Abstract
Objectives
With the objective of bringing clinical decision support systems to reality, this article reviews histopathological whole-slide imaging informatics methods, associated challenges, and future research opportunities.
Target audience
This review targets pathologists and informaticians who have a limited understanding of the key aspects of whole-slide image (WSI) analysis and/or a limited knowledge of state-of-the-art technologies and analysis methods.
Scope
First, we discuss the importance of imaging informatics in pathology and highlight the challenges posed by histopathological WSI. Next, we provide a thorough review of current methods for: quality control of histopathological images; feature extraction that captures image properties at the pixel, object, and semantic levels; predictive modeling that utilizes image features for diagnostic or prognostic applications; and data and information visualization that explores WSI for de novo discovery. In addition, we highlight future research directions and discuss the impact of large public repositories of histopathological data, such as the Cancer Genome Atlas, on the field of pathology informatics. Following the review, we present a case study to illustrate a clinical decision support system that begins with quality control and ends with predictive modeling for several cancer endpoints. Currently, state-of-the-art software tools only provide limited image processing capabilities instead of complete data analysis for clinical decision-making. We aim to inspire researchers to conduct more research in pathology imaging informatics so that clinical decision support can become a reality.
Keywords: pathology imaging informatics, whole-slide images, computer-aided diagnosis, cancer prediction, decision support systems
Introduction
Pathology imaging informatics refers to the analytical and computational methods for handling, analyzing, and exploring histopathological images and their associated clinical data in order to achieve a medical goal, for example, diagnostic or prognostic applications.1–6 Histopathological analysis is a common clinical procedure for diagnosing the presence, type, and progression of diseases such as cancer. While diagnosing cancer patients using biopsy-derived tissue slides, pathologists manually identify the most progressed regions and examine nuclear morphology, among other tissue and cellular properties. However, manual examination and decision-making using tissue slides that may potentially contain millions of cells can be time-consuming and subjective. Researchers have thus proposed clinical decision support systems (CDSS) and informatics methods that can help in decision-making by objectively quantifying morphological properties in histopathological images. Many of these systems and informatics methods still focus on images that represent only limited, manually selected regions of tissue slides rather than on whole-slide images (WSI).5 By including an element of manual selection in these CDSS, researchers have ensured higher quality and disease-relevant input images while decreasing computational complexity.7 However, manually selected tissue slide regions do not capture the complete information available to pathologists during initial microscopic analysis. Moreover, they are subject to biases related to the knowledge of the pathologist that selected the image regions.7 Therefore, we focus on WSI analysis methods that can potentially maximize the amount of information extracted from tissue slides for decision-making and maximize the objectivity and reproducibility of analysis. In particular, we review methods for quality control, representation of WSI using various types of quantitative image features, predictive modeling, and visualization and exploratory analysis (figure 1). This review is by no means a comprehensive description of WSI informatics. However, compared to recent reviews on WSI informatics4 6 8 that highlight general challenges and applications, we discuss state-of-the-art analytical methods in the key components of WSI-based CDSS.
Figure 1.

An example clinical decision support system for quantitative analysis of whole-slide images (WSI) of tissue biopsy samples. This system has the following key components: quality control to ensure only high-quality data are processed, image description to convert WSI into quantitative features, prediction modeling to develop quantitative diagnostic models, and exploratory analysis to interpret the image feature space. We include two case studies as examples of predictive modeling and exploratory analysis. ROI, region of interest.
The importance of quantitative and objective analysis of tissue biopsy WSI has led to several commercial software tools for WSI analysis including GENIE (Aperio, Vista, California, USA), HALO (Indica Labs, Corrales, New Mexico, USA), AQUA Analysis (HistoRx, Branford, Connecticut, USA), and Visiopharm (Hoersholm, Denmark). However, all of these tools provide limited image processing capabilities. In most cases, pathologists manually select the regions of interest (ROI) and make diagnoses based on feedback from these commercial tools. Usually, an expert user calibrates these systems for each laboratory-specific experimental setup. To the best of our knowledge, none of these tools provides complete data analysis for clinical decision-making that includes all of the steps illustrated in figure 1.
Patient-level prediction modeling and exploratory analysis is important for a number of clinical applications including diagnostics and therapeutics.9 The importance of accurate image-based disease diagnosis and the development of novel pathology informatics techniques has led to the establishment of databases such as the NCI Cooperative Prostate Cancer Tissue Resource,10 the NIH Cancer Genome Atlas (TCGA),11 and the Human Protein Atlas.12 Such databases provide a large number of high-quality histopathological images and associated clinical data, further stimulating the development of novel informatics methods. Some of these databases also provide matched genomic and proteomic data, enabling multimodal studies that associate ‘–omic’ data with histopathological image features. We use WSI from TCGA in a case study to demonstrate a CDSS that identifies and eliminates image artifacts such as tissue folds, extracts image features using piecewise analysis, identifies biologically relevant WSI regions, and combines image features from selected WSI regions to predict several clinical endpoints.
Quality control
The quality of histopathological WSI is usually affected by artifacts acquired during image acquisition and batch effects resulting from variations in experimental protocol. Both of these issues can affect the results of downstream clinical applications. Data quality is especially challenging in collaborative repositories, such as TCGA, where a large amount of high-throughput data is collected from multiple institutions.
