Abstract
Background
Computer-aided diagnosis for screening utilizes computer-based analytical methodologies to process patient information. Glaucoma is the leading irreversible cause of blindness. Due to the lack of an effective and standard screening practice, more than 50% of the cases are undiagnosed, which prevents the early treatment of the disease.
Objective
To design an automatic glaucoma diagnosis architecture automatic glaucoma diagnosis through medical imaging informatics (AGLAIA-MII) that combines patient personal data, medical retinal fundus image, and patient's genome information for screening.
Materials and methods
2258 cases from a population study were used to evaluate the screening software. These cases were attributed with patient personal data, retinal images and quality controlled genome data. Utilizing the multiple kernel learning-based classifier, AGLAIA-MII, combined patient personal data, major image features, and important genome single nucleotide polymorphism (SNP) features.
Results and discussion
Receiver operating characteristic curves were plotted to compare AGLAIA-MII's performance with classifiers using patient personal data, images, and genome SNP separately. AGLAIA-MII was able to achieve an area under curve value of 0.866, better than 0.551, 0.722 and 0.810 by the individual personal data, image and genome information components, respectively. AGLAIA-MII also demonstrated a substantial improvement over the current glaucoma screening approach based on intraocular pressure.
Conclusions
AGLAIA-MII demonstrates for the first time the capability of integrating patients’ personal data, medical retinal image and genome information for automatic glaucoma diagnosis and screening in a large dataset from a population study. It paves the way for a holistic approach for automatic objective glaucoma diagnosis and screening.
Keywords: medical imaging informatics, patient data, Medical Retinal Image, Genome information, multiple kernel learning, glaucoma
Background
Glaucoma and early diagnosis
Glaucoma is a disease of the optic nerve. It is the second leading cause of blindness, and will affect approximately 80 million people by 2020.1 Glaucoma is a chronic and irreversible neurodegenerative disease in which the optic nerve is progressively damaged, leading to deterioration in vision and quality of life.2 The disease is usually asymptomatic and patients are commonly unaware of the disease until noticeable visual loss occurs at a later stage, giving rise to its nickname the ‘silent thief of sight’. The estimated progression of optic nerve fiber loss in glaucoma can range from 9% to 63% over a 5-year period.3–5 In view of this, early detection of glaucomatous changes is crucial for timely treatment before the onset of permanent functional visual loss. However, studies in Singapore and other countries have shown that 50–90% of glaucoma cases remain undetected in the population.6–11
Early treatment by lowering the intraocular pressure (IOP) by medication or surgery can halt or slow disease progression. The American Academy of Ophthalmology has strongly recommended screening for glaucoma as part of comprehensive adult medical eye evaluation, with screening frequency depending on an individual's age and other glaucoma risk factors.
Glaucoma can be detected from raised IOP and visual field loss. IOP measurement using non-contact tonometry is found to be neither specific nor sensitive enough to be an effective screening tool as glaucoma can be present with or without increased IOP.4 Functional testing through vision loss is likely to miss most patients with early glaucoma damage12 as these patients do not have many visual symptoms. Assessment of the damaged optic nerve is more promising and superior to IOP measurement or visual field testing, as optic nerve damage precedes vision loss and can be used to detect glaucoma earlier with higher sensitivity. Optic nerve assessment can be performed by a trained professional. However, as such a manual assessment is subjective, time consuming and expensive, there remains a strong need for an efficient and objective way to screen for glaucoma.
Previous work in medical imaging informatics for computer-aided detection
In recent years, there has been increasing use of data mining and informatics in biomedical and medical imaging applications due to the wealth and expanse of available data.13 This has led to a greater understanding and interpretation of the complex relationships in such data. In general, data used in medical informatics can be broadly classified as personal data, imaging data and genetic data.
Personal data and other ocular measurements have been used to develop models of prediction for glaucoma. The Ocular Hypertension Study Group14 designed a predictive model through the use of baseline age, IOP, central corneal thickness, vertical cup-to-disc ratio, and visual field pattern SD to estimate the 5-year risk of glaucoma. Similarly, Hattenhauer et al15 calculated the probability of blindness from new diagnoses of glaucoma using a Kaplan–Meier model, based on perimetric measurements. It should be noted that these models use input parameters obtained from highly specialized instruments found in tertiary care institutions, which may not be suitable or readily available for screening.
Current glaucoma screening
A high IOP in the eye has been traditionally regarded as a principal risk factor for glaucoma.16 Increased IOP is particularly evident in closed angle glaucoma, in which the increase in IOP has been attributed to blockage of the trabecular meshwork. However, for open angle glaucoma, the use of IOP for screening is limited. Results from studies have shown that IOP is unable to differentiate between normal or increased pressure, or low tension and high tension glaucoma.17 Due to the fluctuation of IOP over time and throughout the day, repeated IOP measurements are required to detect any anomalous increases in IOP. Another challenge is that IOP is positively correlated with blood pressure and other cardiac risk variables.18 This has made the use of IOP challenging for glaucoma screening. In one study, increased IOP was found only to have a sensitivity of 47% and a specificity of 92% for diagnosing glaucoma.19
Our objective is to develop an automatic glaucoma diagnosis and screening architecture, automatic glaucoma diagnosis through medical imaging informatics (AGLAIA-MII), which combines patient personal data, medical retinal fundus image features, and genome information for glaucoma prediction.
