome instability (Petermann and Helleday, 2010). Since the modes of dealing with stalled replication forks is restart without recombination, or the formation of double-strand breaks (Labib and Hodgson, 2007), and Myc deregulation leads to DNA damage including DSBs (Karlsson et al., 2003), we propose that Myc-dependent stalled forks are more prone to generate DSBs. Thus, it is possible that by increasing origin firing, Myc overwhelms processes that ensure proper fork progression, fork restart at stalled forks, or fork termination. For example, enzymes such as DNA topoisomerases, nucleases required for replication fork progression (MRN complex), or DNA helicases involved in resolution of aberrant replication intermediates could become limiting when too many origins are activated at once. Indeed, there is evidence that Myc overexpression challenges the normal physiology of DNA replication forks because loss of the WRN RecQ helicase, a protein that is critical for replication fork stability and restart (Ammazzalorso et al., 2010; Sarkies et al., 2012; Sidorova et al., 2008), sensitizes cells to Myc expression and triggers cellular senescence (Grandori et al., 2003; Moser et al., 2012; Robinson et al., 2009).
CMG and Myc-Dependent DNA Replication Stress
Cdc45 and GINS are essential for DNA replication: they are part of the complex that unwinds DNA at each replication fork and tethers DNA polymerase to the initiation complex (Gambus et al., 2006; Hashimoto et al., 2012; Jares and Blow, 2000; Mimura and Takisawa, 1998; Pacek et al., 2006). In contrast to MCMs, which are loaded in excess (Hyrien et al., 2003), Cdc45 is rate limiting in yeast (Edwards et al., 2002; Tanaka et al., 2011) and is thought to be a better marker for active replication forks (Hashimoto et al., 2012). However, our present study and others (Edwards et al., 2002) indicate that Cdc45 may not be rate limiting in Xenopus. Furthermore, excess Cdc45 is known to activate dormant origins (Wong et al., 2011).
Myc overexpression results in increased binding of Cdc45 on chromatin (Dominguez-Sola et al., 2007), and the increased dosage of Cdc45 allows late origins to fire earlier in the S phase (Wong et al., 2011). In this study, we have demonstrated that Cdc45 is a critical regulator of Myc-driven DNA replication stress. Cdc45 overexpression, and to a lesser extent overexpression of GINS, recapitulates all the phenotypes of Myc overexpression: increase in early origin firing, decreased interorigin distance, increase in asymmetrical forks, and subsequent DNA damage. Our data are in agreement with studies demonstrating that increasing Cdc45 levels leads to increased origin usage (Wong et al., 2011; Wu and Nurse, 2009). Importantly, when Cdc45 or GINS are depleted such that normal replication is allowed but these factors become rate limiting, Myc overexpression no longer alters DNA replication timing and fork stability, nor does it trigger DNA damage. The abrogation of Myc-dependent replication stress phenotypes in a Cdc45- or GINS-deficient setting indicates that the CMG complex acts downstream of Myc and is required for the DNA replication stress generated by Myc. Furthermore, and consistent with Myc and Cdc45 operating in a single pathway, we show that coexpression of Myc and Cdc45 does not exacerbate the phenotypes of Myc or Cdc45 expression alone. Therefore, our work suggests that Myc overexpression enhances Cdc45 recruitment to replication origins in a manner that does not require transcription; excess Cdc45 on chromatin then hyperactivates origin firing, which, in turn, results in fork stalling that could lead to genomic instability. Our studies are consistent with the idea that CDK2/cyclin E is a limiting factor for origin activation, and that the threshold of CDK2/cyclin E activity required for origin firing is, at least in part, controlled by Myc. Our studies further raise the possibility that Cdc45 and possibly the CMG complex could collaborate with Myc overexpression in tumor development or, when overexpressed or amplified, could act as oncogenes.
EXPERIMENTAL PROCEDURES
All animal experiments were approved by the Institutional Animal Care and Use Committee at Columbia University Health Sciences Center under the supervision of the Institute of Comparative Medicine.
