Advances in DNA sequencing technology and bioinformatics methodology have led to a revolution in our understanding of the microorganisms that inhabit the human body. Distinctive populations of organisms belonging to each of the 3 domains of life, Archaea, Bacteria, and Eukarya, inhabit each body site.1 The communities of microorganisms are referred to as the microbiota. When considering the organisms and all of their related genomes, the term microbiome is used. The human gut microbiome is particularly unique in that it is home to an enormous number of bacteria, approximately 100 trillion bacteria cells, outnumbering the human cells by an estimated 10 fold and the human genome by 150 fold. We have co evolved to exist with our gut microbiota largely in a mutualistic relationship where we as hosts rely on these organisms for a number of key functions related to nutrition, education of the immune system, and prevention of infection by pathologic species. In turn, we provide our gut microbiota with a unique niche in which to live where we provide a source of nutrition to the microbiota in the form of mucus. Alteration of the microbiota composition associated with disease, known as dysbiosis, has been described in the gut for numerous disease processes including inflammatory bowel diseases (IBDs), metabolic disorders, cancer, and infection, particularly with Clostridium difficile infection (CDI), to name a few.
Advances in Methods to Study the Gut Microbiome
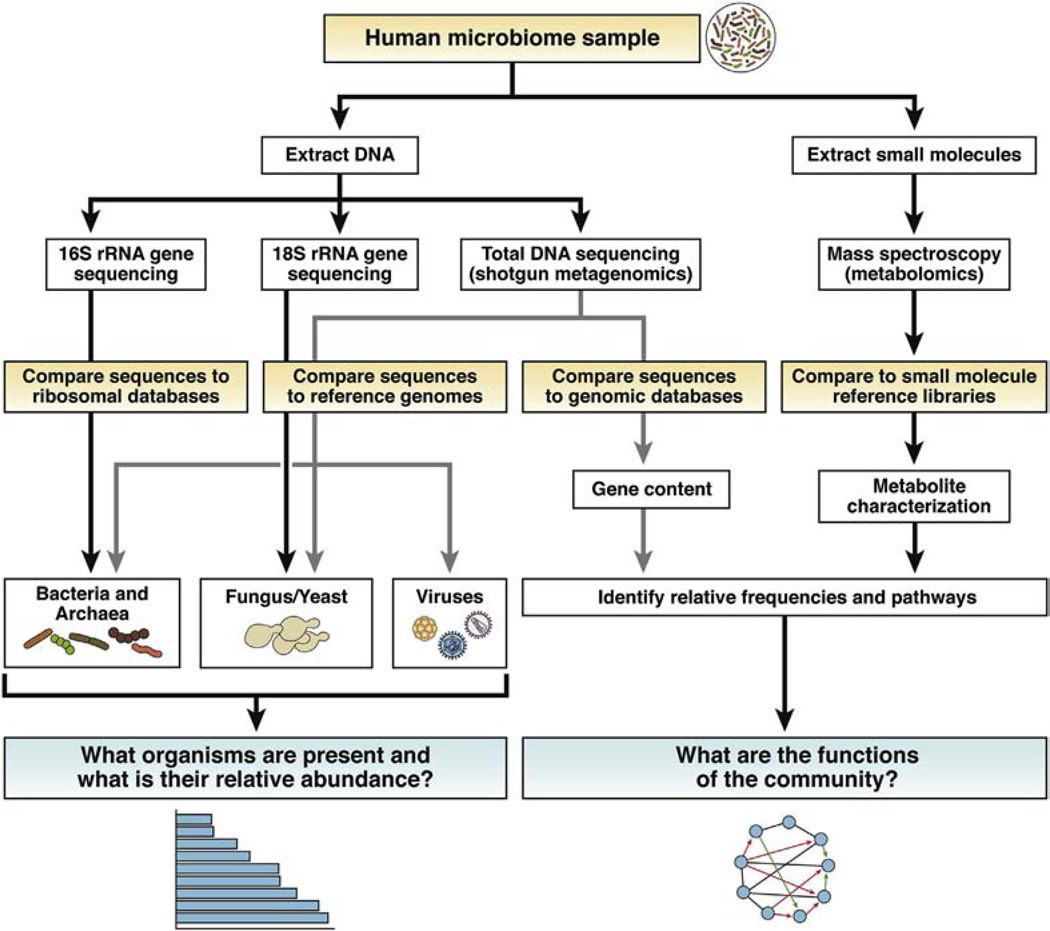
The expression of ribosomal genes are required for all life forms. The 16S genes are found in all bacteria and archaea, the latter of which are prokaryotic microorganisms that, in the mammalian gut, are primarily methanogens responsible for the production of methane. However, similar to human beings, microeukaryotes such as fungi and yeast have the 18S ribosomal gene. Over the past decade, dramatic technical advances in DNA sequencing technologies, sometimes described as high throughput, massively parallel, or deep sequencing, have allowed scientists to determine the DNA sequence of specific regions in either the 16S or 18S gene that, when matched to a ribosomal DNA sequence database, permits the identification of microorganisms that reside within a sample from which DNA was isolated1 (Figure 1). Furthermore, the number of sequence reads for a specific organism is roughly proportional to its abundance within a sample. The use of DNA sequencing technology is a major advance in the characterization of complex microbial communities because it is a culture independent method that avoids the difficulty of growing the majority of obligate anaerobic microorganisms in the gut microbiota. By using even newer technologies capable of sequencing billions of DNA base pairs in a single run at an affordable cost, shotgun metagenomic sequencing can be performed in which community DNA is sequenced in totality, permitting not only an evaluation of microbial community structure but also allowing an evaluation of the genomic representation of the community (Figure 1).1 The