Image artifacts
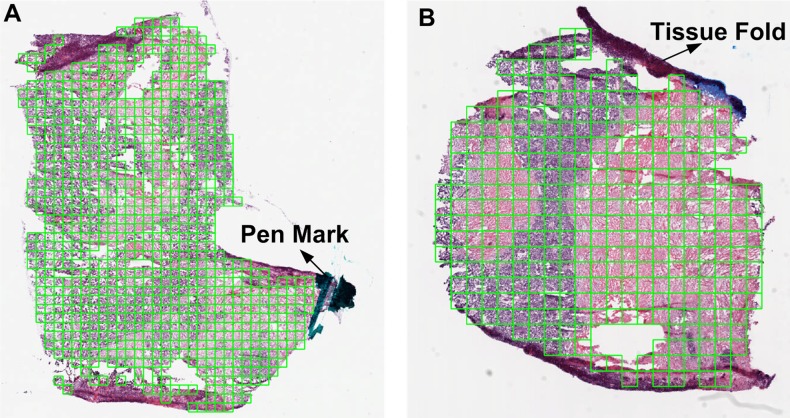
Errors in biopsy slide preparation or in microscope parameters may lead to anomalies, known as image artifacts, in WSI. Common image artifacts include tissue folds, blurred regions, pen marks, shadows, and chromatic aberrations.6 8 Image artifacts have unpredictable effects on image segmentation and other quantitative image features. Therefore, it is essential either to eliminate or correct these artifacts. Tissue-fold artifacts, caused by layering of non-adherent tissue on the slide, can be eliminated using methods based on color saturation and intensity.13–15 Figure 2 illustrates some results for eliminating tissue folds and pen marks in WSI using color properties.13 14 Briefly, we detect tissue folds by using an unsupervised method to cluster the pixels in an image representing the difference between saturation and intensity values for every pixel.14 Because of its unsupervised nature, this method has two limitations: it has low sensitivity for an image with different types of tissue folds and it has low specificity for an image with no tissue folds. Blurred regions, caused by loss of microscope focus, can be detected using a supervised model based on texture properties such as gradient, Laplacian, local grayscale statistics, and wavelet response.16 However, the success of such models depends on good quality annotated data for training. Chromatic aberrations occur when light dispersion through the microscopic lens varies with colors, leading to ghost colors along the edges of objects or discontinuities in an image. Wu et al17 suggest a method that quantifies the amount of color dispersion at the object edges and realigns color components to correct chromatic aberration. Although artifact correction and elimination is essential for robust downstream analysis, literature on the topic is relatively sparse. Moreover, most proposed methods have only been tested on a limited set of images as a proof of concept.
Figure 2.
Eliminating tissue-fold artifacts and pen marks in a whole-slide image of a NIH Cancer Genome Atlas ovarian serous carcinoma biopsy.
Batch effects
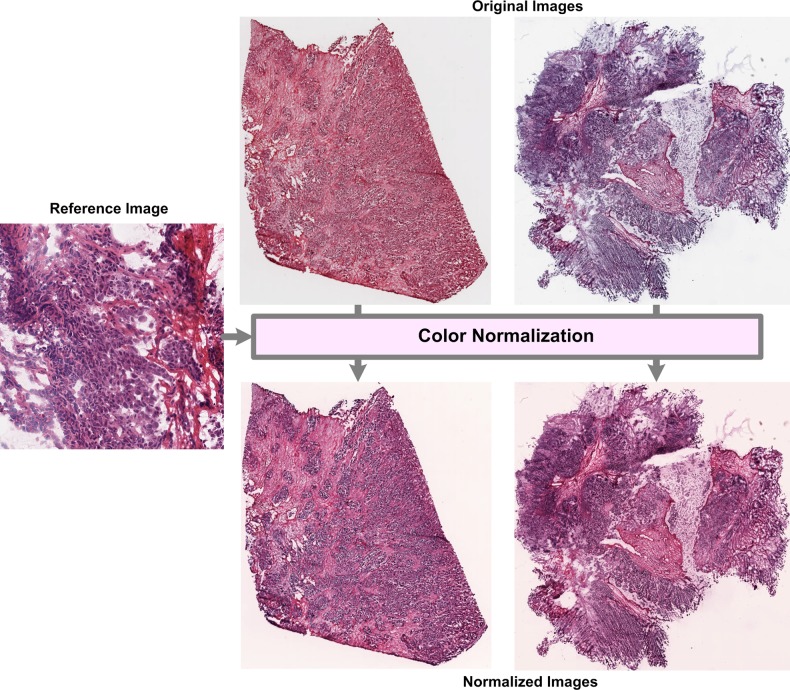
Differences in slide preparation, microscope, and digitizing device between two batches of data may lead to differences in image properties between the two batches. These differences, called batch effects, can bias the performance estimates of predictive models. Histopathological images often suffer from color and scale batch effects. Color batch effects can be addressed by normalizing the color of an image to a reference image18–20 or by converting the image to a color space (eg, CIELAB) that is not affected by color batch effects.21–23 Figure 3 illustrates results for normalizing the color map of two ovarian samples (obtained from TCGA) using color-map quantile normalization.18 Color normalization can be performed either at the pixel level using a single model for a complete image18 or at the stain level using a different model for each stain.20 Pixel-level normalization is affected by differences in morphology between the reference and test images while stain-level normalization is affected by the accuracy of stain segmentation. Unlike color batch effects, which affect only color properties of an image, scale batch effects can affect a variety of image features such as object size, topology, and texture. However, scale batch effects may be difficult to detect or correct because biological factors such as cancer grade or subtype may induce changes in scale. Such batch effects may be detected by examining the differences in distribution of image features between batches. For example, Kothari et al24 detected and proposed a method for correcting scale batch effects by examining the distributions of nuclear areas. Studies suggest that batch effects, if left uncorrected, can severely reduce the performance of genomic prediction models.25 26 Even though preliminary investigations suggest that batch effects are present in histopathological images, most researchers validate their diagnostic models on a single image dataset collected during a single experimental set-up. For clinical application of these systems, it is essential to validate diagnostic models on multiple datasets and to develop effective batch-effect removal methods.
