Abstract
Objective
With the increased routine use of advanced imaging in clinical diagnosis and treatment, it has become imperative to provide patients with a means to view and understand their imaging studies. We illustrate the feasibility of a patient portal that automatically structures and integrates radiology reports with corresponding imaging studies according to several information orientations tailored for the layperson.
Methods
The imaging patient portal is composed of an image processing module for the creation of a timeline that illustrates the progression of disease, a natural language processing module to extract salient concepts from radiology reports (73% accuracy, F1 score of 0.67), and an interactive user interface navigable by an imaging findings list. The portal was developed as a Java-based web application and is demonstrated for patients with brain cancer.
Results and discussion
The system was exhibited at an international radiology conference to solicit feedback from a diverse group of healthcare professionals. There was wide support for educating patients about their imaging studies, and an appreciation for the informatics tools used to simplify images and reports for consumer interpretation. Primary concerns included the possibility of patients misunderstanding their results, as well as worries regarding accidental improper disclosure of medical information.
Conclusions
Radiologic imaging composes a significant amount of the evidence used to make diagnostic and treatment decisions, yet there are few tools for explaining this information to patients. The proposed radiology patient portal provides a framework for organizing radiologic results into several information orientations to support patient education.
Keywords: patient education, radiology, natural language processing, computer assisted image processing
Introduction
The number of patients accessing health information online continues to rise,1 and being diagnosed with cancer has been shown to increase the amount of time an individual searches for information.2–4 However, the popularity of a website is not always indicative of its quality.5 The dearth of quality material online is reflected in the Health Information National Trends Survey (HINTS), which found that Americans feel that online cancer information is inadequate. Of those surveyed, 69% did not have a website they especially liked for cancer information, emphasizing the need for trusted information resources.6 With the quality of sources in question, patients thus often bring up information they find online with their doctor; one study found that up to 90% of respondents who look up health information online verified it with their physicians.7
Several benefits of tailored information within patient portal applications have been demonstrated,8–10 including equipping patients with vetted, higher quality information regarding their disease or condition; and facilitating access to their underlying medical records. However, little work has been done to make the full range of radiology content—imaging and text—available to patients in an understandable format. This lack is in spite of the fact that radiology reports and images constitute a significant amount of the evidence used in diagnosis and treatment assessment. Even though radiology test results are one of the most difficult portions of the clinical record for lay people to understand,11 they are one of the most frequently accessed pieces of information via patient portals when available.12 This suggests the need for new methods of sharing radiology information with patients.
One possibility for bridging consumers’ understanding of illness with professional disease models is the use of an ‘interpretive layer’ between clinically-generated information and consumer-centric disease explanations. Such a layer would potentially enable lay patients to construct more accurate mental models of health, form effective search queries, navigate medical information systems, understand the information found within health documents, and apply the information to their personal situations appropriately.13 In this work we utilize the concept of interpretive layers, and describe a methodology for automatically combining radiology data with educational information for the patient, presented through a web-accessible portal.
Background and significance
Towards satisfying patients’ wishes for access to records and better knowledge resources, government policy has been developed to provide incentives for institutions utilizing patient portals in order to promote usage.14 The US Department of Health and Human Services believes that such portals will not only increase patient access to information, but allow patients to become more active in their care. This sentiment is also reflected in a recent Institute of Medicine Report, which emphasizes the importance of patient portals in a continuously learning healthcare system.15 With this additional motivation, patient portal deployment and use is expected to become commonplace.16 In point of fact, the Health Level 7 (HL7) International Context-Aware Knowledge Retrieval standard now provides a technical specification for integrating electronic health records and personal health records with external information resources, and is increasingly being adopted by vendors and information providers.17–19
Previous studies have found that despite the rising tendency of patients to search for and access health information online, they are often discouraged by the information they find as it is frequently too general to elucidate the specifics of an individual's disease or treatment.1 2 5–7 Notably, patients’ information needs are not limited to general knowledge, but also encompass access to their underlying medical records and the content within them.20 Indeed, receiving (accurate) information relevant to one's cancer diagnosis has been shown to increase patient involvement in decision-making,8 and to enhance satisfaction with treatment options.9 Additionally, giving patients access to personalized health information can improve communication between family members, and between patients and providers.9 10 The latter is especially important as it has been estimated that patients remember approximately only half of the information presented in a conversation with their physician.21
Prior work shows that patient-oriented language is preferred by patients when receiving abnormal radiology results,22 but professional tools to explain medical concepts use expert language, much of which patients do not understand.23 As such, patients who do request copies of radiology reports and images generally receive this information with little or no additional explanatory material, and turn to their healthcare providers for explanations. This scenario is sub-optimal in that some of the resultant questions could be answered with a suitable online information resource. Also, such an information resource could be adapted to the specifics of a patient's case, providing targeted details and lessening the cognitive burden on the patient to reconcile the content of his medical report with generalized information resources designed for a broad spectrum of patients (eg, search engines, MedlinePlus, WebMD). For example, prior research indicates that presenting medical information to patients accompanied by pictures can increase attention, recall, and comprehension of medical concepts.24 25 This observation suggests that showing patients illustrations of imaging or disease concepts specifically related to their studies may provide them with an appreciation of the (causative) reasoning between their symptoms/sequelae and required treatments. While efforts exist to create interfaces that support sharing radiologic imaging across healthcare providers,26 current solutions are not designed specifically to educate patients.27 28
Methods
We implemented an electronic portal for patients with primary brain tumors (eg, gliomas, meningiomas, etc.), a population associated with a high degree of information needs and large amounts of initial and follow-up radiologic imaging. Our system includes explanatory layers of information between the patient and the source clinical data, with each layer offering a lay explanation and overview of the layer immediately below, forming a hierarchy of progressively more specific information views that ultimately link to individual source reports’ findings and associated imaging studies. These layers help to mediate between professional and patient health perspectives, using concepts, illustrations, and key radiology images designed for a consumer audience. Similar notions of augmenting medical information have been discussed previously in the literature.29 30The portal utilizes several information orientations, some of which have been previously explored in the literature, including: a problem orientation to summarize findings in radiologic interpretations31 32; a temporal orientation that shows the evolution of disease via imaging; and a source orientation that allows patients to review an annotated version of their radiology reports.33–36 These three perspectives allow a user to navigate their radiologic information, allowing for the selective drilling down to the original image interpretations.
