Abstract
Objective
We explored two strategies for query expansion utilizing medical subject headings (MeSH) ontology to improve the effectiveness of medical image retrieval systems. In order to achieve greater effectiveness in the expansion, the search text was analyzed to identify which terms were most amenable to being expanded.
Design
To perform the expansions we utilized the hierarchical structure by which the MeSH descriptors are organized. Two strategies for selecting the terms to be expanded in each query were studied. The first consisted of identifying the medical concepts using the unified medical language system metathesaurus. In the second strategy the text of the query was divided into n-grams, resulting in sequences corresponding to MeSH descriptors.
Measurements
For the evaluation of the system, we used the collection made available by the ImageCLEF organization in its 2011 medical image retrieval task. The main measure of efficiency employed for evaluating the techniques developed was the mean average precision (MAP).
Results
Both strategies exceeded the average MAP score in the ImageCLEF 2011 competition (0.1644). The n-gram expansion strategy achieved a MAP of 0.2004, which represents an improvement of 21.89% over the average MAP score in the competition. On the other hand, the medical concepts expansion strategy scored 0.2172 in the MAP, representing a 32.11% improvement. This run won the text-based medical image retrieval task in 2011.
Conclusions
Query expansion exploiting the hierarchical structure of the MeSH descriptors achieved a significant improvement in image retrieval systems.
Introduction
Recent years have seen a huge increase in the amount of information that can be found in digital format in the medical domain. This enormous amount of information requires automatic processing systems and effective search systems to improve the work of users.1 2
By and large, information retrieval systems have focused efforts to improve accessibility on textual information. Nevertheless, there is a growing interest in optimizing access to visual information. The field of image retrieval has received less attention than that of text retrieval. Essentially there are two approaches to the problem of image retrieval, one based on the visual content of the image (content-based image retrieval) and the other based on textual information associated with the image (text-based image retrieval). In this paper we present a study taking the textual approach.
There are various techniques for correlating the language used in collections (medical reports, medical papers, etc) with user queries, chief among which is lexical expansion. In order to perform this expansion, it is quite common to use external resources that significantly improve the performance of information retrieval systems.3 4 Great efforts are currently being made to refine these resources, particularly in medical contexts, with the aim of helping the end user to manage this huge quantity of information. Ontologies are among the most used resources. Within the medical field, there is a large number of thesauri and ontologies. These are habitually used in tasks involving query expansion, semantic indexing of documents, and organizing search results. Nevertheless, due to the huge amounts of information they provide, in most cases their application to information retrieval tasks is no easy matter. In Jimeno et al,5 the authors propose an algorithm for refining ontologies used in information retrieval tasks. Among the most used in the field of biomedical information retrieval are GO,6 medical subject headings (MeSH)7 and the unified medical language system (UMLS).3 In particular, this study used the MeSH ontology to carry out query expansion.
The collection used in the study and the experimentation described in this paper was that used for the medical image retrieval task in the 2011 ImageCLEF (http://www.imageclef.org/) competition.8 In this corpus, the information associated with each image (metadata) is composed of the figure captions and the titles of the papers to which they belong. The paper identifier is also included to facilitate access to the full text.
The remainder of the paper is structured as follows. The Background section reviews the most relevant studies in the field. The Methods section provides a description of the techniques of expansion that were developed and the corresponding external resources used; it also outlines the process of constructing the index, details the document collection used in the study and experimentation, and specifies the measures used for carrying out the evaluation. The Results section summarizes the results obtained in the various experiments. These results are considered more fully in the Discussion section and the most significant are given close analysis. Finally, the Conclusions and directions for further studies are presented.
Background
Various systems for the retrieval of images and related documents are currently available. The majority of these are specially designed for use in the biomedical domain. Examples of such systems include Yale Image Finder,9 ARRS Goldminer,10 BioText11 and the National Library of Medicine's OpenI system.12
Image retrieval from the documents is usually based on the title text, figure captions, abstracts or full text. However, because of the nature of the collections, these systems frequently lose efficiency and fail to return the desired results.
Various works report on studies into the effect of using the MeSH ontology for query expansion. For example, in Gobel et al13 the authors base expansion on the hierarchical structure of MeSH. When this technique locates a MeSH descriptor in the query, it ascends the tree to higher levels to search for more general descriptors and adds those it finds to the user query.