Materials and methods
The rapid development of medical imaging informatics offers new insights into the computer-aided diagnosis of diseases. Integrating automatic medical image analysis with the utilization of informatics methodologies from patients’ personal data and genome single nucleotide polymorphisms (SNP) provides a promising direction for disease diagnosis.
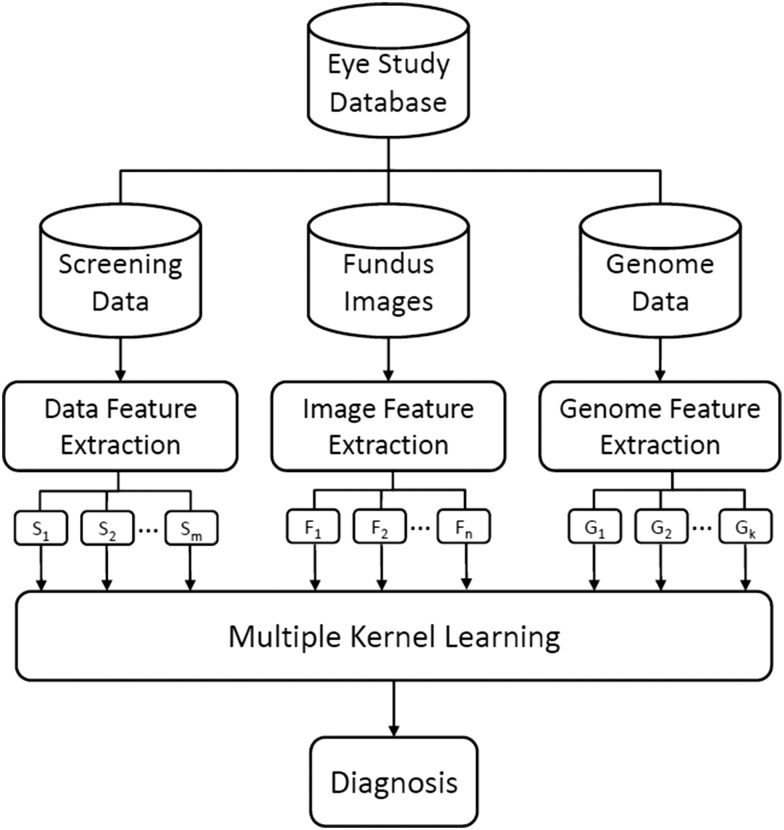
The AGLAIA-MII architecture shown in figure 1 uses information from multiple sources, including subjects’ personal data, imaging information from retinal fundus image, and patients’ genome information. Features from each data source will be extracted automatically. Subsequently, these features will be passed to a multiple kernel learning (MKL) framework to generate a final diagnosis outcome.
Figure 1.
Flowchart of the automatic glaucoma diagnosis through medical imaging informatics (AGLAIA-MII) architecture.
Data description
AGLAIA-MII is trained and evaluated on data from the Singapore Malay Eye Study (SiMES) database. SiMES is a population-based study conducted from 2004 to 2007 to assess the causes and risk factors of blindness and visual impairment in the Singapore Malay community. The study was approved by the institutional review board of Singapore Eye Research Institute. The database contains 3280 subjects, with complete or partial personal data, retinal fundus image data and genome information for each subject. The personal data in SiMES contains demographic data such as age, gender and height, ocular examination data, such as IOP and cornea thickness, as well as historical medical data. In addition, diagnostic information of glaucoma is available and is used as the class labels in our experiments. Image data for glaucoma analysis in SiMES is based on an optic disc centered retinal fundus image. These images were acquired using a 45° FOV Canon CRDGi retinal fundus camera with a 10D SLR backing, at an image resolution of 3072×2048 pixels. The patients were also genotyped on Illumina 610quad arrays, and the data generated form our genome data information.
Quality control was performed on the personal data, retinal image data and genome information to remove incomplete data. Personal data categories with more than 5% missing values were first removed. Subsequently, subjects with more than 5% missing values with the remaining features were also excluded. Subjects with low quality retinal fundus images that are ungradable manually are removed to ensure a clean image dataset. The genome dataset is made up of selected autosomal SNP after a stringent quality control procedure. These three datasets are then merged via subject matching. The final dataset contains 2258 subjects with clean screening data, image data and genome data. Among the 2258 individuals, 100 were diagnosed to have glaucoma and the rest were normal. The distribution of glaucoma subjects in the final dataset is representative of the glaucoma prevalence in the population.
Personal data
The personal data of each subject is collected and used as one type of feature for glaucoma assessment in AGLAIA-MII. Table 1 lists the 46 variables used for each subject.
Table 1.