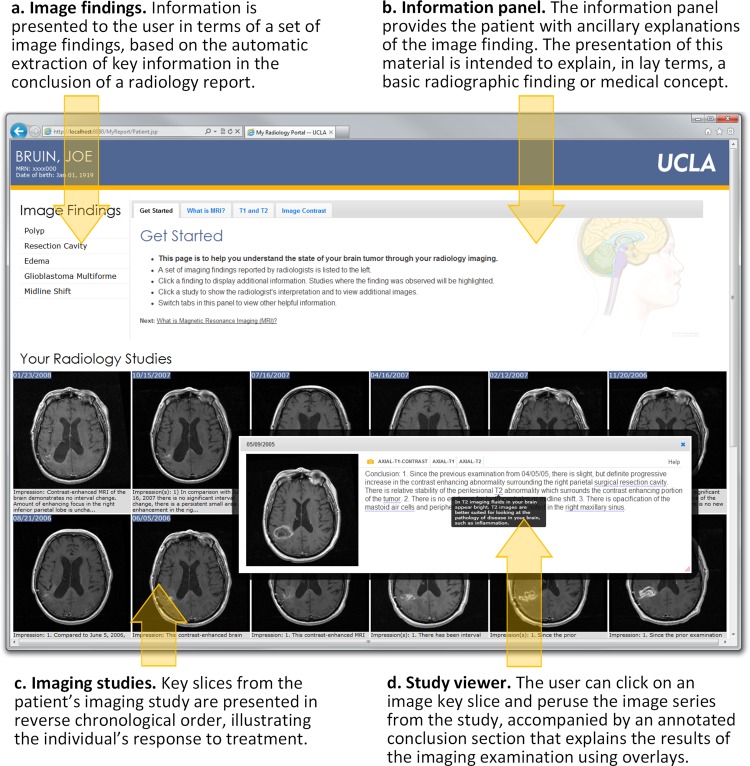
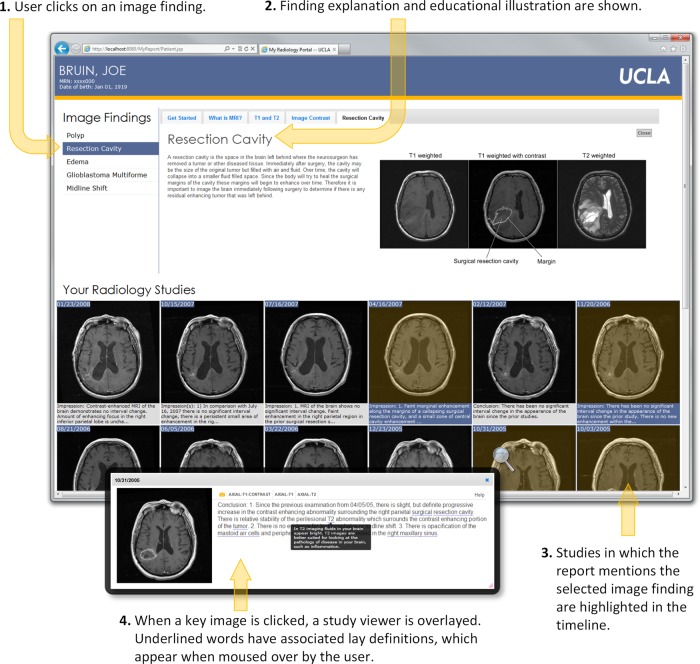
Figure 1 shows the four main components of our radiology portal interface: (1) a panel showing a patient's ‘salient’ imaging findings, organized in reverse chronological order (figure 1A); (2) an information panel providing patient-oriented explanations of imaging techniques, disease concepts, and salient image findings (figure 1B); (3) an interactive panel showing only key slices from patient imaging studies and associated extracted findings from radiology reports (figure 1C); and (4) a study viewer displaying the full image series with an annotated conclusion section from the corresponding report (figure 1D). Interactions with the portal are designed to be driven by the imaging findings list. From the list, a user may click on a finding of interest, which triggers the information panel to display a lay description of the finding with an annotated illustration. In addition, clicking a finding ‘activates’ imaging studies in a patient's record by graphically highlighting studies where the finding was noted by the radiologist. At any point, a user may click on a key slice from an imaging study to launch the study viewer.
Figure 1.