In Lu et al14 the authors explore a strategy for query expansion using a process of advanced queries known as automatic term mapping in PubMed. The study employed a collection of 64 queries and approximately 160 000 MEDLINE citations, which were used in the 2006 and 2007 TREC genomics track. One of the main results was an increase in the F measure15 of 21.5% and 23.3% in the 2006 and 2007 collections, respectively, through the use of query expansion. The researchers conclude that query expansion through MeSH in PubMed can improve the effectiveness of retrieval, but that in real situations the improvement may not prove to be significant for PubMed users.
In Gobeill and colleagues16 17 the authors also employ MeSH ontology to expand both the collection and queries. The approach uses two strategies for expanding the document collection. The first of these extracts the set of MeSH descriptors from the image captions and article titles by means of a classifier, which returns a list of descriptors in order of relevance. The image captions, article titles and the first five descriptors returned by the classifier are then indexed. The second strategy indexes not only the above information, but also the MeSH descriptors associated with the corresponding article in MEDLINE. The query is expanded by extracting the set of MeSH descriptors using the classifier. Only those descriptors belonging to the A and C branches (diseases and anatomical concepts) of the MeSH tree are selected, from which only the three most relevant are added to the query.
In the work by Diaz et al,18 the authors describe experiments performed in the medical information retrieval task in ImageCLEF 2008. They created three different types of collections, the first comprising image captions and paper titles, the second comprising the captions, titles and the text from the section containing the image, and the third comprising the full text of the paper. They performed query expansion using MeSH and UMLS ontologies, achieving the best results with query expansion using MeSH and indexing the captions and titles of the papers. The improvement in the mean average precision (MAP) over the baseline score was 12.5%. Both this study and those of Diaz et al19 and Díaz-Galiano et al20used entry terms (alternative expressions, synonyms and terms related the descriptors) to carry out the expansion via the MeSH ontology.
As these studies show, various authors have taken advantage of the MeSH ontology in order to expand queries and improve information retrieval systems. In doing so, they have utilized the different cross-reference systems provided by the ontology (synonyms, entry terms, associative relationships among descriptors etc). In this study we present a query expansion strategy that uses the MeSH tree structure. Our proposal focuses on the choice of terms to be expanded and demonstrates that the expansion is most efficient when the UMLS metathesaurus is used, in controlled fashion, for determining which terms are expanded.
Methods
The aim of this work was to evaluate the effectiveness of an image retrieval system employing two strategies for query expansion using MeSH ontology. As mentioned above in the Background section, this ontology offers multiple possibilities for performing query expansion. Most researchers who utilize MeSH make use of the different information elements that the ontology makes available in order to find alternative means, synonyms and terms associated with the query terms. Our proposal is based on the way in which MeSH organizes its descriptors.
The MeSH descriptors are arranged into 16 categories: anatomical terms (category A); organisms (category B); illnesses (category C), etc. Each category is divided into subcategories. Within each subcategory, the descriptors are hierarchically ordered from the more general to the more specific, up to a maximum of 11 levels, resulting in a structure similar to that of a tree. The aim of this hierarchical structure is to facilitate the search process in medical documents. For example, papers relating to the query ‘Streptococcus pneumoniae’ are grouped under the descriptor ‘Streptococcus Pneumoniae’ rather than the descriptor ‘Streptococcus’, which is a more general term. Conversely, a paper that refers to a new ‘Streptococcus’ bacteria and that has yet to be included in the hierarchy, will be classified under the descriptor ‘Streptococcus’. Consequently, the user can search the structure from the more general to the more specific and vice versa. In Huang et al21 the authors recommend the use of MeSH terms in order to annotate biomedical articles. In that work, the researchers offer new methods for indexing biomedical documents based on the MeSH terms found in them.
We believe that we can exploit the characteristics of this structure to improve the expansion of search terms in information retrieval systems. In this work we present two strategies for query expansion based on the structural hierarchy of the MeSH descriptors.
Figure 1 shows a short extract from the MeSH tree diagram in which it can be seen that the descriptor ‘Brain’ includes seven more specific descriptors (children) while the descriptor ‘Central Nervous System’ includes only three.
Figure 1.
Extract from the MeSH tree diagram.
The expansion strategies developed in this paper are governed by the following criteria for expanding search terms:
If the search term is a MeSH descriptor and contains more specific descriptors, it is expanded using these.
If the search term is a MeSH descriptor but does not contain more specific descriptors, no expansion is performed.