Figure 3.
Normalization of color batch effects in ovarian samples provided by the NIH Cancer Genome Atlas.
Quantitative image description
WSI data may be described by experimental and clinical-level features (eg, acquisition-related specifications and patient diagnoses) as well as content-based image properties. Content-based features, which are informative for quantitative prediction modeling and for exploratory analysis, can be categorized into three levels—pixel, object, and semantic-level features—based on the amount of raw data captured by the features and the biological interpretability of the features (figure 4).27 28
Figure 4.
Representation of a (A) NIH Cancer Genome Atlas whole-slide image (WSI) of a kidney renal clear cell carcinoma biopsy using various quantitative features extracted from (B) a single image tile. Quantitative features include pixel-level features, ie, (C) color histogram and (D) Gabor filter response; object-level features, ie, (E) segmented shapes and (F) graph-based topology; and semantic-level features, ie, (G) percentage of high-level clinical properties.
Pixel-level features
Pixel-level image features are in the lowest level of the information hierarchy because they are the least interpretable in terms of biology. Pixel-level image features do not focus on any specific set of pixels in a WSI. Rather, they consider all image pixels and capture properties such as color and texture. Color features quantify color spread, prominence and co-occurrence using statistics and frequencies of color histograms in different color spaces including red-green-blue,29–31 hue-saturation-value,32 CIELUV,33 and CIELAB22 34 (figure 4C). Texture features quantify image sharpness, contrast, changes in intensity, and discontinuities or edges by measuring properties derived from gray-level intensity profiles,30 Haralick gray-level co-occurrence matrix (GLCM) features,23 30 35 36 wavelet and multiwavelet submatrices,30 35–37 Gabor filter responses23 30 36 (figure 4D), and fractals.30 36
Despite the lack of biological interpretability, pixel-level features are used extensively in data-driven models because they are simple to extract and are useful (at times sufficient) to describe the images. For example, features from eight color spaces were successfully used for skin melanoma classification,38 gray-level multiwavelet features for prostate grading,37 and color texture (GLCM) properties for follicular lymphoma grading.39 Figure 4 illustrates some pixel-level features including red-green-blue color histograms and Gabor filter textures at various scales.
Object-level features
Object-level features are in a higher level of the information hierarchy compared to pixel-level features because they describe properties of the cellular structures—such as nuclei, cytoplasm, and glands—in a WSI. To extract object-based features, it is essential first to segment cellular structures. As cellular structures appear in different colors in a stained histopathological sample, researchers have proposed color-based methods for segmentation. The literature supports both semi-automatic methods, with some user interaction,35 40 as well as completely automatic methods18 41–43 for segmentation. To increase the accuracy of segmentation, some researchers consider the pixel neighborhood properties using graph cut,39 object graph,44 and Markov models.45 The accuracy of image segmentation methods greatly affects the robustness of downstream analysis. Figure 4E illustrates a pseudo-colored segmentation mask, in which blue, pink, and white represent nuclear, cytoplasmic, and no-stain/gland regions, respectively.18 Object-level features describe the shape, texture, and spatial distribution of cellular structures in a WSI.
Shape-based features can be broadly categorized into contour and region-based features (figure 4E).46 Contour-based features include the properties of shape boundary such as perimeter, boundary fractal dimension, and bending energy. They also include coefficients of parametric shape models such as Fourier shape descriptors and elliptical models.47 Region-based features include area, solidity, and Zernike moments.48 Among all shape features, the properties of elliptical shape models of a nuclear boundary are most prevalent in pathology informatics because they are simple to extract and interpret, and informative for cancer endpoints.39 48–51
Object-level texture features are similar to pixel-level texture features, except that they capture the texture of only a subset of image pixels associated with a tissue object.30 Nuclear texture is reported to be very informative for separating malignant regions,51 subtyping cancer,49 and grading cancer.30
Topological or architectural features can capture the spatial distribution of cellular structures in a tissue sample. Researchers have found spatial graphs (eg, Deluanay triangulations, Voronoi diagrams, and minimum spanning trees), in which graph nodes are centers of cellular (nuclear or cytoplasmic) structures, to be useful for extracting topological features (figure 4F). Common topological features include properties of spatial graphs such as edge length, connectedness, and compactness. Besides graph-based properties, topological properties include object density, average distance between neighbors, and the number of objects within a given neighborhood. Architectural features are useful for cancer endpoints such as grading,30 39 classifying tumor versus non-tumor regions,52 53 classifying low versus high lymphocytic infiltration regions,54 and predicting patient prognosis.55 56
In comparison to pixel-level features, object-level descriptors can be much more computationally expensive to extract due to their dependence on image segmentation. Therefore, in light of the diagnostic benefit and biological interpretability of object-level features, more research is necessary to improve the computational speed of object-level feature extraction using methods such as parallel computing and graphical processing units.
Semantic-level features
Most pixel and object-level features are difficult to interpret biologically and are susceptible to noise. In contrast, semantic-level features easily capture interpretable high-level concepts such as the presence or absence of nucleoli, necrosis, and lymphocytes (figure 4G). A semantic feature is usually a classification or statistical rule based on a subset of low-level features (eg, low-level properties such as nuclear texture, color, and gray-level distribution may capture the high-level concept of nucleolus presence in a nucleus). Because not all low-level features may be useful for capturing high-level biological concepts, CDSS often use feature-preprocessing methods to select a subset of the original or transformed features. Among these preprocessing methods, the bag-of-features method is the one most commonly used for semantic features.57–59 As semantic-level features require a large amount of annotated training data, only a few systems use these features.60–62 There is thus limited research on semantic-level descriptors for histopathology. However, with the large amount of biological variations in WSI because of the heterogeneity of cancer biology, it will be especially beneficial to continue developing and refining semantic-level image descriptors.