Personal data used in AGLAIA-MII
| List of demographic and clinical variables included in personal data | ||||
|---|---|---|---|---|
| Age | Blood creatinine | Albumin-creatinine ratio | Can read | Chronic kidney disease indicator |
| Age group | Blood glucose | Diabetes I | Can write | Hyperlipidemia |
| Gender | Blood HbA1c categories | Diabetes II | Alcoholic drink categories | Metabolic syndrome |
| Height | Blood glycosylated hemoglobin | Job categories | Ever smoke | Microalbuminuria |
| Weight | Blood total cholesterol | Race | Current smoker | Incremental metabolic variable |
| Diastolic blood pressure | Blood LDL cholesterol | Marital categories | Angina* | Lens status |
| Systolic blood pressure | Blood HDL cholesterol | Income categories | Heart attack * | |
| Pulse pressure | Triglycerides | Type of place living in | Stroke* | |
| Mean arterial pressure | Hypertension | Place of birth | Hypercholessterolemia* | |
| BMI | Hypertension treatment and control | Education categories | Thyroid condition* | |
*Self-reported.
AGLAIA-MII, automatic glaucoma diagnosis through medical imaging informatics; BMI, body mass index; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Univariant analysis is conducted for all demographic and clinical variables that compose the personal data. p Values are obtained by conducting Student's t test for numerical variables and χ2 test for categorical variables. Table 2 lists the mean, SD and p values for all numerical variables. ‘Age’ is highly related to glaucoma with p values smaller than 0.001. Systolic blood pressure, pulse pressure, and triglycerides are also associated with glaucoma. For categorical variables, three are found to be associated with glaucoma: ‘age group’ (p<0.001), ‘race’ (p=0.01) and ‘job categories’ (p=0.047).
Table 2.
Univariant analysis for demographic and clinical variables
| Normal (2158) | Glaucoma (100) | ||||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | p Value | |
| Age (years) | 57.52 | 10.5 | 63.43 | 10.55 | <0.001 |
| Height (cm) | 158.89 | 9.0 | 157.63 | 10.25 | 0.226 |
| Weight (kg) | 66.74 | 13.6 | 64.55 | 15.34 | 0.161 |
| Diastolic blood pressure (mm Hg) | 80.04 | 11.4 | 78.20 | 10.43 | 0.087 |
| Systolic blood pressure (mm Hg) | 146.30 | 23.5 | 151.44 | 24.14 | 0.040 |
| Pulse pressure (mm Hg) | 66.26 | 18.0 | 73.24 | 20.68 | 0.001 |
| Mean arterial pressure (mm Hg) | 102.13 | 14.1 | 102.61 | 13.10 | 0.725 |
| Body mass index (kg/m2) | 26.42 | 5.0 | 25.98 | 6.10 | 0.482 |
| Blood glycosylated Hemoglobin (mmol/l) | 6.42 | 1.5 | 6.42 | 1.38 | 0.987 |
| Blood total cholesterol (mmol/l) | 5.61 | 1.2 | 5.55 | 1.17 | 0.604 |
| Blood LDL cholesterol (mmol/l) | 3.53 | 1.0 | 3.56 | 1.07 | 0.835 |
| Blood HDL cholesterol (mmol/l) | 1.34 | 0.3 | 1.39 | 0.37 | 0.266 |
| Triglycerides (mmol/l) | 1.62 | 1.3 | 1.34 | 0.99 | 0.007 |
HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Genome information
Vithana et al20 recently reported three SNP markers (rs11024102 in PLEKHA7, rs3753841 in COL11A1 and rs1015213), which showed significant association with glaucoma. Besides these three SNP, other SNP that have demonstrated moderate significance, or have been reported in previous findings (tables 1, 3, 4, 6 and 7 of Vithana et al)20 have also been used in our model. In total, 178 SNP are selected in this study, as listed in supplementary table S1 (available online only). These 178 SNP are used as genetic features for glaucoma assessment in AGLAIA-MII. For each single SNP, its value 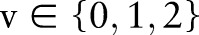 represents the genotype, with the values of
represents the genotype, with the values of 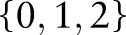 reflecting the number of minor alleles in the SNP pair.
reflecting the number of minor alleles in the SNP pair.
Medical retinal image and features
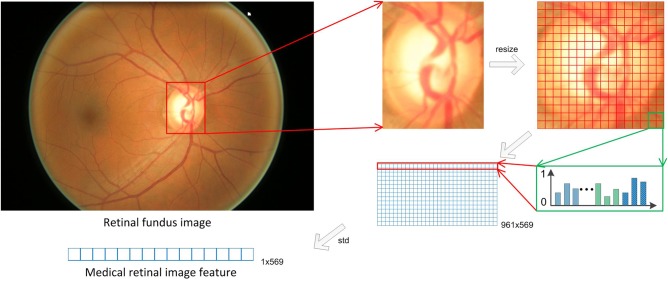
As illustrated in figure 2, for each retinal image, the optic disc is first segmented by using the method in Wong et al.21 In this method, the approximate location of the optic disc is detected using the intensity of equal-sized patches from the retinal image. Next, a region of interest approximately twice the size of the typical optic disc size is then extracted from the candidate patch location. To localize the optic disc, we used a variational level-set approach, which seeks to minimize an energy functional consisting of contextual parameters in the image domain.
Figure 2.
Medical retinal image feature extraction.