If the search term is not a MeSH descriptor, no expansion is performed.
In many cases a descriptor comprises more than one term, and performing the expansion at the level of the term is not so efficient. For example, if the search for ‘Mitral Valve’ treats each term independently, neither the term ‘Mitral’ nor the term ‘Valve’ corresponds to a descriptor. Nevertheless, the two terms in combination correspond to the descriptor ‘Mitral Valve’, a biomedical concept.
The main difference between the two expansion strategies developed in this work consists of the means of identifying the search terms to be expanded. In the first, the medical concepts of each query are identified, while in the second the query is divided into n-grams.
External resources
Ontologies represent a particular knowledge domain in the form of a set of concepts and relations between them. There are many terminological and ontological resources available in the biomedical domain, along with a wide range of applications in natural language processing: information retrieval, question answering, automatic summarization and classification among others. The two resources used in this work were the MeSH ontology and the UMLS metathesaurus using the source vocabulary SNOMED-CT.22
MeSH is a controlled vocabulary used for indexing Medline papers. It is composed of sets of terms, or descriptors, organized into a hierarchical structure to allow searches at various levels of specificity. At present, MeSH encompasses 26 142 descriptors or main headings. This vocabulary is what is used to index Medline citations. Alternative forms, synonyms and terms related to the descriptors are known as entry terms. There are over 177 000 entry terms in MeSH.
The medical concepts approach
The main aim of this approach is that of identifying the medical concepts contained in the queries with the idea of restricting the expansion. Various resources are available for identifying significant concepts within a domain. This technique is known as recognition of named entities.23 In order to discover the medical concepts within the queries, in this study we have used the National Library of Medicine's MetaMap Transfer program24 (MMTx) using the source vocabulary SNOMED-CT of the UMLS metathesaurus V.2011AA.
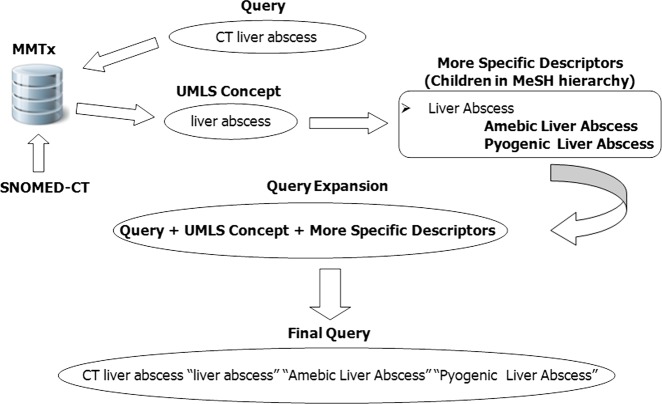
The concepts labelled in this phase were mapped to the MeSH hierarchy in order to perform an expansion of each one. Each labelled concept is sought within the MeSH tree. If the concept is a descriptor, its children are retrieved and added to the query according to the general scheme described above. In this approach, in addition to the terms expanded via the MeSH tree, the UMLS concepts identified are added, as illustrated in figure 2, which provides a schematic representation of the expansion process for the query ‘CT liver abscess’.
Figure 2.
Example of query expansion process. UMLS, unified medical language system. Access the article online to view this figure in colour.
The n-gram approach
This strategy divides all queries into n-grams with the aim of exploring all textual possibilities deriving from the query. In this work only those n-grams consisting of more than one term (n>1) are considered, as those comprising a single term return an overly broad expansion, which introduces too much noise into the query. Each n-gram is compared with the descriptors in the MeSH tree in order to perform the expansion according to the general scheme. The example below shows the composition of an n-gram for a query:
Query: ‘Microscopic giant cell’
N-grams
Microscopic
Microscopic giant
Microscopic giant cell
Giant
Giant cell
Cell
where n-grams 5 and 6 are MeSH descriptors. In this case only n-gram 5 is expanded as n-gram 6 consists of a single term.
Experimental document collection
The collection of documents used in carrying out the evaluation of the system was supplied by the ImageCLEF organization, in particular, the collection provided for the 2011 medical image retrieval task. That year, the collection was made up of more than 230 000 images and 30 queries. Participation was widespread, with 14 research groups from around the world registering for the task and a total of 130 runs being received. Several folders in XML format were included in the collection: the article collection, the queries (topics) and the full text of the articles.