Preparation of Xenopus Interphase Extracts
Xenopus extracts were prepared as described earlier (Srinivasan and Gautier, 2011). Briefly, eggs were crushed by centrifugation to obtain the cytosol/membrane fractions and supplemented with cycloheximide (20 μg/ml), creatine phosphate, creatine phosphokinase, and protease inhibitors cocktail before use.
Preparation of Sperm Chromatin
Sperm chromatin was prepared as described previously (Srinivasan and Gautier, 2011). For radioactive replication assays, 10,000 nuclei were used for a 10 μl reaction. For replication assays in DNA-combing experiments, 40,000 nuclei were used per 20 μl of extract.
Isolation, Induction, and Labeling of B Cells
B cells were isolated from spleens of nonimmunized, 3-month-old, wild-type or λ-Myc mice, using the B cell Isolation Kit (Miltenyi Biotec). B-cell-enriched fractions were cultured (0.5 × 106 cells/ml) in RPMI (Life Technologies) with 10% fetal bovine serum (Hyclone), 53 nM B-mercaptoethanol, 1% sodium pyruvate, 1% glutamine, 10 mM HEPES, and antibiotics. Cells were induced by 20 μg/ml lipopolysaccharide (Sigma-Aldrich) and 5 ng/ml interleukin-4 (RBD Systems), grown for 2 days, and then labeled with 5-chloro-2-deoxyuridine (green) (2 hr) followed by 5-iodo-2-deoxyuridine (Sigma-Aldrich) (2 hr). Labeled cells were processed for DNA combing and immunofluorescence staining.
Radioactive Replication Assay I
The reaction was set up with 10 μl of extract, 0.1 μl of P32 dCTP, and 10,000 sperm nuclei. Where indicated, recombinant protein (Myc, Cdc45, p27, or GINS) was added. The samples were processed, electrophoresed, and analyzed as described previously (Srinivasan and Gautier, 2011).
Replication Assay II and DNA Combing
Twenty microliters of undepleted, mock-depleted, or partially depleted extracts was supplemented with 2,000 sperm nuclei/μl and 0.4 μl digoxigenin dUTP (1 mM stock Roche) and incubated at 21°C for 1 hr. After the first hour, biotin dUTP (0.4 μl of 1 mM stock) was added, and the reaction was incubated at 21°C for 1 hr; reaction was stopped with ice-cold 1× PBS. After terminating the reaction, DNA was processed and combed onto silanized coverslips. The incorporated digoxigenin (green) and biotin (red) were detected by immunofluorescence staining. Technical details have been carefully described earlier (Marheineke et al., 2009). For DNA combing, we used a custom-built DNA-combing device that immersed silanized coverslips in the MES/DNA solution for 5 min and lifted them at the rate of 300 μM per second. For the analysis of replication, we used DNA molecules that were 40 kb or longer and had incorporated either digoxigenin (green) or both digoxigenin and biotin (reddish-orange). Only DNA fibers that exhibited continuous staining or had gaps <2 kb were analyzed. Experiments were repeated at least three times.
Immunofluorescence Staining
Detection of digoxygenin-dUTP and biotin-dUTP (extracts) and of the IdU and CldU incorporation (B cells) is described earlier (Marheineke et al., 2009).
Protein Preparation and Purification
Preparation of Myc:Flag-tagged recombinant Xenopus Myc protein (both wild-type and mutant) was prepared as previously described (Dominguez-Sola et al., 2007). Recombinant Myc protein was used at a final concentration of 75 nM in overexpression experiments.
Preparation of Cdc45 Protein
Recombinant Cdc45 was prepared by amplifying the Cdc45-encoding baculovirus in Sf9 cells and infecting High Five cells to produce recombinant protein. Cells were harvested, and protein was purified using standard nickel column purification techniques (Life Technologies).