latter can be used to help understand the functions encoded by the genomes of the gut microbiota.1 Shotgun metagenomic sequencing also can be used to characterize the abundance of viruses, or the virome, biological entities that lack ribosomal genes yet are among the most abundant organisms in the biosphere. Although currently not feasible as a routine clinical test, profiling of the gut microbiota in fecal samples may ultimately have utility in the clinical setting with the caveat that the microbiota in fecal samples is somewhat distinct from the microbial communities adherent to the intestinal mucosa. As the technology of DNA sequencing continues to advance, costs continue to decrease significantly. Together with the automation of bioinformatic tools made available online, the accessibility of these technologies for more widespread clinical and research use is becoming apparent.
Figure 1.
Analytic approach to study both the structure and function of the human gut microbiome. rRNA, ribosomal RNA.
The Role of the Gut Microbiota in the Maturation of the Mucosal Immune System and Its Link With Immune-Mediated Diseases
Interactions between the microbiota and the intestinal mucosa play a critical role in the maturation of the host immune system. For example, certain bacteria and their products play a role in the development of tolerogenic anti inflammatory T regulatory cells while others lead to the development of more proinflammatory Th17 cells as well as the programming of B lymphocytes to produce secretory IgA that is secreted at mucosal surfaces. Further emphasizing this host microbial mutualism, the intestinal epithelium interacts with the gut microbiota through the production of nutrients in the form of mucus to support bacterial metabolism, as well as antimicrobial peptides that help to shape the structure of the gut microbiota. Genetic loci associated with the development of IBD include pathways that impact each of these processes, showing that alterations in the delicately balanced homeostatic relationship between the gut microbiota and the host can lead to unrestrained inflammation, the hallmark of IBD.2
The interaction between host genetics and environmental factors in the development of complex immunologically mediated diseases such as asthma, IBD, type 1 diabetes, and others, have led to the notion that the rapidly increasing incidence of these diseases over the past few decades is caused, in part, by an alteration in the microbial environment. Indeed, many aspects of our environment have been changed dramatically over the past few decades concurrent with the increasing incidence of these disease processes. Elements of the modern lifestyle that have been postulated to result in changes in the gut microbiota include improved sanitation, vaccinations, increased antibiotic use, decline in parasite infections, caesarean section, decline in Helicobacter pylori, smaller family size, refrigeration, less crowded living conditions, sedentary life styles, food processing, and dietary changes.
Dysbiosis of the Gut Microbiota: Cause or Effect?