Predictive modeling
Predictive modeling is an important part of pathology imaging informatics because it is applicable to a number of diagnostic clinical endpoints. Three important steps of WSI prediction modeling include: ROI selection and tile-based WSI representation; informative feature selection and reduction; and classification. We discuss ROI selection and tile-based WSI representation in detail in the following section. As the number of image features is generally much larger than the number of available samples, predictive modeling in pathology imaging informatics faces similar algorithmic challenges as that of other informatics fields. As described in the supplementary methods (available online only), feature selection, feature reduction, and classification methods address the problem of robust model building based on high-dimensional data.
ROI selection and tile-based WSI representation
A high-resolution scan of a tissue biopsy slide results in a very large WSI (eg, up to 40 000×60 000 pixels). Such WSI contain a large amount of biologically related spatial variation including regions of high-grade tumor, low-grade tumor, necrosis, and stroma. When pathologists examine a WSI, they identify regions that are most important or relevant for the final prognostic decision (eg, the region with the highest cancer grade). Similarly, an informatics system aims to identify a ROI in the WSI before developing a predictive model. Several researchers have developed supervised models for identifying ROI in WSI, but these methods require previous annotation for training.13 63–65 Researchers have recently proposed unsupervised knowledge-based methods for identifying ROI.66 67
Because of limitations in computer memory and processing time, WSI are often cropped into smaller tiles (eg, 512×512-pixel tiles), and then features are extracted from each tile in parallel.13 22 65 67–69 Representation of WSI by combining data from multiple WSI tiles is an emerging area of research with limited published results, especially in the context of clinical prediction.22 49 After identifying tiles corresponding to ROI, an informatics system can either combine the tiles to represent the WSI in a prediction model49 or predict the label for individual tiles and then combine labels to represent the final prediction result of the WSI.22 In the former method, outlier features might dominate WSI properties. In the latter method, annotation of individual tiles, instead of the WSI, might be necessary for training models. In the case study, we demonstrate a simple method for combining features from multiple tiles and show that this method yields reasonable clinical prediction results. A related topic to piecewise analysis of WSI is multiresolution or multiscale analysis, in which a WSI is processed at various scales/resolutions to achieve different modeling objectives.22 23 67 70 The basic concept of multiscale analysis is that a coarse level of prediction—such as tumor and non-tumor classification—can be achieved at a low resolution, when WSI are smaller and processing time is shorter. In contrast, for more complex problems such as grade prediction, WSI need to be processed at higher resolution.
Most WSI are millions of pixels in size and capture a large amount of biological heterogeneity. It is thus necessary to develop automatic methods for accurately selecting ROI in WSI. Without accurate ROI selection, the prediction performance of decision support systems for WSI may suffer compared to that for manually selected image portions. In order to achieve automatic ROI selection, we need to develop representation methods that capture high-level biological heterogeneity in WSI (ie, regions of high/low-grade cancer or regions of tissue necrosis). These methods can be as simple as capturing pathologists’ annotations for biological heterogeneity, then using these annotations to train automatic ROI selection methods. Such models will not only aid in WSI-based patient prediction modeling but will also aid in exploratory analysis for discovering factors that lead to differential clinical outcomes.
Visualization and exploratory analysis
Pathology imaging informatics has traditionally focused on predictive modeling. However, the research focus has evolved into a combination of predictive modeling and exploratory analysis for two reasons. First, large-scale studies such as TCGA aim to reveal new insights about aggressive cancer endpoints and to discover new prognostically different subtypes. Second, predictive modeling with high-dimensional data is very difficult and requires tools for interpreting the biological relevance of features and quantitative models.
Unsupervised clustering and high-dimensional feature patterns
Patterns in image features can be captured in simple two or three-dimensional visualizations such as scatter plots, surface plots, and distribution curves.35 39 51 54 56 71 However, if the number of descriptors is very large (>50), such visualizations may be difficult to implement or interpret. Therefore, unsupervised clustering methods are useful for reducing the feature space before visualization. Common clustering techniques in pathology imaging informatics include hierarchal clustering, self-organizing maps, and k-means. Hierarchal clustering is useful for patient stratification and visualization.49 68 72–74 Self-organizing maps are commonly used for feature interpretation,75 patient stratification76 and segmentation39 77 78 in pathology imaging informatics systems. k-Means is mostly used for color segmentation79 and for image classification and visualization as part of the bag-of-features representation.58 80 All of these methods are useful for visualizing the underlying structure of high-dimensional representations of histopathological data.
Virtual microscope and spatial patterns
With the availability of large histopathological data repositories such as TCGA, ‘virtual microscope’ software applications have emerged that enable the spatial exploration of high-resolution digital WSI.1 68 81 82 Without such applications, it is a challenge to share or even to view these images in real time. In addition, researchers have developed compression methods specifically for WSI.83 84 The popularity of the Google Maps interface for exploring satellite images at many different detail levels has inspired similar tools for exploring whole-slide tissue images.82 85 86 In addition to viewing a WSI, some systems can highlight ROI (eg, regions of high-grade cancer or regions with lymphocyte infiltration).56 64 65 67 70 87 Moreover, some visualizations annotate histopathological images with semantic labels such as necrosis, glands, and lymphocytes,61 62 or highlight the spatial distributions of proteins, image features, or biomarker expression across the histopathological image.13 71
Both spatial and patient-level exploratory analysis of WSI is an open area of research that requires interdisciplinary collaborations among pathologists, biologists, and computer scientists. Such collaboration is necessary to tackle the difficult problem of discovering and interpreting novel patterns in histopathological data that may lead to improved patient care. Moreover, it is necessary to develop novel quantitative metrics for assessing the stability and reproducibility of patterns related to both spatial and patient-level analysis to ensure that these patterns are biologically relevant. The supplemental case study illustrates a method for exploring spatial patterns in WSI.