The information (metadata) associated with each paper in this corpus comprises the paper identifier, its URL, a link to the full text, and the identifier for each image occurring in it along with the corresponding caption. Figure 3 provides an example of a record for the article collection.
Figure 3.
Example of ImageCLEF article record.
The information provided with each query comprises the query identifier and the type and text of the query written in English alongside a translation into French and German. In this work, only queries in English were used. Figure 4 provides an example of a record for the queries folder.
Figure 4.
Example of ImageCLEF topic record.
The relevant judgments for each query are supplied so as to enable the evaluation of systems. This information is provided to the participants once the competition has finished and is made available for future studies and experiments.
Index construction
In order to evaluate the two query expansion strategies developed in this work, an index of all the image captions in the collection was constructed. Previous studies into image retrieval found that indexing captions was more effective than indexing the full text. For example, in Kahn and Rubin25 the authors propose semantic indexing of figure captions as a means of improving the retrieval of radiological images. For the indexing, a pre-process was used consisting of the elimination of stop words and the stemming of all the image captions. To build the index for this study we used Lucene.26 This is a library implementing all the characteristics of an information retrieval engine. It is of use to any application requiring indexing and text search facilities. It runs under Java and is open source.
Information retrieval metrics
The evaluation of the system was performed using TREC_EVAL software, developed by the TREC (Test REtrieval Conference), which enables the calculation of the most used measurements in order to evaluate the retrieval efficiency. As usual in the ImageCLEF competition, the most relevant measurement for the organization was the MAP. This measurement consists of a single value that takes into account the ranking of the relevant retrieved documents. For a set of queries, this is calculated by:
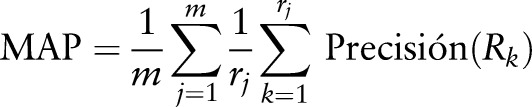 |
where m is the number of queries, rj is the number of documents relevant to the query j, and Rk is the kth retrieved relevant document, in the retrieved document ranking.
Results
This section gives details of the results obtained from the experiment. Some of the measurements provided by TREC_EVAL have been included in order to perform the comparison. In addition to the MAP, the values for precision at 10 and 20 documents were also included, as well as the number of relevant images retrieved for each query.
Table 1 shows a comparison for the first method of query expansion. In order to carry out this comparison the average scores of all participants for each of the measurements were calculated (AV ImageCLEF 2011). So as to ensure a greater degree of relevance for the results obtained, the median for each of the measurements was also calculated (MV ImageCLEF 2011).
Table 1.
Results of medical concepts approach
| Topic | Relevant images retrieved | MAP | P10 | P20 |
|---|---|---|---|---|
| 1 | 71 | 0.0781 | 0 | 0.15 |
| 2 | 11 | 0.0740 | 0 | 0 |
| 3 | 126 | 0.4863 | 1 | 0.85 |
| 4 | 16 | 0.3911 | 0.5 | 0.40 |
| 5 | 30 | 0.0952 | 0.3 | 0.15 |
| 6 | 55 | 0.0727 | 0.3 | 0.40 |
| 7 | 26 | 0.1180 | 0.2 | 0.15 |
| 8 | 28 | 0.1080 | 0.2 | 0.25 |
| 9 | 141 | 0.4051 | 0.8 | 0.75 |
| 10 | 4 | 0.3628 | 0.2 | 0.20 |
| 11 | 71 | 0.1762 | 0.5 | 0.45 |
| 12 | 1 | 0.0313 | 0.1 | 0.05 |
| 13 | 112 | 0.3080 | 0.7 | 0.65 |
| 14 | 40 | 0.0641 | 0.1 | 0.10 |
| 15 | 30 | 0.1044 | 0.3 | 0.15 |
| 16 | 98 | 0.8541 | 1 | 1 |
| 17 | 31 | 0.0141 | 0 | 0 |
| 18 | 6 | 0.0945 | 0.1 | 0.05 |
| 19 | 7 | 0.1898 | 0.1 | 0.05 |
| 20 | 6 | 0.4169 | 0.4 | 0.30 |
| 21 | 5 | 0.0526 | 0 | 0.05 |
| 22 | 20 | 0.1351 | 0.2 | 0.15 |
| 23 | 79 | 0.1600 | 0.5 | 0.50 |
| 24 | 131 | 0.1123 | 0.7 | 0.40 |
| 25 | 143 | 0.2411 | 0.5 | 0.55 |
| 26 | 4 | 0.0015 | 0 | 0 |
| 27 | 28 | 0.0363 | 0.3 | 0.20 |
| 28 | 4 | 0.6107 | 0.3 | 0.15 |
| 29 | 4 | 0.3423 | 0.2 | 0.10 |
| 30 | 143 | 0.3784 | 0.9 | 0.85 |
| Medical concepts approach | 1471 | 0.2172 | 0.3467 | 0.3017 |
| AV ImageCLEF 2011 | 1285.125 | 0.1644 | 0.3003 | 0.2663 |
| MV ImageCLEF 2011 | 1341 | 0.1668 | 0.3033 | 0.2633 |
MAP, mean average precision.