Preparation of Recombinant p27 Protein
The pGEX-p27 plasmid (a kind gift from Dr. R. Yew) was used to transform BL21 cells; p27 is expressed as a GST-fusion protein. The BL21 cells were cultured at 27°C and induced with isopropyl β-D-1-thiogalactopyranoside for 6 hr before harvesting. Standard GST protein purification methods were used to isolate p27 protein.
Depletion of Xenopus Extracts
Cdc45 serum (a generous gift from Dr. J. Walter) was used to deplete the extract of Cdc45. Protein A beads were incubated with Cdc45 serum overnight. Xenopus extract was added to the washed beads, incubated for 15 min at 4°C, and recovered by low-speed centrifugation through a cellulose column. One round of depletion was sufficient to render the extract Cdc45 deficient; two rounds of depletion for 15 min each was performed to obtain extracts devoid of Cdc45. Depletion of Myc and GINS was performed using Myc and GINS (Psf2, a kind gift from Dr. Mendez) antibodies.
Western Blotting and Detection of Chromatin-Bound Proteins
Depletion of Cdc45/GINS or Myc in the extracts was confirmed by western blotting. To detect chromatin-bound proteins, reactions were assembled with 50 μl of extract and 10,000 sperm nuclei/μl and recombinant protein wherever indicated. Chromatin was isolated through a sucrose cushion; bound proteins were resolved by SDS-PAGE and detected with indicated antibodies (Srinivasan and Gautier 2011).
Microscopy
The Zeiss Palm Microbeam Fluorescent Microscope with a 100× objective and 10× eye piece was used to capture images. Images were taken in a series using an automated stage with 10% overlap and then stitched to generate a comprehensive image.
Data analysis
Data analysis was carried out using ImageJ 1.46d software (http://imagej.nih.gov/ij/index). The pixel to micrometer ratio used was 0.09 for multiple-tiled images and 0.06 for individual images and was in accordance with the calibration of the microscope. A conversion factor of 1 μm = 2 kb was applied to obtain DNA length. All error bars represent SD of the mean. The two-tailed t test was used to determine significance.
Supplementary Material
Acknowledgments
We thank Dr. J. Walter from Harvard Medical School for generously providing the Cdc45 antibody; Dr. J. Mendez from Spanish National Cancer Research Centre for the generous gift of the GINS proteins and antibodies, Dr. R. Yew for p27Xic1 construct, Drs. C. Ying, A. Sfeir and T. Aparicio for helpful discussions; Drs. T. Swayne and A. White for help with Microscopy/data analysis; members of the Hyrien laboratory for training and advice on the DNA-combing technique. We gratefully acknowledge funding support from the National Institutes of Health (CA92245 and CA167826) to J.G., (5K99 CA151827) to D.D.-S. and the US Department of Defense (W81XWH-09-1-0504) to S.V.S.