Annotated screenshot of the web-based patient radiology portal showing the different components of the application. (A) The imaging findings list displays salient concepts extracted from the conclusion section of radiology reports (eg, ‘edema’). (B) The information panel is used to display explanations of imaging findings and imaging techniques (eg, ‘What is edema?’ or ‘What is MRI?’). (C) The same key slice from each imaging study is displayed chronologically with the conclusion section from the corresponding radiology report to illustrate the individual's response to treatment. (D) The interactive study viewer allows the user to view entire image series (ie, all slices) within the study and to read the conclusion section of the radiology report where complex terms (underlined words) have been annotated with mouse-over definitions (dark box overlaying the text).
System architecture and components
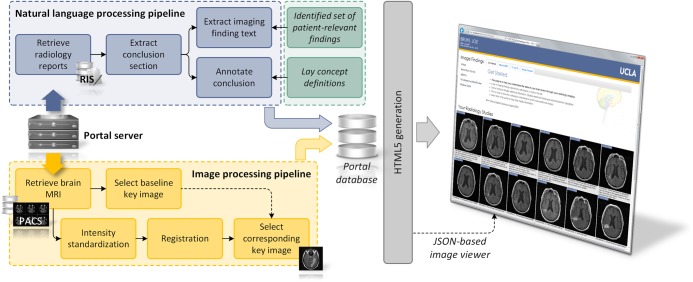
The system architecture is shown in figure 2. Patients seen at the oncology clinic are pre-identified by a clinician, and on request, our portal server fetches the required patient information (images from the institutional picture archive and communications system (PACS); reports from the radiology information system). The application is not intended to make new information available to the patient before prior practitioner–patient communication. Rather, it is meant to review information already disclosed to a patient by his healthcare provider, in order to limit both the stress of encountering new information without professional guidance and that of attempting to recall detailed information after talking with practitioners. The retrieved data is fed into image and natural language processing (NLP) modules, with the resultant analyses stored in a database on the portal server. When a user accesses a given patient portal page, a Java Server Pages application dynamically generates a set of HTML5 views from the raw and analyzed data, and serves up the web-based portal application. The modules and portal components are now described in detail.
Figure 2.
System architecture showing natural language processing and imaging processing modules that generate several information orientations. The portal is implemented as a Java-based web application, with key slices in the imaging timeline shown as JPEGS, and an HTML Canvas image viewer that displays complete series using pixel data passed in a JSON object.
Salient findings panel
The salient findings panel provides patients with a list of pertinent observations made over time and as documented through radiologists’ interpretations. To define salient findings, a superset of candidate concepts was automatically extracted from the conclusion section of neuroradiology reports (brain MRI studies), using the Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES) NLP software operating with the Systemized Nomenclature of Medicine Clinical Terms (SNoMED-CT) terminology.37 Negated concepts (eg, ‘no evidence of hydrocephalus’) were discarded using the cTAKES integrated negation detector, which is based on NegEx.38 Although negated concepts can be particularly important (eg, ‘no edema present’), a design decision was made to focus only on concepts that were observed by the radiologist and therefore visible in the imaging study. In total, 2883brain MRI reports from 277 patients (based on all brain MRI studies and the related radiology report, from all patients) were processed, resulting in the extraction of 448 unique concepts. Although patients have a desire to understand the significance of their radiology findings, they lack the clinical expertise to define the set of specific imaging concepts that fulfills this information need (eg, hydrocephalus, midline shift, necrosis, etc). Therefore, the selection of the concept subset was performed with guidance from: (1) clinical experts, who have experience answering patient questions; and (2) literature, which indicates that patients are concerned with understanding their different diagnoses (eg, glioblastoma), procedures (eg, craniotomy), and symptoms (eg, edema).39–41 First, the investigators removed a large number of concepts that were detected as a result of a radiologist's mention of a patient's historical disease or co-morbidity (ie, concepts without a visual representation in corresponding imaging) from the set. Next, manual reconciliation of concepts that would be considered synonymous by a patient (eg, ‘malignant neoplastic disease’ and ‘malignant neoplasm of brain’) was conducted, as was the removal of erroneous concepts from the set (eg, syncope, as in ‘to faint’, is often mapped, when a radiologist mentions ‘faint contrast enhancement’). This process resulted in 52 terms that comprised the final set of salient finding concepts, which were stored in a lookup table. The 15 most common concepts from this set are given in table 1. Using the NLP module (see figure 2), concepts automatically extracted from a patient's conclusion section were referenced in the lookup table; matches were retrieved and sorted in reverse chronological order for display in the salient findings panel.
Table 1.
Fifteen most frequent concepts extracted from the conclusion sections of radiology reports
| UMLS Concept Unique Identifier (CUI) | Term |
|---|---|
| C0027651, C0006118 | Brain neoplasms |
| C0728940 | Excision |
| C0013604 | Edema |
| C0010280 | Craniotomy |
| C1510420, C0333343, C1515091 | Surgical resection cavity |
| C1627358 | Contrast enhancement |
| C0017636 | Glioblastoma |
| C0229985 | Surgical margins |
| C0027540 | Necrosis |
| C0543478 | Residual tumor |
| C0019080 | Hemorrhage |
| C0020255 | Hydrocephalus |
| C0576481 | Midline shift |
| C0020564 | Hypertrophy |
| C0178874F | Tumor progression |
Given the context of brain cancer imaging, some concepts were combined and presented to the user in synonymous fashion.
UMLS, Unified Medical Language System.