The concept expansion approach exceeded the average obtained in the competition for all the parameters included in table 1. The average MAP score achieved by this approach was 0.2172, an increase of 32.11% over the competition MAP average, and 30.21% over the median for the same. In addition, this method retrieved a significantly larger number of relevant images than the rest of the participants.
As can be seen in figure 5, the medical concepts approach achieves a good MAP in comparison with the rest of the participants. In particular, if it is compared with the average score of all participants, the MAP obtained using this method surpasses 21 of the 30 queries. In comparison with the median, the approach surpasses 23 of the 30 queries. Table 2 illustrates the same comparison for the second method of query expansion described in this paper.
Figure 5.
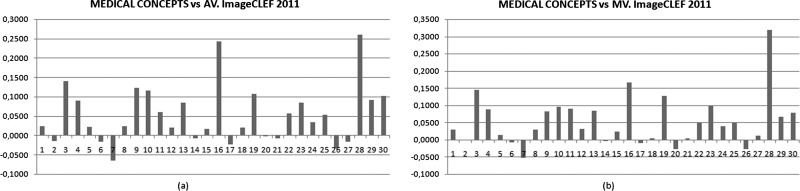
Mean average precision evaluation for the 30 queries using the medical concepts approach. (A) The approach is compared with the average scores for all participants. (B) The comparison is with the median for all participants.
Table 2.
Results of n-grams approach
| Topics | Relevant images retrieved | MAP | P10 | P20 |
|---|---|---|---|---|
| 1 | 71 | 0.0781 | 0 | 0.15 |
| 2 | 11 | 0.0740 | 0 | 0 |
| 3 | 126 | 0.4863 | 1 | 0.85 |
| 4 | 16 | 0.2535 | 0.5 | 0.35 |
| 5 | 30 | 0.0952 | 0.3 | 0.15 |
| 6 | 55 | 0.0727 | 0.3 | 0.40 |
| 7 | 26 | 0.1180 | 0.2 | 0.15 |
| 8 | 28 | 0.1080 | 0.2 | 0.25 |
| 9 | 141 | 0.4051 | 0.8 | 0.75 |
| 10 | 4 | 0.3628 | 0.2 | 0.20 |
| 11 | 60 | 0.0644 | 0 | 0.10 |
| 12 | 0 | 0 | 0 | 0 |
| 13 | 112 | 0.3080 | 0.7 | 0.65 |
| 14 | 40 | 0.0641 | 0.1 | 0.10 |
| 15 | 30 | 0.1340 | 0.3 | 0.30 |
| 16 | 98 | 0.6819 | 0.3 | 0.50 |
| 17 | 31 | 0.0241 | 0.2 | 0.10 |
| 18 | 6 | 0.0945 | 0.1 | 0.05 |
| 19 | 7 | 0.1898 | 0.1 | 0.05 |
| 20 | 6 | 0.4554 | 0.3 | 0.25 |
| 21 | 5 | 0.0768 | 0.1 | 0.10 |
| 22 | 20 | 0.2094 | 0.3 | 0.15 |
| 23 | 79 | 0.1600 | 0.5 | 0.50 |
| 24 | 131 | 0.1123 | 0.7 | 0.40 |
| 25 | 145 | 0.1909 | 0.1 | 0.10 |
| 26 | 4 | 0.0015 | 0 | 0 |
| 27 | 28 | 0.0363 | 0.3 | 0.20 |
| 28 | 4 | 0.5746 | 0.2 | 0.15 |
| 29 | 4 | 0.2089 | 0.2 | 0.10 |
| 30 | 143 | 0.3712 | 0.9 | 0.80 |
| Average n-gram approach | 1461 | 0.2004 | 0.2967 | 0.2617 |
| Average ImageCLEF 2011 | 1285.125 | 0.1644 | 0.3003 | 0.2663 |
| MV ImageCLEF 2011 | 1341 | 0.1668 | 0.3033 | 0.2633 |
MAP, mean average precision.