The detected disc is then resized to 256×256 to extract medical retinal features as follows. First, the optic disc is divided into 16×16 grids, which are half-overlapping with its neighbors, thus obtaining 16×16+15×15+2×16×15=961 grids. In each 16×16 grid, a 256-dimensional histogram of the green channel and a 256-dimensional histogram of the local binary pattern22 on the grey channel are extracted. In addition, mode: red green blue (RGB) colors, mode LAB colors (a color-opponent space with dimension L for lightness and A and B for the color-opponent dimensions), color moments of the hue saturation value (HSV) colors (1st to 4th moments),23 image moment invariants RGB colors,24 statistical texture descriptors24 from RGB channels (average grey level, average contrast, measure of smoothness, third moments, uniformity and entropy) are also calculated, resulting in a 569 dimensional descriptor for each grid. The final medical image features are obtained as the standard derivation of the descriptors for all grids, which represents the variance of each local part of the retinal image.
In AGLAIA-MII, three sources of data from different domains are used for glaucoma assessment, that is, personal data, genome information and medical retinal image feature. Before applying learning algorithms, each feature dimension is normalized to the range of [0 1] in order to avoid magnitude differences and bias among the dimensions, and a support vector machine (SVM) based MKL framework is utilized to train the classifier for glaucoma assessment.
SVM-based MKL
SVM25 26 are powerful tools for classification. Given a set of training examples, each marked as belonging to one of two categories in a binary classification application, a SVM training algorithm builds a model that assigns new examples into one category or the other. SVM cast the data into a higher dimensional space called the ‘kernel-induced feature space’ where the data are separable. Different kernel functions correspond to different embeddings of the data and thus can be viewed as capturing different notions of similarity. Standard SVM methods build a single kernel function on different types of input data throughout the algorithm and may have compromises on learning with fusion of heterogeneous data types. To utilize fully the data acquired from various sources and boost the overall system performance, a classifier can be trained using the increasingly popular MKL27 approach. Many applications have shown that using multiple kernels instead of a single one can enhance the interpretability of the decision function and improve performance.27
Given a training set of instance-label pairs 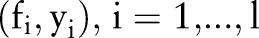 where
where 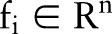 is the whole feature of a subject, and
is the whole feature of a subject, and 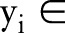 {−1,1} denotes its label, that is, 1 denotes glaucoma and −1 denotes normal, feature fi is mapped into a higher dimensional space by using a mapping function
{−1,1} denotes its label, that is, 1 denotes glaucoma and −1 denotes normal, feature fi is mapped into a higher dimensional space by using a mapping function 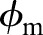 28 In the case of SVM-based MKL, a convenient approach is to consider thekernel K (fi, fi) as a convex combination of the basis kernels:
28 In the case of SVM-based MKL, a convenient approach is to consider thekernel K (fi, fi) as a convex combination of the basis kernels:
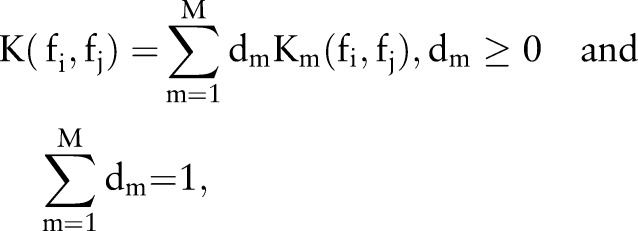 |
where M is the total number of basis kernels, 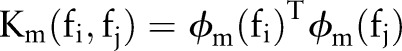 is mth basis kernel and dm is its weight to be determined. Each basis kernel Km may either use the full set of features describing samples or subsets of features stemming from different data sources.27 Within this MKL framework, the problem of data representation through the kernel is then transferred to the selection of weights dm. In AGLAIA-MII, we use basis kernels based on each single feature from only one source, which can demonstrate that the combination of multiple features from different sources is better than using a single feature. For efficiency, one linear kernel is initialized for each feature type. There are many MKL solver toolboxes that are publicly available, such as SimpleMKL29 and Group Lasso.30 In AGLAIA-MII, the LIBLINEAR toolbox31 is used to train linear SVM models for each individual feature, and the Group Lasso30 toolbox is used to train MKL models.
is mth basis kernel and dm is its weight to be determined. Each basis kernel Km may either use the full set of features describing samples or subsets of features stemming from different data sources.27 Within this MKL framework, the problem of data representation through the kernel is then transferred to the selection of weights dm. In AGLAIA-MII, we use basis kernels based on each single feature from only one source, which can demonstrate that the combination of multiple features from different sources is better than using a single feature. For efficiency, one linear kernel is initialized for each feature type. There are many MKL solver toolboxes that are publicly available, such as SimpleMKL29 and Group Lasso.30 In AGLAIA-MII, the LIBLINEAR toolbox31 is used to train linear SVM models for each individual feature, and the Group Lasso30 toolbox is used to train MKL models.
Experimental methods for AGLAIA-MII
In AGLAIA-MII, for the purpose of demonstrating that the combination of multiple source data can boost up the diagnosis, we report and compare the diagnosis performance of eight methods using different features and their combinations: (1) personal data (referred to as P); (2) genetic info(referred to as G); (3) low-level direct image features (referred to as I); (4) P+G; (5) P+I; (6) G+I; (7) P+G+I (AGLAIA-MII); (8) IOP, which is the current glaucoma assessment method.