Case study: tile-based ROI selection for WSI prediction modeling
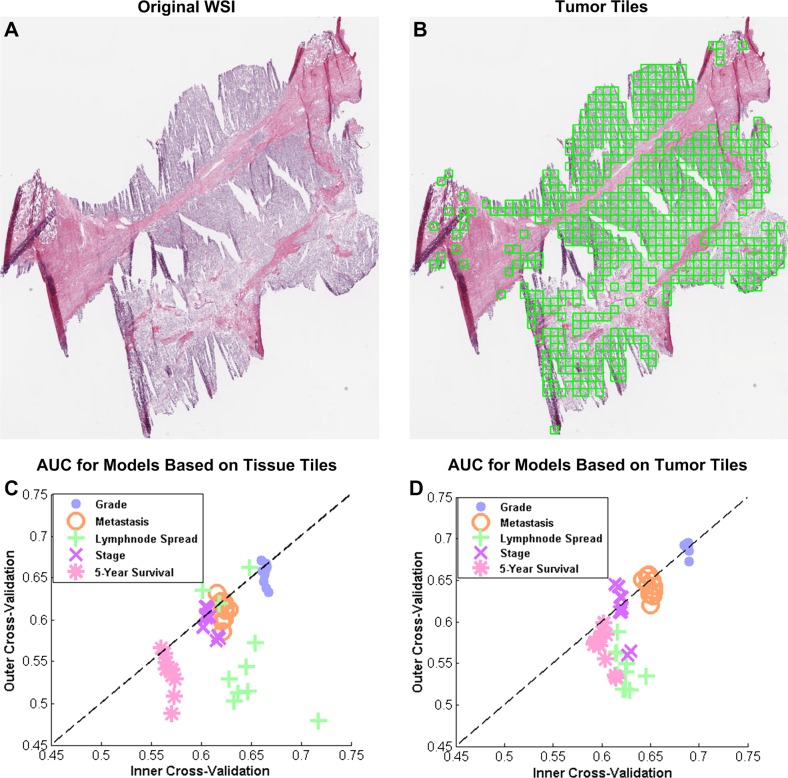
In this case study, we examine the effect of WSI ROI selection on the prediction performance of clinical endpoints. We use 906 WSI of tumor samples from 451 kidney renal clear cell carcinoma (KiCa) patients from TCGA.11 As described in supplement 1 (available online only), information extraction from quality-controlled WSI include the following steps: tile segmentation; image feature extraction; tumor detection; and patient representation using tissue (tumor and non-tumor tiles) or tumor tiles. Using the clinical data from TCGA, we develop WSI-based decision models for five binary endpoints (table 1). Prediction models use classifiers based on discriminant analysis—linear, quadratic, spherical and diagonal—and minimum-redundancy, maximum-relevance (mRMR) feature selection.88 We optimize feature size in the range of 1 to 100 and classifier parameters using five-fold, 10 iterations of nested cross-validation. The optimized models have average feature size in the range of 28 to 74 (table 2). Among all feature subsets, the nuclear shape subset is statistically overrepresented for most endpoints, which implies that nuclear shape features are most informative for these endpoints (table 2).30
Table 1.
Prediction performance for whole-slide image-based binary endpoints
| Class 1 | Class 2 | AUC for outer cross-validation | |||||
|---|---|---|---|---|---|---|---|
| Endpoint | Description | No. of patients | Description | No. of patients | Tissue (incl. non-tumor tiles) | Tumor | p Value |
| Histological grade | Grade 1 or 2 | 204 | Grade 3 or 4 | 239 | 0.66±0.01 | 0.69±0.01 | 0.0000 |
| Metastasis | No spread to other organs | 381 | Spread to other organs | 68 | 0.61±0.01 | 0.64±0.01 | 0.0001 |
| Stage | Stage I or II | 267 | Stage III or IV | 182 | 0.60±0.02 | 0.61±0.03 | 0.0562 |
| Five-year survival | <5 years | 126 | ≥5 years | 101 | 0.54±0.02 | 0.57±0.02 | 0.0151 |
| Lymphnode spread | No spread to nearby lymph nodes | 210 | Spread to nearby lymph nodes | 17 | 0.56±0.06 | 0.54±0.02 | 0.5148 |
AUC, area under the curve.
Table 2.
Statistically overrepresented feature subsets in models based on tumor tiles
| Endpoint | Average feature size | Statistically overrepresented feature subsets (Fishers test, p value=0.05) |
|---|---|---|
| Histological grade | 74 | Nuclear shape (0.013) |
| Metastasis | 28 | Nuclear shape (0.000) |
| Stage | 54 | Nuclear shape (0.013) and Basophilic-object shape (0.002) |
| Five year survival | 37 | Basophilic-region texture (0.000) |
| Lymph node spread | 34 | Nuclear shape (0.000) |
Figure 5C,D illustrates scatter plots of area under the curve for inner cross-validation and outer cross-validation performance for models based on tissue (tumor and non-tumor tiles) and tumor tiles, respectively. Each point in the scatter plot is an average performance for one cross-validation iteration. We can observe that the performance in both cases—models based on tumor and tissue tiles—is close to the diagonal, which indicates that inner cross-validation can predict the performance of outer cross-validation. We can also observe that the models based on tumor tiles perform equivalent to or better than the models based on all tissue tiles. We report the average and SD of outer cross-validation performance for all endpoints in table 2. For the histological grade and metastasis prediction models, prediction performances based on tumor tiles are more than those based on all tissue tiles with statistical significance (evaluated using a t test). Although this case study adopts a robust analytical pipeline, the classification performances are lower compared to the performances observed in the literature for manually curated sections. Two causes for low prediction performance are various quality issues with TCGA data, that is, tissue folds, pen marks, and out-of-focus regions that are inherent to WSI, and difficulty in predicting clinical endpoints, that is, patient survival, which are not normally targeted by pathologists. Therefore, automatic image quality control, ROI selection in WSI, and clinically informative feature extraction are still open challenges in the field of pathology imaging informatics. Despite these challenges, such CDSS will provide an objective and fast means for clinical diagnosis with minimal user intervention. Moreover, such systems can be trained to diagnose rare subtypes of cancer that are often missed in traditional diagnosis.89 The knowledge extracted by these systems may also contribute to a holistic diagnostic platform by integration with data from other imaging modalities as well as with data from genomic and proteomic experiments.90
Figure 5.