As can be seen, the results for this expansion strategy are somewhat inferior to those obtained with the medical concepts approach. In terms of the MAP, the results are better than the average and median for the competition (21.89% and 20.14%, respectively). With respect to precision at 10 and 20 documents, the values obtained were similar to those obtained by the rest of the participants. Nevertheless, this approach retrieved a far greater number of relevant images than the rest of the participants.
As can be seen in figure 6, the n-gram approach also achieves a good MAP score in comparison with the average and median. In this case, if a comparison is made with the average score for all participants, the MAP obtained by this approach surpassed 19 of the 30 queries, and in comparison with the median, it surpassed 18 of the 30 queries.
Figure 6.
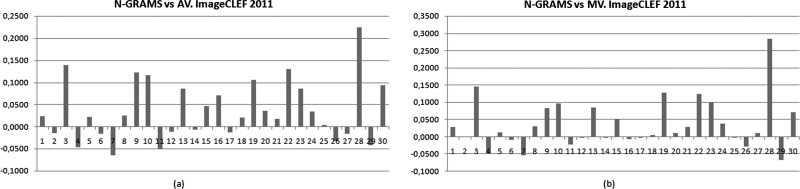
Mean average precision evaluation for the 30 queries using the n-grams approach. (A) The approach is compared with the average score for all participants. (B) The comparison is with the median for all participants.
Discussion
In the above section it was seen that the results obtained by the medical concepts approach were very good in comparison with those obtained by the n-gram approach and the other competition results. Several of the queries expanded by this approach are analyzed below.
Focusing on queries 28, 4 and 29, we can consider the three possibilities for expansion that this approach performs. Query 28, ‘Mitral valve prolapse’, is mapped to a medical concept. Within the MeSH hierarchy, the concept lacks a more specific descriptor. As a result, the expansion consisted of no more than the addition of the medical concept, with the following outcome: ‘Mitral valve prolapse “Mitral valve prolapse”’.
Query 4 ‘chest CT images with emphysema’, is mapped to the UMLS concept ‘emphysema’. When the expansion is performed, this concept is not added to the query as, consisting of a single word, it is already included in the original query. The concept has two more specific descriptors in the MeSH hierarchy: ‘Mediastinal emphysema’ and ‘Subcutaneous emphysema’. Consequently, the final expansion of this query is ‘chest CT images with emphysema ‘Mediastinal emphysema’ ‘Subcutaneous emphysema’’.
Query 29, ‘pulmonary embolism all modalities’, combines the two previous possibilities. The UMLS concept is ‘pulmonary embolism’, and MeSH also contains a more specific descriptor, ‘pulmonary infarction’, for the medical concept. Therefore, the final expansion for this query is ‘pulmonary embolism ‘pulmonary embolism’ ‘all modalities ‘pulmonary infarction’’.
As can be seen in figure 5, these queries improve their results with the expansion. The result obtained in the case of query 28 is particularly impressive, given that the only addition to the original query was that of the medical concept.
Although the overall results of this approach are very good, in certain queries the strategy did not work out as expected. For example, in query 17, ‘microscopic pathology images of the kidney’, the medical concept mapped is ‘pathology’. The search for this concept in MeSH returned a final expansion of ‘microscopic pathology images of the kidney ‘Forensic Pathology’ ‘Clinical Pathology’ ‘Molecular Pathology’ ‘Surgical Pathology’ ‘Telepathology’’. In this case, the medical concept is too generic and the expansion adds too many terms to the query, resulting in noise rather than fine-tuning.
With respect to query 20, ‘CT liver abscess’, the corresponding medical concept is ‘liver abscess’, and with the addition of the more specific terms returned by MeSH, the expanded query becomes ‘CT liver abscess ‘liver abscess’ ‘Amebic Liver Abscess’ ‘Pyogenic Liver Abscess’’. A comparison between this expansion and that performed by the n-gram approach reveals that the two are exactly the same, with the exception that this latter does not incorporate the medical concept (‘CT liver abscess ‘Amebic Liver Abscess’ ‘Pyogenic Liver Abscess’’). Despite this, the results of the n-gram approach are better. This is because, in this particular case, the addition of the concept ‘liver abscess’ gives greater relevance to the retrieval of images containing this concept, whereas the key to this query is the term ‘CT’, which describes the type of image to be retrieved. Therefore, the addition of the concept caused more images relating to ‘liver abscess’ to be retrieved, but not necessarily of the sort created by CT; x-rays and other types of image were equally likely, and so the expansion actually impaired the results.