To evaluate the automatic extraction of salient findings, we generated a gold standard set and compared it to results from the NLP module. First, two annotators (the first and second author) jointly annotated a random sample of 50 radiology conclusion sections from our dataset under the guidance of a neuroradiologist (the third author). Next, the annotators separately annotated 150 impression sections, which had an average length of 85 words. Following Hripcsak and Rothschild42 and Fleiss,43 we calculated the positive specific agreement between the annotators to be 0.91, where the annotated spans for a term were required to overlap. Discordances were adjudicated by a neuroradiologist to generate the final gold standard. In total, 684 instances of the 52 salient terms were identified. Using this gold standard, the NLP module achieved an accuracy of 73% at identifying mentions of a concept, with an F1 score of 0.67 (precision 0.63, recall 0.73).44
Information panel
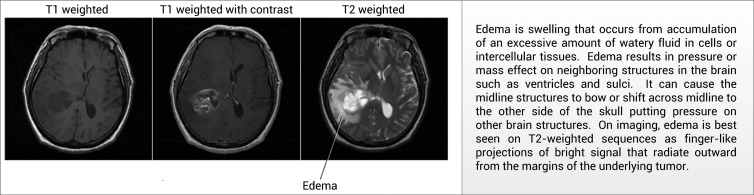
Using a combination of graphical pictures and text, the information panel explains the salient concepts detected by NLP in the conclusion section of radiology reports. We first attempted to employ existing concept explanations from the National Institutes of Health (NIH) MedlinePlus repository. However, we found these definitions, as well as those present in the Unified Medical Language System, to be too generic for use in this application. For example, when extracted from a brain magnetic resonance (MR) report from a patient with brain cancer, there is a high degree of certainty as to the meaning of a radiologist's mention of ‘edema’ (ie, excess accumulation of water in the brain). Therefore, instead of using a general definition of edema, we present a more specific explanation to support the patient in understanding the relative importance and context of cerebral edema. These explanations were developed by our clinical investigators based on their experience in providing such clarifications to patients in real life. When available, explanations were augmented with information from existing resources, such as the patient version of definitions from the National Cancer Institute. Figure 3 shows an example explanation with an accompanying pictorial illustration.
Figure 3.
An example definition for the disease finding ‘edema’ used by the system, including textual and visual descriptions of the concept.
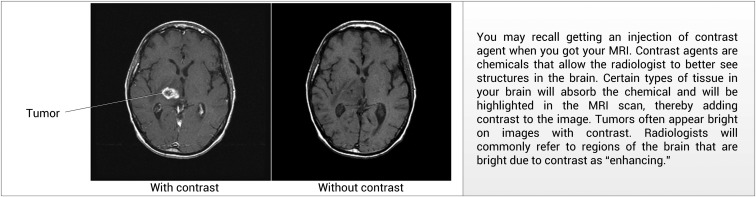
In addition to providing descriptions of disease concepts, the information panel contains explanations of MRI pulse sequences and contrast agents with the goal of helping patients understand why a given imaging study was performed, and how to interpret images in the portal. By way of illustration, figure 4 shows the explanation for the function of contrast agents in MRI brain tumor imaging.
Figure 4.
An example definition for the imaging technique of ‘contrast’ used by the portal, including textual and visual descriptions of the concept.
Imaging studies panel
For oncology patients, imaging is frequently used to assess response to treatment. For neuro-oncology, MR is the predominant modality, given its ability to highlight key pathophysiology concomitant to the tumor's progression/regression. Multiple variations of MR sequences are performed, with each series acquired to provide unique evidence on the state of a tumor: T1-weighted imaging highlights anatomy and therefore provides a good view of a tumor's structure; whereas T2-weighted images highlight water and are useful for observing edema surrounding a tumor (and potential fluid accumulation indicating possible increasing intracranial pressure). Additional series, such as apparent diffusion coefficient maps (used for monitoring the diffusion of water within a tumor), may also be acquired, but are not performed with the same regularity.
At our institution, each brain cancer patient typically receives a T1-weighted scan; a T1-weighted scan with contrast; a T2-weighted scan; and a FLAIR (fluid attenuated inversion recovery) or PD (proton density) scan (FLAIR and PD scans are used to look for lesions and edema proximal to brain ventricles). These scans are conducted every 4–6 weeks while receiving chemotherapy, and then every few months if the cancer is in remission. The frequency of imaging and the fact that many medical decisions are predicated on imaging results makes patients naturally curious as to what their images mean. However, clinical MRI data is not ideal for unsupervised presentation to patients. First, it exists in DICOM (Digital Imaging and Communications in Medicine) format, which cannot be easily viewed without specialized software. Next, scans are acquired as two-dimensional slices and viewed in stacks that must be scrolled through to build a mental three-dimensional view of the brain. Patients are not accustomed to interpreting images in this manner; nor will they have sufficient knowledge to comprehend the neuro-anatomy seen in such studies. Finally, as scans are acquired at different times, there are variations across studies. For instance, patients’ heads may be tilted at different angles in the scanner, the field of view may change, pixel intensity may vary, and the number of slices in a study is often different. Ultimately, all of these factors confound the non-expert in viewing and comparing imaging studies.