Role of region of interest (ROI) selection on the performance of whole-slide image (WSI)-based prediction models. (A) An example WSI. (B) Tiles in the tumor region (ROI) of the WSI highlighted with green boxes. Scatter plots between the prediction performance (area under the curve; AUC) of inner and outer loop of nested cross-validation for (C) models based on features from tissue tiles, including tumor and non-tumor (normal, necrosis, and stroma) regions; and for (D) tiles in the tumor region only.
Table 3.
Summary of key methods in each component of a WSI-based clinical decision support system
| Section | Subsection | Key methods |
|---|---|---|
| Quality control | Image artifacts | Tissue folds,13–15 blurred regions,16 and chromatic aberration17 |
| Batch effects | Color normalization,18–20 batch-invariant color space,21–23 and scale normalization24 | |
| Image description | Pixel-level features | Color,38 gray-level intensity profiles,30 Haralick features,23 30 35 36 wavelet and multiwavelet submatrices,30 36 37 Gabor filter responses,23 30 36 and Fractals30 36 |
| Object-level features | Shape47 48 and graph-based topology52–56 | |
| Semantic-level features | Bag-of-features60 and spatial hidden Markov model61 | |
| Prediction modeling | ROI selection and tile-based WSI representation | ROI selection: supervised13 63–65 and unsupervised66 67; tile combination: feature49 and prediction22; multiscale analysis22 23 67 70 |
| Informative feature selection and reduction | Feature selection: filter,30 51 53 57 sequential search (wrapper),22 36 91 and random forest (embedded)65; Feature reduction: PCA,32 39 91 graph embedding,54 ISOMAP,80 and MDS92 | |
| Classification | Multiple classifiers,22 29 36 39 80 boosting,23 80 93 ensemble methods,22 and active learning3 94–96 | |
| Visualization and exploratory analysis | Unsupervised clustering and high-dimensional feature patterns | Hierarchal clustering,49 68 72–74 Self-organizing maps,75 76 k-means,58 79 80 and expectation maximization22 |
| Virtual microscope and spatial patterns | Image compression,83 84 Google map interface,82 85 86 highlight ROI,56 64 65 67 70 87 annotation,61 62 and spatial variation of features13 71 |
MDS, ; PCA, ; ROI, region of interest; WSI, whole-slide image.
Conclusion
With the emergence of WSI technology, high-resolution scans of complete tissue biopsy slides are becoming a common clinical practice. Despite the benefits of WSI for histopathological diagnosis, the literature reports that existing CDSS primarily use only rectangular sections of WSI. Moreover, commercial software tools for WSI analysis are also limited because they are typically trained for only a single experimental set-up and only focus on segmenting tissue structures and quantifying a limited set of image descriptors to aid manual histopathological analysis. Based on these systems, we learned that quantitative image features are able to model cancer diagnosis and prognosis. However, the development of CDSS for WSI has been impeded by several informatics challenges: quality control; robust and fast image segmentation; knowledge (semantic-level) models for WSI; and ROI selection. Researchers have developed methods to address these challenges (table 3). However, most studies validate their methods on a limited number of samples and cancer endpoints. To make CDSS for pathology a reality, it is necessary to develop a generalizable system (such as the system described in the case study) that can be applied to multiple cancer endpoints and that is validated using large multibatch datasets. With the availability of large WSI datasets for multiple cancer endpoints in public repositories such as TCGA, the data required to make the necessary advances in pathology imaging informatics research have now become more accessible.
Supplementary Material
Footnotes
Contributors: SK researched the literature about pathology imaging informatics methods, contributed to the design of the structure of the paper, implemented methods for quality control, color normalization, and image feature extraction, and drafted the manuscript. JHP contributed to the design of the structure of the paper and image feature extraction methods and implemented machine-learning methods for the case study. THS designed and implemented methods for supplemental case study. MDW initiated the literature survey of the pathology imaging informatics methods, acquired funding to sponsor this effort, and directed the development of the case studies and publication. JHP, THS and MDW reviewed and revised the manuscript. All authors read and approved the final manuscript.