Twofold cross-validation is adopted in our experiments. All subjects are randomly divided into non-overlapping set A and B with equal size. Because of imbalanced positive (glaucomatous) and negative (normal) subjects, we use all the positive subjects and the same number of randomly selected negative subjects from set A as the training set. The trained model is used for testing in set B. After that, a similar procedure is conducted to use all positive subjects and the same number of randomly selected negative subjects from set B as the training set. The trained model is then used for testing in set A. It is important to clarify that the optimal parameters in the above training are obtained through a similar second round twofold cross-validation within the training set. This strategy is commonly used.32
We conducted 10 sets of independent testing using the cross-validation and data-balanced strategy as outlined above. Therefore, 20 sets of testing results were obtained. This approach was adopted for each of the learning-based methods that is, 1–7. For the IOP data in method 8, the measured IOP values corresponding to the individual eye were directly used without further processing.
Analysis methods used for AGLAIA-MII
Sensitivity and specificity are often used as measures of accuracy in classification tests. Sensitivity is defined as the proportion of true positives, in our study glaucoma cases, which are correctly identified by the test. In contrast, specificity refers to the proportion of true negatives, or normals, which are correctly determined by the test. Both are used to obtain an assessment of the diagnostic accuracy of a test. The positive predictive value (PPV), defined as the proportion of true cases from the total number of positive detections, is often used as another measure of reliability of the test. The use of PPV is suitable in our study set, because the ratio of the true positives and true negatives in the study dataset are representative of the prevalence in the population. The F-measure score was also calculated from the results and is included as an additional metric in our analysis.
We further considered two scenario setpoints for testing the diagnostic accuracy. At the screening setpoint, we maintained a baseline specificity of 0.850 to limit the rate of false negatives, and determined the corresponding performance of the various source combinations. At the second setpoint, we determined the knee point of the receiver operating characteristic (ROC) curve and compared the diagnostic metrics at this point. The knee point is defined as the point on the ROC curve nearest to the ideal accuracy of specificity (1.000) and sensitivity (1.000). The latter approach seeks to compare the different source configurations at their optimal settings, while the former is based on screening settings.
There is a tradeoff between the specificity and sensitivity of a test. This is usually expressed on a ROC curve, which plots sensitivity against 1-specificity for all possible values. Often, the area under the curve (AUC) is also calculated. While the individual specificities and sensitivities are measures of diagnostic accuracy, the AUC is often regarded as the overall measure of the diagnostic strength of a test. In our analysis, we compared the different sources combinations through their AUC using the method of Hanley and McNeil.33
Results and discussion
Table 3 presents the specificity, sensitivity, PPV and the AUC for the different source configurations. At the screening setpoint with specificity set at 0.850, we observed that the use of only genome information resulted in a better sensitivity (0.543), PPV (0.143) and F-measure (0.227) than that obtained from only retinal fundus image features (sensitivity: 0.420; PPV: 0.115; F-measure: 0.180) or only personal data (sensitivity: 0.197; PPV: 0.057; F-measure: 0.089). When sources were combined, the use of all data sources in AGLAIA-MII (genome information, retinal fundus image features, and personal data) resulted in the best performing sensitivity, PPV and F-measure metrics for screening over any other combinations or its individual components. Furthermore, we found that the combination of any two data sources resulted in a better performance than that of its individual components. The majority of these results outperformed the current practice of using IOP for glaucoma screening. In particular, more than a twofold increase in performance over IOP was observed when all data sources were used.
Table 3.
Key diagnostic metrics (specificity, sensitivity, PPV, F-measure, AUC) as a result of using the various individual and combined sources
| Screening setpoint | Optimal ROC knee setpoint | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Specificity | Sensitivity | PPV | F-measure | Specificity | Sensitivity | PPV | F-measure | AUC | ||
| Single source | Genome information (G) | 0.850 | 0.543 | 0.143 | 0.227 | 0.744 | 0.760 | 0.122 | 0.183 | 0.810 |
| Retinal fundus image features (I) | 0.850 | 0.420 | 0.115 | 0.180 | 0.720 | 0.662 | 0.101 | 0.166 | 0.722 | |
| Personal data (P) | 0.850 | 0.197 | 0.057 | 0.089 | 0.596 | 0.554 | 0.060 | 0.108 | 0.561 | |
| Combined source | Genome information + retinal image features (G+I) | 0.850 | 0.647 | 0.166 | 0.264 | 0.764 | 0.812 | 0.139 | 0.192 | 0.856 |
| Personal data + genome information (P+G) | 0.850 | 0.639 | 0.164 | 0.261 | 0.750 | 0.824 | 0.134 | 0.186 | 0.853 | |
| Personal data + retinal image features (P+I) | 0.850 | 0.454 | 0.123 | 0.193 | 0.714 | 0.718 | 0.107 | 0.171 | 0.753 | |
| AGLAIA-MII: Personal data + genome information + retinal fundus image features (P+G+I) | 0.850 | 0.671 | 0.171 | 0.273 | 0.786 | 0.816 | 0.153 | 0.201 | 0.866 | |
| Current glaucoma screening using IOP | 0.850 | 0.304 | 0.086 | 0.134 | 0.614 | 0.560 | 0.064 | 0.118 | 0.604 | |
For specificity, sensitivity and PPV, two conditions are considered: screening setpoint by setting specificity constant at 0.85, and at the optimal ROC knee point. The best results are highlighted in italics.