By using the latest DNA sequencing technologies, scientists now are able to characterize alterations in the composition of the gut microbiota associated with various disease processes, otherwise known as dysbiosis. Dysbiosis could have value as biomarkers of disease such as atherosclerosis or in predicting response to drugs. However, more meaningful would be a demonstration that the dysbiotic microbiota play a role in disease pathogenesis and that restoration of the normal healthy micro biota is an effective therapy. The only disease process in which this has been shown to be the case is CDI, in which the consumption of antibiotics dramatically, but transiently, alters the composition of the gut microbiota, providing a niche in which C difficile can expand. Dramatic alteration of the gut microbiota by direct transfer of an entire community from a healthy donor, a process known as fecal microbial transplantation (FMT), is highly effective in the treatment of refractory disease. The fecal microbiota typically have been administered via enema, colonoscopy, or a nasogastric, nasoduodenal, or nasojejunal tube. The major distinction between probiotics, defined by the World Health Organization as “live microorganisms which when administered in adequate amounts confer a health benefit on the host,”3 and FMT is that FMT entails transfer of entire communities in the same relative abundance as occurs within a healthy host. Although not proven, this ability of a complex community of microorganisms to engraft in the host also may contribute to the dramatic efficacy of FMT for refractory and recurrent CDI, with approximately 90% success rates.4 Supporting this notion, the failure of individual probiotics to form resilient gut communities would explain why they have not been beneficial in the treatment of CDI.5
Despite its effectiveness in the treatment of CDI, FMT may be associated with unforeseen long term health consequences. For example, recent studies have linked the composition of the human gut microbiota to the risk of metabolic syndrome, car diovascular disease, and alteration of drug metabolism. As such, we believe that FMT should be used only after traditional approaches have failed and with the appropriate regulatory oversight. The use of FMT for the treatment of any other disease process should be offered only to patients within the context of a clinical trial so that appropriate informed consent can be obtained and patient’s health status after transplantation can be monitored. Similar for any new medical technology, clinicians who perform FMT must remain informed of scientific advances in the field of gut microbiota research so that they can appropriately council their patients about the benefits and potential risks of FMT. Given the fast pace of the scientific advances along with the highly technical nature of the subject matter, this undoubtedly will be a significant challenge for both physicians and their patients. Ultimately, we anticipate that defined microbial communities, which can be manufactured in a laboratory environment with carefully characterized biological properties, will provide a safer and more convenient alternative to FMT.
Although it is relatively easy to establish a causal association between the gut microbiome and acute onset of infectious diseases such as CDI, establishing a causal association with chronic diseases is more difficult. An important step to inferring causality can include showing that the microbial changes precede the onset of disease. However, for rare diseases, it is inefficient to collect microbiome data in large cohorts before the onset of disease. Therefore, most of the existing research has used cross sectional designs. As such, the observed associations could be causal, or merely a consequence of the disease. Inflammatory diseases such as Crohn’s disease and ulcerative colitis are examples of chronic diseases hypothesized to be linked to the composition of the gut microbiome, but in which intestinal inflammation could contribute to the observed dysbiosis. Causality also can be inferred by showing efficacy of therapeutic strategies that alter the microbiome. However, it should be recognized that for some diseases, the microbiome could contribute to the etiology while not being the principal driver of disease perpetuation. In such settings, therapeutic manipulation of the microbiome may not be an efficacious therapy. Thus, as with many aspects of human pathophysiology, showing causality likely will require combining the results of in vitro and in vivo experiments to define potential causal pathways, followed by translational observational and experimental studies in human beings. IBD represents an ideal disease process in which such a model can be implemented.