Funding: This research has been supported by grants from NIH (U54CA119338, 1RC2CA148265, and R01CA163256), Georgia Cancer Coalition Award to MDW, Hewlett Packard, and Microsoft Research.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Gabril MY, Yousef GM. Informatics for practicing anatomical pathologists: marking a new era in pathology practice. Mod Pathol 2010;23:349–58 [DOI] [PubMed] [Google Scholar]
- 2.Wetzel A. Computational aspects of pathology image classification and retrieval. J Supercomput 1997;11:279–93 [Google Scholar]
- 3.Fuchs TJ, Buhmann JM. Computational pathology: challenges and promises for tissue analysis. Comput Med Imaging Graph 2011;35:515–30 [DOI] [PubMed] [Google Scholar]
- 4.Amin W, Chandran U, Parwani Anil V, et al. Biomedical informatics for anatomic pathology. In: Cheng L, Bostwick DG, eds. Essentials of anatomic pathology. New York: Springer, 2011:469–80 [Google Scholar]
- 5.Gurcan MN, Boucheron L, Can A, et al. Histopathological image analysis: a review. IEEE Rev Biomed Eng 2009;2:147–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadimin ET, Foran DJ. Pathology imaging informatics for clinical practice and investigative and translational research. N Am J Med Sci (Boston) 2012;5:103–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho J, Parwani AV, Jukic DM, et al. Use of whole slide imaging in surgical pathology quality assurance: design and pilot validation studies. Hum Pathol 2006;37:322–31 [DOI] [PubMed] [Google Scholar]
- 8.Pantanowitz L, Valenstein PN, Evans AJ, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform 2011;2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunkle R. Role of image informatics in accelerating drug discovery and development. Drug Discovery 2003;4:75–82 [Google Scholar]
- 10.Melamed J, Datta MW, Becich MJ, et al. The cooperative prostate cancer tissue resource: a specimen and data resource for cancer researchers. Clin Cancer Res 2004;10:4614–21 [DOI] [PubMed] [Google Scholar]
- 11.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol 2010;28:1248–50 [DOI] [PubMed] [Google Scholar]
- 13.Kothari S, Phan JH, Osunkoya AO, et al. Biological interpretation of morphological patterns in histopathological whole-slide images. Proceedings of the 3rd ACM Conference on Bioinformatics, Computational Biology and Biomedicine. 2012:218–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palokangas S, Selinummi J, Yli-Harja O. Segmentation of folds in tissue section images. Conference Proceedings of the IEEE Engineering in Medicine and Biology Society. 2007:5642–5 [DOI] [PubMed] [Google Scholar]
- 15.Bautista PA, Yagi Y. Detection of tissue folds in whole slide images. Conference Proceedings of the IEEE Engineering in Medicine and Biology Society. 2009:3669–72 [DOI] [PubMed] [Google Scholar]
- 16.Gao D, Padfield D, Rittscher J, et al. Automated training data generation for microscopy focus classification. Med Image Comput Comput Assist Interv 2010;13:446–53 [DOI] [PubMed] [Google Scholar]
- 17.Wu HS, Murray J, Morgello S, et al. Restoration of distorted colour microscopic images from transverse chromatic aberration of imperfect lenses. J Microsc 2011;241:125–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kothari S, Phan JH, Moffitt RA, et al. Automatic batch-invariant color segmentation of histological cancer images. Proceedings of the IEEE International Symposium on Biomedical Imaging. 2011:657–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macenko M, Niethammer M, Marron JS, et al. A method for normalizing histology slides for quantitative analysis. Proceedings of the IEEE International Symposium on Biomedical Imaging. 2009:1107–10 [Google Scholar]
- 20.Magee D, Treanor D, Crellin D, et al. Colour normalisation in digital histopathology images. Proceedings of the Optical Tissue Image analysis in Microscopy, Histopathology and Endoscopy (MICCAI Workshop), 2009:100–11 [Google Scholar]
- 21.Kong H, Gurcan M, Belkacem-Boussaid K. Partitioning histopathological images: an integrated framework for supervised color-texture segmentation and cell splitting. IEEE Trans Med Imaging 2011;30:1661–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong J, Sertel O, Shimada H, et al. Computer-aided evaluation of neuroblastoma on whole-slide histology images: classifying grade of neuroblastic differentiation. Pattern Recognition 2009;42:1080–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle S, Feldman M, Tomaszewski J, et al. A boosted bayesian multi-resolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans Biomed Eng 2010;59:1205–18 [DOI] [PubMed] [Google Scholar]
- 24.Kothari S, Phan JH, Wang MD. Scale normalization of histopathological images for batch invariant cancer diagnostic models. Conference Proceedings of the IEEE Engineering in Medicine and Biology Society. 2012:4406–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Grennan K, Badner J, et al. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS ONE 2011;6:e17238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo J, Schumacher M, Scherer A, et al. A comparison of batch effect removal methods for enhancement of prediction performance using maqc-ii microarray gene expression data. Pharmacogenomics J 2010;10:278–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Z, Yu W. Stable feature selection for biomarker discovery. Comput Biol Chem 2010;34:215–25 [DOI] [PubMed] [Google Scholar]
- 28.Hsu W, Lee Mong L, Zhang J. Image mining: trends and developments. J Intell Inf Syst 2002;19:7–23 [Google Scholar]
- 29.Tabesh A, Teverovskiy M, Ho-Yuen P, et al. Multifeature prostate cancer diagnosis and gleason grading of histological images. IEEE Trans on Med Imaging 2007;26:1366–78 [DOI] [PubMed] [Google Scholar]
- 30.Kothari S, Phan JH, Young AN, et al. Histological image feature mining reveals emergent diagnostic properties for renal cancer. Proceedings of the IEEE International Conference on Bioinformatics Biomedicine. 2011:422–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs T, Wild P, Moch H, et al. Computational pathology analysis of tissue microarrays predicts survival of renal clear cell carcinoma patients. In: Metaxas D, Axel L, Fichtinger G, Székely G, eds. Medical image computing and computer-assisted intervention. Berlin/Heidelberg: Springer, 2008:1–8 [DOI] [PubMed] [Google Scholar]