Following our paradigm of creating explanatory layers around such complex clinical data, we developed an image processing module that generates key slices, which are displayed over time in a single view (figure 1C). The process works as follows:
Key slice selection: When an individual is first added to the patient portal system, one axial reference study is chosen by a radiologist, from which a key slice is selected. The key slice reflects as much of the radiologist's description as possible. For example, if a patient has a tumor with midline shift (movement of the brain across the sagittal plane as a result of the space-occupying tumor), a key slice containing the tumor and the shifted brain ventricles will be selected. Typically, a key diagnostic or post-surgical resection study will be chosen. This key slice and study serves as a baseline reference point for subsequent imaging studies. In our dataset, this process took approximately 3 min for a neuroradiologist to complete.
Subsequent image normalization: When a new imaging study is performed, it is retrieved from the institution's PACS and automatically intensity standardized and registered to the reference study selected by the neuroradiologist. Intensity standardization is performed using histogram matching.45 Intra-subject registration is performed using a rigid transformation with nine degrees of freedom (three rotations, three translations, three scalings) using the FLIRT (Functional MRI of the Brain Linear Image Registration Tool) package from FSL (Functional MRI of the Brain Software Library).46 This step aligns anatomy across studies, ensuring that the key slice selected by the radiologist will match the same anatomical slice in other studies, thereby automatically selecting the same key slice in all studies. The normalization process requires approximately 1 min per study, with preprocessed results stored on the portal server for subsequent presentation.
Key slice layout: The normalized key slices are then displayed on a timeline, side-by-side. This layout provides a view of changes occurring within the brain as the result of disease progression and treatment.
The imaging studies panel presents these key slices accompanied by the conclusion section of the corresponding radiology report, allowing a user to track changes visually and through the narrative of the radiologist. Figure 5 presents a sample result from this processing module illustrating the collapse of a resection cavity over time.
Figure 5.
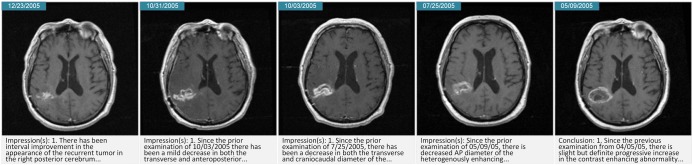
Key slices created by the imaging pipeline showing the temporal evolution of disease. In this example, a surgical resection cavity is collapsing over time, with the most recent image on the left.
Figure 6.
Portal interaction illustrating the use of the image findings list to drive exploration of the radiology information. (1) Imaging findings may be clicked to display a definition in the information panel (2) and highlight relevant imaging studies (3). When a key image is clicked, the study viewer (4) is displayed.
Study viewer
The study viewer allows a user to peruse the original series data from an imaging study alongside further educational materials specific to the conclusion section of the radiology report. The materials presented in this panel introduce concepts that are not necessarily considered relevant in the specific context of a patient's salient imaging findings list (eg, ‘lateral ventricles’), but as they are core medical concepts that are not generally known by laypersons, they require explanation. We first tried to automatically translate the source text to a summary that a patient could understand. Necessarily, this approach required abstraction and simplification, as it is not feasible to define such concepts to the same degree to which they are understood by clinicians. The Open-Access and Collaborative Consumer Health Vocabulary (OAC-CHV) was used to classify concepts found by cTAKES as ‘complex’ terms, which were then replaced with OAC-CHV lay definitions. Following Zeng-Treitler et al,33 terms with a combination familiarity score lower than 0.6 in OAC-CHV were deemed to be unfamiliar to the lay reader and were replaced. This approach had several limitations. Though OAC-CHV is under continual development, it does not contain entries for many neuroradiology concepts (eg, ‘midline shift’) and includes many terms without lay explanations that have uninformative complexity scores (eg, ‘vasogenic cerebral edema’); this latter point is likely due to the scarcity of the concept in the corpus of query logs used to estimate complexity in the OAC-CHV.47 Also, as a result of incorrect term detection by NLP or improper lay definition insertion (eg, incorrectly matching the tense of a sentence), we observed that the automatic translation system introduced undesirable errors, if not misunderstandings. We therefore felt it was more appropriate to leave the radiology conclusion unmodified and instead focused on supplementing it with lay neuroradiology definitions. Similar to the creation of the salient image finding concept set, a set of concepts was defined to augment the conclusion section by manual review, a process that resulted in 247 concepts. Definitions for these concepts were written by the clinical investigators with a consumer audience in mind, an approach followed in the creation of the OAC-CHV. This process resulted in a repository of explanations that are available to the portal to support the conclusion section of a radiology report when a pertinent concept is identified by the NLP module.
Discussion
In contrast to previous work in patient portal development, which focuses on sharing text reports, medications, and laboratory results,12 31 32 48 49 our proposed portal displays and attempts to explain both radiology imaging and reports, information that is known to be difficult for patients to comprehend.11 The presented portal view for neuro-oncology is just one way information can be augmented to provide a (lay) patient with additional context pertinent to a particular disease. In its most general form, this developed radiology portal framework can be used across a multitude of diseases and anatomies. For instance, a view for lung cancer could also apply image registration to CT images to show changes related to disease progression, and the effects of interventions over time. One possible addition to this imaging-centric view is the integration of treatment information (eg, chemotherapy) concurrent with the imaging timeline, providing a clearer picture of treatment effects as observed via radiologic imaging.