Finally, it should be noted that the n-gram approach produced poorer results than the medical concepts approach because, by exploring all the combinations of the query text, the expansion introduces too much noise. In order to solve this problem, we decided not to expand individual terms (unigrams) and to search the MeSH hierarchical structure for more specific terms from bigrams and trigrams upwards. This produced a notable improvement in the overall result, although not sufficient to put it on a par with the medical concept approach, as the expansions it generated still retained more noise than the latter. For example, with regard to query 11, ‘abdominal CT images showing liver blood vessels’, the medical concept approach expands the UMLS concept ‘liver’ via the related MeSH concepts to ‘abdominal CT images showing liver blood vessels ‘Intrahepatic Bile Ducts’’. By contrast, in the n-gram approach, as the concept ‘liver’ is formed by only one term or unigram, it is not considered for expansion. Nevertheless, it calls up the bigram ‘blood vessel’ and the final expansion of the query is ‘abdominal CT images showing liver blood vessels ‘Arteries’ ‘Vascular Endothelium’ ‘Microvessels’ ‘Vascular Smooth Muscle’ ‘Retinal Vessels’ ‘Tunica Intima’ ‘Vasa Nervorum’ ‘Vasa Vasorum’ Veins’, which introduces noise into the query as too many search terms are added in the expansion.
Conclusions
In this paper we have presented a study of two approaches to query expansion making use of the hierarchical structure by which the MeSH ontology organizes its descriptors. The principal objective was to improve the effectiveness of image retrieval systems through textual content.
In the course of our experimentation we gained an understanding of the difficulties of finding an appropriate strategy for performing query expansion. We are nevertheless convinced that there are elements or combinations of elements that can be expanded in the query to improve image retrieval systems substantially.
Our results show that the expansion strategy employing medical concepts alongside the MeSH hierarchy successfully improved the effectiveness of the system. The results achieved with the n-gram strategy are somewhat inferior as it proved less discriminating of the terms to be expanded, resulting in too many being expanded and so diluting the accuracy of the results. The identification of UMLS concepts for carrying out the expansion considerably improved the results as it provided a significantly more controlled expansion.
However, so far our work has only experimented with expanding queries. In future studies we also intend to perform expansion on the medical concepts occurring in the text used for constructing the index. We will also explore the possibilities of expanding other elements in the MeSH structure, for example, descriptors situated at the same level or at more general levels, and not only the more specific descriptors. Finally, we also intend to dedicate future studies to analyzing queries in detail so as to extract information from abbreviations, type of image to search for27 (eg, radiographs, tomographs) and so on. After all, the most essential thing for an image retrieval system to work well is to know exactly what one is searching for.
Footnotes
Contributors: All authors listed in the paper provided substantial contribution to conception and design, acquisition of data or analysis and interpretation of data, drafting the article or revising it critically for important intellectual content, and final approval of the completed manuscript.