AGLAIA-MII, automatic glaucoma diagnosis through medical imaging informatics; AUC, area under the (ROC) curve; IOP, intraocular pressure; PPV, positive predictive value; ROC, receiver operating characteristic.
When the ROC knee point is used to select the diagnostic accuracy metrics, a similar trend as above was observed, in which genome information performed the best out of the single sources. The best performance was again noted to be from AGLAIA-MII fusion of genome information, personal data and retinal image features, resulting in a specificity of 0.786, sensitivity of 0.816, PPV of 0.153 and F-measure of 0.201, generally higher than any other independent or combined data source. Combined source results were also improved over individual data source results.
Figure 3 shows the ROC plots for the different data sources used in the experiments. The corresponding AUC for the ROC curves in figure 2 are presented in the last column of table 3. When the single data sources of AGLAIA-MII are considered individually, results show that genome information gives the best AUC at 0.810, and was significantly better than retinal image features (0.810 vs 0.722, p<0.005) and personal data (0.810 vs 0.561, p<0.005), while the AUC for retinal image features was significantly better than that of personal data (0.722 vs 0.561, p<0.005). Results using genome information and retinal image features were significantly better than the AUC derived from current IOP-based glaucoma screening (p<0.005).
Figure 3.
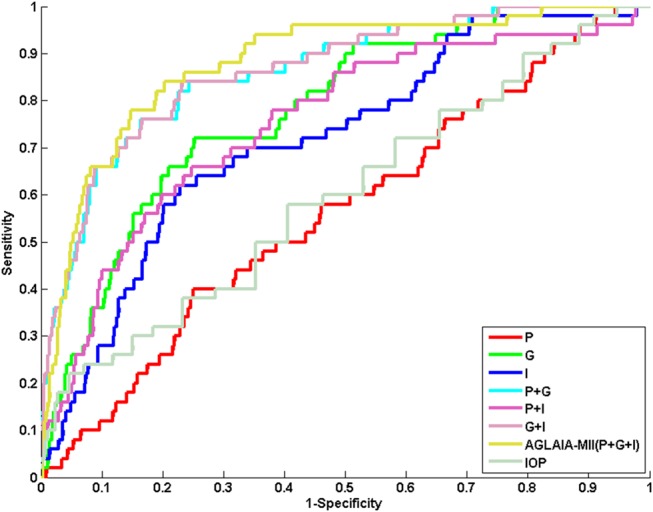
Receiver operating characteristic (ROC) plots for automatic glaucoma diagnosis through medical imaging informatics (AGLAIA-MII) and the other source combinations. The ROC for intraocular pressure (IOP)-based glaucoma screening is also provided for comparison.
The AGLAIA-MII combination of personal data, retinal image features and genome information resulted in the highest AUC (0.866) among any other source, single or combined. Furthermore, the combination of any two sources resulted in a better AUC than their component sources. This observation holds for both the top performing single source genome information, which registered an average 5% improvement when combined, as well as a more substantial improvement for the poorest performing single source based on personal data, showing a substantially significant improvement of at least 34%. Encouragingly, each of the results from the combination of sources, regardless of combination, generated an AUC that was significantly higher (p<0.005) than that of the current clinical standard of care using IOP.
OTable 4 shows the quantitative accuracy improvements in terms of the AUC when combined sources are used compared to single sources. When the AGLAIA-MII combinations of all sources (genome information plus retinal image features plus personal data) are used, the results yield a 5.59%, 20.36% and 60.33% improvement over the individual genome information, retinal image features and personal data sources, respectively. Personal data benefited the most from combining with other sources, yielding an improvement in AUC of at least 37.45%. The addition of genome information to any other single source had the most beneficial effect, boosting the accuracy of retinal image features by 18.84% and that of personal data by 57.93%.
Table 4.
Comparative improvements from the use of combined sources over single sources
| Accuracy improvement over component single sources | |||
|---|---|---|---|
| Combinations | Genome information (G) (%) | Retinal image features (I) (%) | Personal data (P) (%) |
| Genome information + retinal image features (G+I) | 5.68 | 18.56* | |
| Personal data + genome information (P+G) | 4.01 | – | 57.93* |
| Personal data + retinal image features (P+I) | – | 4.29 | 34.22* |
| AGLAIA-MII: personal data + genome information + retinal fundus image features (P+G+I) | 6.91 | 19.94* | 54.37* |
*Significant at p<0.005.
Best results are highlighted in italics.
AGLAIA-MII, automatic glaucoma diagnosis through medical imaging informatics.
In AGLAIA-MII, we tested and compared several methods using different features and combinations. From the results listed in tables 3 and 4 and figure 2, we can make the following observations:
Performance boosting using data from different domains
In AGLAIA-MII, we have used data from three independent sources based on patient data, retinal fundus image features and genome information, each of which has some success in the detection of glaucoma. Through the use of the MKL framework in AGLAIA-MII, we have shown that data from different source domains have a complementary effect in boosting glaucoma diagnosis performance. The experimental results also show the complementary advantages in combining data domains, as any one combination was seen to outperform the performance based on a single domain source.