Footnotes
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.04.002.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Alexandrow MG, Moses HL. c-myc-enhanced S phase entry in keratinocytes is associated with positive and negative effects on cyclin-dependent kinases. J Cell Biochem. 1998;70:528–542. [PubMed] [Google Scholar]
- Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010;29:3156–3169. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic J, Coloma J, Aparicio T, Zhou M, Robinson CV, Méndez J, Montoya G. Molecular architecture of the human GINS complex. EMBO Rep. 2007;8:678–684. doi: 10.1038/sj.embor.7401002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjo S, Murga M, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–59. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- Classon M, Henriksson M, Sümegi J, Klein G, Hammarskjöld ML. Elevated c-myc expression facilitates the replication of SV40 DNA in human lymphoma cells. Nature. 1987;330:272–274. doi: 10.1038/330272a0. [DOI] [PubMed] [Google Scholar]
- Cole MD, Cowling VH. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat Rev Mol Cell Biol. 2008;9:810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- Czajkowsky DM, Liu J, Hamlin JL, Shao Z. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J Mol Biol. 2008;375:12–19. doi: 10.1016/j.jmb.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Davidson IF, Li A, Blow JJ. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell. 2006;24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb-Basu D, Karlsson A, Li Q, Dang CV, Felsher DW. MYC can enforce cell cycle transit from G1 to S and G2 to S, but not mitotic cellular division, independent of p27-mediated inihibition of cyclin E/CDK2. Cell Cycle. 2006;5:1348–1355. doi: 10.4161/cc.5.12.2860. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Schärer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furstenthal L, Swanson C, Kaiser BK, Eldridge AG, Jackson PK. Triggering ubiquitination of a CDK inhibitor at origins of DNA replication. Nat Cell Biol. 2001;3:715–722. doi: 10.1038/35087026. [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet. 2010;11:673–684. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol. 2010;75:143–153. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, et al. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 2003;17:1569–1574. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Puddu F, Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol. 2012;19:17–24. doi: 10.1038/nsmb.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkila R, Schwab G, Wickstrom E, Loke SL, Pluznik DH, Watt R, Neckers LM. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature. 1987;328:445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC? Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- Hydbring P, Bahram F, Su Y, Tronnersjö S, Högstrand K, von der Lehr N, Sharifi HR, Lilischkis R, Hein N, Wu S, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci USA. 2010;107:58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–125. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Blow JJ. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci USA. 2003;100:9974–9979. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, Shima N. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk AL, Qi CF, Torrey TA, Taddesse-Heath L, Feigenbaum L, Park SS, Gerbitz A, Klobeck G, Hoertnagel K, Polack A, et al. Burkitt lymphoma in the mouse. J Exp Med. 2000;192:1183–1190. doi: 10.1084/jem.192.8.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Hodgson B. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol Biol Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Walter JC. New Myc-anisms for DNA replication and tumorigenesis? Cancer Cell. 2007;12:102–103. doi: 10.1016/j.ccr.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Lucas I, Hyrien O. Hemicatenanes form upon inhibition of DNA replication. Nucleic Acids Res. 2000;28:2187–2193. doi: 10.1093/nar/28.10.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I, Chevrier-Miller M, Sogo JM, Hyrien O. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J Mol Biol. 2000;296:769–786. doi: 10.1006/jmbi.2000.3500. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- Marheineke K, Goldar A, Krude T, Hyrien O. Use of DNA combing to study DNA replication in Xenopus and human cell-free systems. Methods Mol Biol. 2009;521:575–603. doi: 10.1007/978-1-60327-815-7_33. [DOI] [PubMed] [Google Scholar]
- Méchali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser R, Toyoshima M, Robinson K, Gurley KE, Howie HL, Davison J, Morgan M, Kemp CJ, Grandori C. MYC-driven tumorigenesis is inhibited by WRN syndrome gene deficiency. Mol Cancer Res. 2012;10:535–545. doi: 10.1158/1541-7786.MCR-11-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Robinson K, Asawachaicharn N, Galloway DA, Grandori C. c-Myc accelerates S-phase and requires WRN to avoid replication stress. PLoS ONE. 2009;4:e5951. doi: 10.1371/journal.pone.0005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar N, Kadeppagari RK, Thimmapaya B. c-Myc-induced aberrant DNA synthesis and activation of DNA damage response in p300 knockdown cells. J Biol Chem. 2009;284:15193–15205. doi: 10.1074/jbc.M900776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan SV, Gautier J. Study of cell cycle checkpoints using Xenopus cell-free extracts. Methods Mol Biol. 2011;782:119–158. doi: 10.1007/978-1-61779-273-1_10. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011;21:2055–2063. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- Wong PG, Winter SL, Zaika E, Cao TV, Oguz U, Koomen JM, Hamlin JL, Alexandrow MG. Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PLoS ONE. 2011;6:e17533. doi: 10.1371/journal.pone.0017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AM, Göhler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission Yeast. Cell. 2009;136:852–864. doi: 10.1016/j.cell.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.