Our preliminary results indicate the feasibility of the NLP module at identifying salient terms; however the problems of using cTAKES’ annotators without customization are evident in our performance metrics. Common errors included the selection of findings that are not negated, but are also not present in the image. Representative instances of this type of error include, ‘…following neurosurgical resection of the left lateral posterior frontal lobe mass…,’ and, ‘the pneumocephalus as well as the layer of acute blood previously seen within the resection cavity have resolved’. In the first example the mass has been resected and therefore will not appear in the imaging. Similarly, in the second example pneumocephalus and acute blood have resolved from the resection cavity and therefore will not be visualized. Such expressions illustrate the complexity of identifying the findings from a radiology report that were actually observed in the corresponding imaging study by the radiologist, and suggest the need for training contextual sequence models of both words and reports over time. Further supporting the need for temporal models, there are few standards for reporting in the domain of neuroradiology and therefore a patient may observe inconsistencies in the coverage of concepts across reports over time due to contrasting styles between radiologists, or reports that describe only incremental changes, rather than all findings. A sufficiently robust temporal model may be able to recognize such gaps in concept coverage and interpolate the missing findings for the patient. Our future work includes exploring the integration of domain-specific ontologies (eg, RadLex50) to improve the identification of findings and anatomical concepts, and creating a gold standard to evaluate the performance of the NLP module at identifying terms for explanation in the study viewer. Additionally, assessing the readability of the clinician-generated definitions is a future goal, with previous work in the area using cloze scoring and metrics specific to health-related content.51 52 Finally, we note that as the NLP module targets only those concepts that have been manually validated, it is possible that relevant concepts in unseen radiology conclusion sections may be missed. This problem is mitigated by having a large dataset on which to base concept selection, and may be further minimized by performing regular updates. However, if applied at a new institution, a comprehensive review of radiology reports would likely be necessary to account for institution-specific reporting practices. To receive feedback from a diverse group of healthcare professionals, the system was exhibited at a demonstration booth at the Radiological Society of North America 2011 annual meeting. The application and its use were presented, with the authors available for support. Many people recognized the portal's ability to educate a patient on their disease state through their record, provide a means by which a patient may review diagnosis and treatment history, and allow a patient to share their record with family members and other supporting individuals. However, there was also concern that despite the mechanisms for structuring and explaining, the application thrusts information onto the patient that may be past the average individual's comprehension. Indeed, there are documented ‘mismatches’ between lay and professional definitions of terms13 and furthermore, the language and concepts used by patients is reflective of their ‘cultural, social, and experiential knowledge’.53 Thus, no augmentation can allot for and correct all patient misconceptions. Ensuing misunderstandings could in turn result in extra work for the physician, who would be left with the burden of answering questions that would have not otherwise arisen. Such viewpoints resulted in the suggestion that the application be used as a tool during office visits, allowing for practitioners to be present while patients viewed the content in order to provide information support.
These critiques and comments were taken under consideration. The application is intended to facilitate the sharing and explanation of radiology reports, providing users with new information in the form of increased comprehension of their medical imaging procedures and results. However, the portal is not designed to convey information beyond this scope (eg, there is no personalized prognostic information offered). Furthermore, the system is designed to release test results only after approval by a physician. For instance, a neuro-oncologist may control when a patient has access to a study through the portal, which is likely only after an in-person clinic consultation. And while steps can be taken to prevent issues regarding patients misunderstanding and clinician workload increases, the potential for these issues cannot be completely eliminated.11 24 54 55 There are, and will always be, risks in allowing patients direct access with their records, but evidence indicates that these potential risks are outweighed by the benefits provided by such a system to an engaged patient.9 10 33 56
Conclusion
As part of having a health-literate patient population that is engaged and informed in its own care, it is of growing importance to establish educational portals that deliver customized, understandable radiology content to patients. Despite concerns, studies measuring the impact of patient portals have not found an increase in consumer misconceptions about health information.57 58 In contrast, patients who reviewed their data via a portal reported that it led them to more accurate information and better prepared them for upcoming clinical visits,32 as well as made them more able to cope with the anxiety associated with diagnoses when they receive information on disease progression and treatment.10 48 49 56 Moreover, increased access to information for cancer patients, including their medical records, has been shown to increase satisfaction with treatment choices, increase confidence in care providers, and improve adherence to medical advice.48 59 Portal access also allows the consumer to consolidate information that has historically been dispersed across sources, an important concern in medical imaging.60 The presented system offers a novel solution to sharing radiology information with consumers, and is driven by imaging informatics tools to transform clinically-generated information into educational views that may be customized by patient and disease.
Footnotes
Contributors: CWA and MM were responsible for all aspects of this work, including conceptualization, design, implementation, and evaluation. SE-S, RKT, and AATB performed conceptualization and design. SC performed implementation and evaluation. CWA, MM, SE-S, and AATB participated in the writing and editing of the manuscript.
Funding: This work was supported by NIH/NLM grant number R01 LM011333 and NIH/NCI grant number R01 CA1575533.
Competing interests: None.