Funding: This work was partially funded by the Spanish Ministry of Science and Innovation, the Spanish Government Plan E and the European Union through ERDF, grant number TIN2009-14057-C03-03.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Karamanis N. Text mining for biology and biomedicine. Comput Linguist 33 2007;1:135–40 [Google Scholar]
- 2.Müller H, Deselaers T, Martin Deserno T, et al. Overview of the ImageCLEFmed 2006 Medical Retrieval and Medical Annotation Tasks. LNCS 2006;4730:595–608 [Google Scholar]
- 3.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32:267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson SJ, Johnston D, Humphreys BL. Relationships in medical subject headings. Relationships in the Organization of Knowledge New York: Kluwer Academic Publishers, 2001:171–84 [Google Scholar]
- 5.Jimeno A, Berlanga R, Rebholz D. Ontology refinement for improved information retrieval. Inf Process Manag 46 2010;4:426–35 [Google Scholar]
- 6.Stevens R, Goble CA, Bechhofer S. Ontology-based knowledge representation for bioinformatics. Brief Bioinform 1 2000;4:398–414 [DOI] [PubMed] [Google Scholar]
- 7.Nelson S, Schopen M, Savage A, et al. The MeSH translation maintenance system: Structure, interface design, and implementation. Proceedings of the 11th World Congress on Medical Informatics. San Francisco, CA: IOS Press 2004;67–9 [PubMed] [Google Scholar]
- 8.Kalpathy-Cramer J, Müller H, Bedrick S, et al. Overview of the CLEF 2011 Medical Image Classification and Retrieval Tasks, 2011;19–22 http://clef2011.org/resources/proceedings/Overview_ImageCLEF_Medical_Retrieval_Clef2011.pdf (accessed 31 Aug 2012)
- 9.Xu S, McCusker J, Krauthammer M. Yale Image Finder (YIF): a new search engine for retrieving biomedical images. Bioinformatics 24 2008;17:1968–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahn CH, Thao C. GoldMiner: a radiology image search engine. AJR Am J Roentgenol 2007;188:1475–8 [DOI] [PubMed] [Google Scholar]
- 11.Hearst M, Divoli A, Guturu H, et al. BioText Search Engine: beyond abstract search. Bioinformatics 23 2007;16:2196–7 [DOI] [PubMed] [Google Scholar]
- 12.Demner-Fushman D, Antani SA, Thoma GR. Multimodal biomedical information retrieval. Proceedings of the Fourth International Symposium on Languages in Biology and Medicine (LBM 2011). 14 -15 December 2011, Singapore. Singapore: Nanyang Technological University, 2011 [Google Scholar]
- 13.Gobel G, Andreatta S, Masser J, et al. A MeSH based intelligent search intermediary for consumer health information systems. Int J Med Inform 2001;64:241–51 [DOI] [PubMed] [Google Scholar]
- 14.Lu Z, Kim W, Wilbur W. Evaluation of query expansion using MeSH in PubMed. Inf Retr Boston 2009;12:69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Rijsbergen CJ. Information eetrieval, 2nd edn London, UK: Butterworth, 1979. [Google Scholar]
- 16.Gobeill J, Theodoro D, Patsche E, et al. Taking benefit of query and document expansion using MeSH descriptors in medical imageCLEF 2009. http://clef.isti.cnr.it/2009/working_notes/gobeill-papermedImageCLEF2009.pdf (accessed 31 Aug 2012)
- 17.Gobeill J, Ruch P, Zhou X. Query and Document Expansion with Medical Subject Headings Terms at Medical ImageCLEF 2008. LNCS 2009;5706:736–43 [Google Scholar]
- 18.Díaz MC, García MA, Martín MT, et al. Query expansion on medical image retrieval: MeSH vs. UMLS. Evaluating systems for multilingual and multimodal information access. LNCS 2009;5706:732–5 [Google Scholar]
- 19.Díaz MC, Martín MT, Ureña LA. Query expansion with a medical ontology to improve a multimodal information retrieval. Comput Biol Med 2009;4:396–403 [DOI] [PubMed] [Google Scholar]
- 20.Díaz-Galiano MC, García-Cumbreras MA, Martín-Valdivia MT, et al. Integrating MeSH ontology to improve medical information retrieval. In: Peters C, Jijkoun V, Mandl T, et al. eds. CLEF 2007. LNCS 2008;5152:601–6 [Google Scholar]
- 21.Huang M, Névéol A, Lu Z. Recommending MeSH terms for annotating biomedical articles. J Am Med Inform Assoc 2011;18:660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.SNOMED Clinical Terms. International Health Terminology Standards Development Organisation (IHTSDO) http://www.ihtsdo.org/snomed-ct/ (accessed 7 Mar. 2012. )
- 23.Borthwick A. A Maximum Entropy Approach to Named Entity Recognition. PhD thesis, New York University, New York, USA, 1999 [Google Scholar]
- 24.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001;17–21 [PMC free article] [PubMed] [Google Scholar]
- 25.Kahn CE, Rubin DL. Automated semantic indexing of figure captions to improve radiology image retrieval. J Am Med Inform Assoc 2009;16:380–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cutting D, Busch M, Cohen D, et al. Apache Lucene. 2008. http://lucene.apache.org/ (accessed 7 Mar 2012)
- 27.Rahman M, Antani S, Fushman D, et al. Biomedical image retrieval using multimodal context and concept feature spaces, in: medical content-based retrieval for clinical decision aupport. LNCS 2012;7075:24–35 [Google Scholar]