Animal models of IBD have shown an important role for the gut bacteria and viruses in the initiation and perpetuation of intestinal inflammation. This has been translated to human beings in a number of cross sectional studies. The characteristics of the dysbiotic microbiota associated with IBD have been highly reproducible, including an enrichment of bacterial taxa belonging to the Proteobacteria and Actinobacteria phyla, a decrease in representation of Firmicutes, and a reduction in microbial richness, the latter being an indication that there are less total microbial species.6 The notion that the dysbiotic IBD associated microbiota are supported by the over representation of injurious bacterial taxa such as Proteobacterial Enterobacteriaceae and a reduction in protective Firmicutes such as Fecalibacterium prauzsnitzii based on murine models of intestinal inflammation. Furthermore, with respect to the former, Enterobacteriaceae family members have been shown to be colitogenic in wild type mice when transferred maternally in cross fostering mouse experiments. The mechanism by which IBD associated dysbiosis develops is not well understood but may involve alterations in the expression of antimicrobial peptides produced by the intestinal epithelium or an increase in the oxidative environment of the gut owing to hyperemia, bleeding, and/or the oxidative nature of the inflammatory response. Indeed, Proteobacteria tend to be more aerotolerant than species associated with other bacterial phyla. Thus, the dysbiosis associated with IBD could be a consequence of the IBD itself or could be initiated by an acute event such as infectious gastroenteritis and in turn trigger the onset of IBD in a susceptible host. Even if IBD associated dysbiosis is not an initiator of disease, the dysbiotic microbiota may be important in perpetuating the disease. Additional insights into these issues will play a critical role in formulating effective microbial based therapeutic approaches for patients with IBD. Finally, it is important to recognize that other microorganisms, including eukaryotic viruses and fungi, also have been hypothesized to play a role in the pathogenesis of IBD.7,8
Dysbiotic microbiota have been associated with a number of other diseases. The challenges in defining the role of dysbiosis in disease pathogenesis and, by extension, the development of effective therapeutic modalities for each of these, will be similar to that described here for CDI and IBD.
Impact of the Gut Microbiota on Disease: Form vs Function
DNA sequencing technology provides information useful in determining the composition of the gut microbiota and, to a degree, function if shotgun metagenomics is performed to determine bacterial gene representation (Figure 1). The production of metabolites by the microbiota also might have a significant effect on the pathogenesis of disease independent of alterations in community structure. For example, diet not only alters the composition of the human gut microbiota but also provides substrates for the production of small molecules that influence disease development. Some examples include: the consumption of phosphatidylcholine provides the gut microbiota substrate for the generation of trimethyl amine from choline that, in animal models, accelerates atherosclerosis, the production of ammonia from host urea by urease activity of the gut microbiota worsens hepatic encephalopathy, and the production of gas by the fermentation of carbohydrates in the small bowel in patients with small bowel bacterial overgrowth. For each of these disease processes, how much of the effects are caused by composition dependent vs composition independent functional alterations in the gut microbiome have yet to be determined. Therefore, the integration of data from both deep DNA sequencing technologies and metabolomics will play an important role in rationally designed gut microbiota based therapeutic strategies (Figure 1).
The Remaining Questions and Challenges in the Future of Gut Microbiome Research
Dramatic advances in DNA sequencing technology as well as the development of ever more sophisticated bio computational tools to analyze massive data sets continue to fuel research into the gut microbiome. The future challenge in this field of research will be to address a number of fundamental questions relevant to human health and disease including the following: (1) is there a microbiome associated with human health the “healthy gut microbiome”; (2) are there causal relationships between the gut microbiome and human disease; (3) is there a role for nonbacterial gut microbes (ie, microeukaryote [yeast], viruses, and archaea) in human health and disease; and (4) what is the optimal method to alter the gut microbiota in a way that is beneficial to human beings with disease?
The answers to these and other fundamental questions in the field of gut microbial ecology await additional studies in human subjects in whom clinical metadata is carefully collected together with continued investigation in animal models in the era of rapidly advancing broad based technologies in addition to DNA sequencing such as transcriptomics, proteomics, and metabolomics to assess not only the structure but also the function of the gut microbiome (Figure 1). Ultimately, the answers to these questions could lead to a fundamental shift in the way that we treat many common diseases.
Acknowledgments
Funding
This work was supported by National Institutes of Health grants UH2/3 DK 083981 (G.D.W. and J.D.L.) and RO1 DK 089472 and R01 GM 103591 (G.D.W.).
Abbreviations used in this paper
- CDI
Clostridium difficile infection
- FMT
fecal microbial transplantation
- IBD
inflammatory bowel disease
Footnotes
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Weinstock GM. Genomic approaches to studying the human micro biota. Nature. 2012;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization and Food and Agriculture Organizaion of the United Nations. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Available at: www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf.
- 4.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 5.Butler M, Bliss D, Drekonja D, et al. Effectiveness of early diagnosis, prevention, and treatment of Clostridium difficile infection. Rockville, MD: Agency for Healthcare Research and Quality (US); 2011. [PubMed] [Google Scholar]
- 6.Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968–984. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
- 7.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C type lectin receptor Dectin 1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadwell K, Patel KK, Maloney NS, et al. Virus plus susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]