AGLAIA-MII framework is able to integrate high feature dimensions successfully to boost performance over fewer features
In our results, we observed a hierarchy of performance, in which the maximal combination of sources in AGLAIA-MII outperformed any two combinations of sources. Similarly, any two source combinations produced results that were better than the individual component source. The MKL approach is able to boost the performance because the use of multiple kernels improves the interpretability of the decision function, resulting in an optimal use of discriminative features to boost detection outcomes, despite the increased feature dimension and complexity.
Effect of feature discriminant power
We observed that using only one type of feature leads to a lower performance compared to the use of MKL-based multiple feature fusion in AGLAIA-MII. However, the use of genome information alone can still result in a relatively high performance. In AGLAIA-MII, we selected 178 SNP out of more than 260 000 SNP based on a strong association with glaucoma disease, as determined from studies on very large datasets comprising of data from various races. This resulted in less noise and redundancy.
Experimental results showed that personal data can be quite noisy and may have a lower association with glaucoma. The medical retinal image features used in AGLAIA-MII focused on the variability of local grids through low level features such as intensity and contrast, and these features may not have as high a discriminant power as genetic information for glaucoma disease. In the future, AGLAIA-MII can be improved by introducing more and better image features, which are more representative of glaucoma under the same MKL framework.
AGLAIA-MII's performance has been proved in a large population study. To boost performance further, we will continue to test the system's performance in other population studies to find more evidence and fine tune the performance of AGLAIA-MII. The same framework and principles can also be used to design automatic diagnostic systems for other ocular diseases such as age-related macular degeneration, cataract and pathological myopia.
Conclusion
A patient's personal data, medical retinal image and genome information are data types of different natures, with each providing a different and potentially complementary view of an indvidual's condition. Combining these data types intelligently and providing a holistic glaucoma diagnosis is a new approach to boost diagnostic accuracy. Accurate early detection of glaucoma is crucial to allow for early treatment before the onset of permanent functional visual loss. AGLAIA-MII demonstrates for the first time the integration of the above three data types for automatic glaucoma screening and diagnosis in a large population dataset. Our experiments have shown that the proposed AGLAIA-MII framework fuses these data from various sources intelligently and effectively, paving the way for a holistic approach for automatic objective glaucoma diagnosis and screening.
In clinics, the combination of multiple measurements is currently often an art rather than a science, and can usually only be mastered by experienced glaucoma specialists. The interpretation can be susceptible to interobserver differences due to variances in training and experience. The promising results demonstrated in this work raise the possibility of using a clinical decision support system such as AGLAIA-MII to run in parallel with existing clinical workflows to offer an objective, evidence-based diagnosis to clinicians as a second opinion. From the public healthcare perspective, a carefully designed glaucoma screening programme based on AGLAIA-MII can provide a faster, more cost-effective and more accurate detection of the disease. This will lead to improvements in glaucoma disease management and cost savings for patients, public healthcare providers and the government.
Footnotes
Contributors: JL, ZZ and DWKW conceived the idea; ZZ and FY extracted and formatted the data; NMT, YX and JC extracted medical retinal image features; YX conducted MKL learning; DWKW and ZZ analysed the results; CKK and DX assisted in the analysis method and MKL learning; YCT, TA and TYW provided clinical validation; JL, ZZ, DWKW, YX, FY, JC and NMT wrote the manuscript, and provided editorial advice.
Funding: This work was supported in part by the Agency for Science, Technology and Research, Singapore under SERC grant 092-148-0073.
Competing interests: None.
Patient consent: Obtained.