Ethics approval: UCLA IRB.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Pew Internet and American Life Project. The Social Life of Health Information 2009 [Google Scholar]
- 2.Bass S, Ruzek S, Gordon TF, et al. The relationship of internet health information use with patient behavior and self efficacy: experiences of newly diagnosed cancer patients who contact the National Cancer Institute's Cancer Information Service. J Health Commun 2006;11:219–36 [DOI] [PubMed] [Google Scholar]
- 3.Castleton K, Fong T, Wang-Gillam A, et al. A survey of internet utilization among patients with cancer. Support Care Cancer 2011;19:1183–90 [DOI] [PubMed] [Google Scholar]
- 4.Metz JM, Devine P, DeNittis A, et al. A multi-institutional study of internet utilization by radiation oncology patients. Int J Radiat Oncol Biol Physiol 2003;56:1201–5 [DOI] [PubMed] [Google Scholar]
- 5.Meric F, Bernstam EV, Mirza NQ, et al. Breast cancer on the world wide web: cross sectional survey of quality of information and popularity of websites. BMJ 2002;324:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Cancer Institute. Cancer Communication Health Information National Trends Survey 2003 and 2005. 2007; NIH Publication No. 07-6214
- 7.Schwartz KL, Roe T, Northrup J, et al. Family medicine patients’ use of the internet for health information: a MetroNet Study. J Am Board Fam Med 2006;19:39–45 [DOI] [PubMed] [Google Scholar]
- 8.Luker KA, Beaver K, Leinster SJ, et al. The information needs of women newly diagnosed with breast cancer. J Adv Nurs 1995;22:134–41 [DOI] [PubMed] [Google Scholar]
- 9.Cawley M, Kostic J, Cappello C. Informational and psychosocial needs of women choosing conservative surgery/primary radiation for early stage breast cancer. Cancer Nurs 1990;13:90–4 [PubMed] [Google Scholar]
- 10.Gravis G, Protiere C, Eisinger F, et al. Full access to medical records does not modify anxiety in cancer patients: results of a randomized study. Cancer 2011;117:4796–804 [DOI] [PubMed] [Google Scholar]
- 11.Keselman A, Slaughter L, Arnott-Smith C, et al. editors. Towards consumer-friendly PHRs: patients’ experience with reviewing their health records. American Medical Informatics Association (AMIA) Annual Symposium Proceedings 2007. 2007;399–403 [PMC free article] [PubMed] [Google Scholar]
- 12.Weingart SN, Rind D, Tofias Z, et al. Who uses the patient internet portal? The PatientSite experience. J Am Med Assoc 2006;13:91–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soergel D, Tse T, Slaughter L. Helping healthcare consumers understand: an ‘interpretive layer’ for finding and making sense of medical information. Int Med Inform Assoc Medinfo Proc 2004;11(Pt 2):931–5 [PubMed] [Google Scholar]
- 14.Office of the National Coordinator for Health Information Technology Electronic health records and meaningful use. 2012. http://healthit.hhs.gov/portal/server.pt?open=512&objID=2996&mode=2
- 15.IOM (Institute of Medicine) Best care at lower cost: the path to continuously learning health care in America Washington, DC: National Academies Press, 2012 [Google Scholar]
- 16.Koch-Weser S, Bradshaw YS, Gualtieri L, et al. The internet as a health information source: findings from the 2007 Health Information National Trends Survey and implications for health communication. J Health Commun 2010;15:279–93 [DOI] [PubMed] [Google Scholar]
- 17.Kemper D, Del Fiol G, Hall L, et al. Getting patients to meaningful use: using the HL7 infobutton standard for information prescriptions. Healthwise, 2010
- 18.Fiol GD, Huser V, Strasberg HR, et al. Implementations of the HL7 context-aware knowledge retrieval (“Infobutton”) standard: challenges, strengths, limitations, and uptake. J Biomed Inform 2012;45:726–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma W, Dennis S, Lanka S, et al. MedlinePlus connect: linking health IT systems to consumer health information. IT Professional 2012;14:22–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckjord EB, Rechis R, Nutt S, et al. What do people affected by cancer think about electronic health information exchange? Results from the 2010 LIVESTRONG Electronic Health Information Exchange Survey and the 2008 Health Information National Trends Survey. J Oncol Pract 2011;7:237–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley P, Whitworth MA, Skilbeck CE, et al. Improving doctor-patient communication in general practice. J Royal Coll Gen Pract 1976;26:720–4 [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson AJ, Easterling D, Nelson R, et al. Access to radiologic reports via a patient portal: clinical simulations to investigate patient preferences. J Am Coll Radiol 2012;9:256–63 [DOI] [PubMed] [Google Scholar]
- 23.Zielstorff RD. Controlled vocabularies for consumer health. J Biomed Inform 2003;36:326–33 [DOI] [PubMed] [Google Scholar]
- 24.Zeng-Treitler Q, Kim H, Hunter M, edis. Improving patient comprehension and recall of discharge instructions by supplementing free texts with pictographs. American Medical Informatics Association (AMIA) Annual Symposium Proceedings 2008; 2008:849–53 [PMC free article] [PubMed] [Google Scholar]
- 25.Houts PS, Doak CC, Doak LG, et al. The role of pictures in improving health communication: a review of research on attention, comprehension, recall, and adherence. Patient Educ Couns 2006;61:173–90 [DOI] [PubMed] [Google Scholar]
- 26.RSNA Image Share Network Reaches First Patients. http://www.rsna.org/NewsDetail.aspx?id=2409.
- 27.lifeIMAGE. https://cloud.lifeimage.com.
- 28.Dell Unified Clinical Archive. http://content.dell.com/us/en/healthcare/healthcare-medical-archiving-unified-clinical-archive.aspx.