Ethics approval: The study was approved by the institutional review board of Singapore Eye Research Institute.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol 2006;90:262–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kass MA, Heuer DK, Higginbotham EJ, et al. The Ocular Hypertension Treatment Study: a randomized trial determines that topical ocular hypotensive medication delays or prevents the onset of primary open-angle glaucoma. Arch Ophthalmol 2002;120:701–13; discussion 829–30. [DOI] [PubMed] [Google Scholar]
- 3.Komulainen R, Tuulonen A, Airaksinen PJ. The follow-up of patients screened for glaucoma with non-mydriatic fundus photography. Int Ophthalmol 1992;16:465–9 [DOI] [PubMed] [Google Scholar]
- 4.Caprioli J. Clinical evaluation of the optic nerve in glaucoma. Trans Am Ophthalmol Soc 1994;92:589–641 [PMC free article] [PubMed] [Google Scholar]
- 5.Shen SY, Wong TY, Foster PJ, et al. The prevalence and types of glaucoma in malay people: the Singapore Malay Eye Study. Invest Ophthalmol Vis Sci 2008;49:3846–51 [DOI] [PubMed] [Google Scholar]
- 6.Ramakrishnan R, Nirmalan PK, Krishnadas R, et al. Glaucoma in a rural population of southern India: the Aravind Comprehensive Eye Survey. Ophthalmology 2003;110:1484–90 [DOI] [PubMed] [Google Scholar]
- 7.Vijaya L, George R, Paul PG, et al. Prevalence of open-angle glaucoma in a rural south Indian population. Invest Ophthalmol Vis Sci 2005;46:4461–7 [DOI] [PubMed] [Google Scholar]
- 8.Varma R, Ying-Lai M, Francis BA, et al. Prevalence of open-angle glaucoma and ocular hypertension in Latinos: the Los Angeles Latino Eye Study. Ophthalmology 2004;111:1439–48 [DOI] [PubMed] [Google Scholar]
- 9.de Voogd S, Ikram MK, Wolfs RC, et al. Incidence of open-angle glaucoma in a general elderly population: the Rotterdam Study. Ophthalmology 2005;112:1487–93 [DOI] [PubMed] [Google Scholar]
- 10.Tielsch JM, Sommer A, Katz J, et al. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey. JAMA 1991;266:369–74 [PubMed] [Google Scholar]
- 11.American Academy of Ophthalmology. Comprehensive adult medical eye evaluation, preferred practice pattern. San Francisco: American Academy of Ophthalmology, 2010 [Google Scholar]
- 12.Bartz-Schmidt KU, Thumann G, Jonescu-Cuypers CP, et al. Quantitative morphologic and functional evaluation of the optic nerve head in chronic open-angle glaucoma. Surv Ophthalmol 1999;44(Suppl. 1):S41–53 [DOI] [PubMed] [Google Scholar]
- 13.Zeeberg BR, Feng W, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol 2003;4:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon MO, Torri V, Miglior S, et al. Validated prediction model for the development of primary open-angle glaucoma in individuals with ocular hypertension. Ophthalmology 2007;114:10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattenhauer MG, Johnson DH, Ing HH, et al. The probability of blindness from open-angle glaucoma. Ophthalmology 1998;105:2099–104 [DOI] [PubMed] [Google Scholar]
- 16.Jonas JB, Budde WM, Panda-Jonas S. Ophthalmoscopic evaluation of the optic nerve head. Surv Ophthalmol 1999;43:293–320 [DOI] [PubMed] [Google Scholar]
- 17.Sommer A, Tielsch JM, Katz J, et al. Relationship between intraocular pressure and primary open angle glaucoma among white and black Americans. The Baltimore Eye Survey. Arch Ophthalmol 1991;109:1090–5 [DOI] [PubMed] [Google Scholar]
- 18.Shiose Y. Intraocular pressure: new perspectives. Surv Ophthalmol 1990;34:413–35. [DOI] [PubMed] [Google Scholar]
- 19.Fleming C, Whitlock EP, Beil T, et al. Screening for primary open-angle glaucoma in the primary care setting. Ann Fam Med 2005;3:167–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vithana EN, Khor CC, Qiao C, et al. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat Genet 2012;44:1142–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong DW, Liu J, Lim JH, et al. Intelligent fusion of cup-to-disc ratio determination methods for glaucoma detection in ARGALI. 31st Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Minneapolis, Minnesota, USA: IEEE 2009;5777–80 [DOI] [PubMed] [Google Scholar]
- 22.Ahonen T, Hadid A, Pietikainen M. Face description with local binary patterns: application to face recognition. IEEE Trans Pattern Anal Mach Intell 2006;28:2037–41 [DOI] [PubMed] [Google Scholar]
- 23.Stricker M, Orengo M. Similarity of color images. In: SPIE Conference on Storage and Retrieval for Image and Video Databases III, San Diego/La Jolla, CA, USA: SPIE 1995;2420:381–92 [Google Scholar]
- 24.Gonzalez RC, Woods RE. Digital image processing. 2nd edn. Boston, MA, USA: Addison-Wesley Longman Publishing Co., Inc., 1992 [Google Scholar]
- 25.Boser BE, Guyon I, Vapnik V. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, New York, USA: ACM 1992;144–52 [Google Scholar]
- 26.Cortes C, Vapnik V. Support-vector network. Machine Learning 1995;20:273–97 [Google Scholar]
- 27.Bach F, Lanckriet G, Jordan M. Multiple kernel learning, conic duality, and the SMO algorithm. International conference on Machine learning. New York, NY, USA: ACM 2004;41–48 [Google Scholar]
- 28.Fan RE, Chen PH, Lin CJ. Working set selection using the second order information for training SVM. J Mach Learn Res 2005;6:1889–918 [Google Scholar]
- 29.Rakotomamonjy A, Bach FR, Canu S, et al. SimpleMKL. J Mach Learn Res 2008;9:2491–524 [Google Scholar]
- 30.Xu Z, Jin R, Yang H, et al. Simple and efficient multiple kernel learning by group Lasso. 27th International Conference on Machine Learning. Haifa, Israel, 2010 [Google Scholar]
- 31.Fan RE, Chang KW, Hsieh CJ, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res 2008;9:1871–4 [Google Scholar]
- 32.Rosenberg LH, Franzén B, Auer G, et al. Multivariate meta-analysis of proteomics data from human prostate and colon tumours. BMC Bioinformatics 2010;11:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 2008;148:839–43 [DOI] [PubMed] [Google Scholar]