- 29.Miller T, Leroy G, Wood E, eds Dynamic generation of a table of contents with consumer-friendly labels. American Medical Informatics Association (AMIA) Annual Symposium Proceedings 2006; 2006:559–63 [PMC free article] [PubMed] [Google Scholar]
- 30.Maddock C, Camporesi S, Lewis I, et al. Online information as a decision making aid for cancer patients: recommendations from the Eurocancercoms project. Eur J Cancer 2011;48:1055–9 [DOI] [PubMed] [Google Scholar]
- 31.Grant RW, Wald JS, Poon EG, et al. Design and implementation of a web-based patient portal linked to an ambulatory care electronic health record: patient gateway for diabetes collaborative care. Diabetes Technol Ther 2006;8:576–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnipper JL, Gandhi TK, Wald JS, et al. Design and implementation of a web-based patient portal linked to an electronic health record designed to improve medication safety: the patient gateway medications module. Inform Prim Care 2008;16:147–55 [DOI] [PubMed] [Google Scholar]
- 33.Zeng-Treitler Q, Goryachev S, Kim H, et al. Making texts in electronic health records comprehensible to consumers: a prototype translator. AMIA Annual Symposium Proceedings. American Medical Informatics Association, 2007 [PMC free article] [PubMed] [Google Scholar]
- 34.Cardillo E, Tamilin A, Serafini L. A methodology for knowledge acquisition in consumer-oriented healthcare. Knowledge discovery, knowledge engineering and knowledge management. Springer, 2011:249–61 [Google Scholar]
- 35.Brennan PF, Downs S, Casper G. Project HealthDesign: rethinking the power and potential of personal health records. J Biomed Inform 2010;43:S3–5 [DOI] [PubMed] [Google Scholar]
- 36.Adnan M, Warren J, Orr M. Enhancing patient readability of discharge summaries with automatically generated hyperlinks. Health Care and Informatics Review Online 2009
- 37.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 39.Davidson JR, Brundage MD, Feldman-Stewart D. Lung cancer treatment decisions: patients’ desires for participation and information. Psycho Oncol 1999;8:511–20 [DOI] [PubMed] [Google Scholar]
- 40.Jenkins V, Fallowfield L, Saul J. Information needs of patients with cancer: results from a large study in UK cancer centres. Br J Cancer 2001;84:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leydon GM, Boulton M, Moynihan C, et al. Cancer patients’ information needs and information seeking behaviour: in depth interview study. BMJ 2000;320:909–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005;12:296–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleiss JL. Measuring agreement between two judges on the presence or absence of a trait. Biometrics 1975:651–9 [PubMed] [Google Scholar]
- 44.Manning CD, Raghavan P, Schütze H. Introduction to information retrieval. Cambridge: Cambridge University Press, 2008 [Google Scholar]
- 45.Nyul LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. Med Imaging IEEE Trans 2000;19:143–50 [DOI] [PubMed] [Google Scholar]
- 46.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23:S208–19 [DOI] [PubMed] [Google Scholar]
- 47.Zeng-Treitler Q, Tse T, Crowell J, et al. edn Identifying consumer-friendly display (CFD) names for health concepts. American Medical Informatics Association (AMIA) Annual Symposium Proceedings 2005;2005:859–63 [PMC free article] [PubMed] [Google Scholar]
- 48.Ross SE, Moore LA, Earnest MA, et al. Providing a web-based online medical record with electronic communication capabilities to patients with congestive heart failure: randomized trial. J Med Internet Res 2004;6:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Earnest MA, Ross SE, Wittevrongel L, et al. Use of a patient-accessible electronic medical record in a practice for congestive heart failure: patient and physician experiences. J Am Med Inform Assoc 2004;11:410–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langlotz CP. RadLex: a new method for indexing online educational materials1. RadioGraphics 2006;26:1595–7 [DOI] [PubMed] [Google Scholar]
- 51.Kim H, Goryachev S, Rosemblat G, et al. Beyond surface characteristics: a new health text-specific readability measurement. American Medical Informatics Association (AMIA) Annual Symposium Proceedings 2007; 2007;418–22 [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor WL. “Cloze procedure”: a new tool for measuring readability. Journalism Quarterly 1953;30:415–33. [Google Scholar]
- 53.Keselman A, Logan R, Smith CA, et al. Developing informatics tools and strategies for consumer-centered health communication. J Am Med Inform Assoc 2008;15:473–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross SE, Lin CT. A randomized controlled trial of a patient accessible medical record. American Medical Informatics Association (AMIA) Annual Symposium Proceedings 2003;2003:990. [PMC free article] [PubMed] [Google Scholar]
- 55.Detmer D, Bloomrosen M, Raymond B, et al. Integrated personal health records: transformative tools for consumer-centric care. BMC Med Inform Decis Mak 2008;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meredith C, Symonds P, Webster L, et al. Information needs of cancer patients in West Scotland: cross sectional survey of patients’ views. BMJ 1996;313:724–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker J, Leveille SG, Ngo L, et al. Inviting patients to read their doctors’ notes: patients and doctors look ahead. Ann Intern Med 2011;155:811–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delbanco T, Walker J, Bell SK, et al. Inviting patients to read their doctors’ notes: a quasi-experimental study and a look ahead. Ann Intern Med 2012;157:461–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baldry M, Cheal C, Fisher B, et al. Giving patients their own records in general practice: experience of patients and staff. BMJ (Clinical Res Ed) 1986;292:596–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pratt W, Unruh K, Civan A, et al. Personal health information management: integrating personal health information helps people manage their lives and actively participate in their own health care. Commun ACM 2006;49:51–5 [Google Scholar]