- 32.Rahman M, Bhattacharya P, Desai BC. A framework for medical image retrieval using machine learning and statistical similarity matching techniques with relevance feedback. IEEE Trans Inf Technol Biomed 2007;11:58–69 [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Tuzel O, Chen W, et al. Pathminer: a web-based tool for computer-assisted diagnostics in pathology. IEEE Trans Inf Technol Biomed 2009;13:291–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovalev V, Dmitruk A, Safonau I, et al. A method for identification and visualization of histological image structures relevant to the cancer patient conditions. In: Real P, Diaz-Pernil D, Molina-Abril H, Berciano A, Kropatsch W, eds. Computer analysis of images and patterns. Berlin/Heidelberg: Springer, 2011: 460–8 [Google Scholar]
- 35.Chaudry Q, Raza S, Young A, et al. Automated renal cell carcinoma subtype classification using morphological, textural and wavelets based features. J Signal Processing Syst 2009;55:15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Po-Whei H, Cheng-Hsiung L. Automatic classification for pathological prostate images based on fractal analysis. IEEE Trans Med Imaging 2009;28:1037–50 [DOI] [PubMed] [Google Scholar]
- 37.Jafari-Khouzani K, Soltanian-Zadeh H. Multiwavelet grading of pathological images of prostate. IEEE Trans Biomed Eng 2003;50:697–704 [DOI] [PubMed] [Google Scholar]
- 38.Celebi ME, Kingravi HA, Uddin B, et al. A methodological approach to the classification of dermoscopy images. Comput Med Imaging Graph 2007; 31:362–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sertel O, Kong J, Catalyurek U, et al. Histopathological image analysis using model-based intermediate representations and color texture: Follicular lymphoma grading. J Signal Processing Syst 2009;55:169–83 [Google Scholar]
- 40.Jun K, Shimada H, Boyer K, et al. Image analysis for automated assessment of grade of neuroblastic differentiation. Proceedings of the IEEE International Symposium on Biomedical Imaging. 2007:61–4 [Google Scholar]
- 41.Meurie C, Lebrun G, Lezoray O, et al. A comparison of supervised pixels-based color image segmentation methods. Application in cancerology. WSEAS Trans Computers 2003;2:739–44 [Google Scholar]
- 42.Mao K, Zhao P, Tan P. Supervised learning-based cell image segmentation for p53 immunohistochemistry. IEEE Trans Biomed Eng 2006;53:1153–63 [DOI] [PubMed] [Google Scholar]
- 43.Ranefalla P, Egevadb L, Nordina B, et al. A new method for segmentation of colour images applied to immunohistochemically stained cell nuclei. Anal Cell Pathol 1997;15:145–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunduz-Demir C, Kandemir M, Tosun A, et al. Automatic segmentation of colon glands using object-graphs. Med Image Anal 2010;14:1–12 [DOI] [PubMed] [Google Scholar]
- 45.Monaco J, Tomaszewski J, Feldman M, et al. High-throughput detection of prostate cancer in histological sections using probabilistic pairwise Markov models. Med Image Anal 2010;14:617–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang D, Lu G. Review of shape representation and description techniques. Pattern Recognition 2004;37:1 [Google Scholar]
- 47.Kothari S, Phan JH, Young AN, et al. Histological image classification using biologically interpretable shape-based features. BMC Medical Imaging 2013;13:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boucheron L. Object-and spatial-level quantitative analysis of multispectral histopathology images for detection and characterization of cancer. PhD thesis, Santa Barbara: University of California, 2008 [Google Scholar]
- 49.Cooper LAD, Jun K, Gutman DA, et al. An integrative approach for in silico glioma research. IEEE Trans Biomed Eng 2010;57:2617–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kothari S, Chaudry Q, Wang MD. Extraction of informative cell features by segmentation of densely clustered tissue images. Conference Proceedings of the IEEE Engineering in Medicine and Biology Society. 3–6 September 2009, 6706–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muthu Rama Krishnan M, Pal M, Paul RR, et al. Computer vision approach to morphometric feature analysis of basal cell nuclei for evaluating malignant potentiality of oral submucous fibrosis. J Med Syst 2012;36:1746–56 [DOI] [PubMed] [Google Scholar]
- 52.Gunduz C, Yener B, Gultekin SH. The cell graphs of cancer. Bioinformatics 2004;20(Suppl. 1):i145–i51 [DOI] [PubMed] [Google Scholar]
- 53.Bilgin CC, Bullough P, Plopper GE, et al. Ecm-aware cell-graph mining for bone tissue modeling and classification. Data Min Knowl Discov 2009;20:416–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basavanhally AN, Ganesan S, Agner S, et al. Computerized image-based detection and grading of lymphocytic infiltration in her2+ breast cancer histopathology. IEEE Trans Biomed Eng 2010;57:642–53 [DOI] [PubMed] [Google Scholar]
- 55.Sudbø J, Marcelpoil R, Reith A. New algorithms based on the voronoi diagram applied in a pilot study on normal mucosa and carcinomas. Anal Cell Pathol 2000; 21:71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sudbo J, Bankfalvi A, Bryne M, et al. Prognostic value of graph theory-based tissue architecture analysis in carcinomas of the tongue. Lab Invest 2000;80:1881–9 [DOI] [PubMed] [Google Scholar]
- 57.Cruz-Roa A, Caicedo JC, González FA. Visual pattern mining in histology image collections using bag of features. Artif Intell Med 2011;52:91. [DOI] [PubMed] [Google Scholar]
- 58.Raza S, Parry R, Moffitt R, et al. An analysis of scale and rotation invariance in the bag-of-features method for histopathological image classification. In: Fichtinger G, Martel A, Peters T, eds. Medical image computing and computer-assisted intervention. Berlin/Heidelberg: Springer, 2011:66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rahman M, Antani S, Thoma G. A learning-based similarity fusion and filtering approach for biomedical image retrieval using svm classification and relevance feedback. IEEE Trans Inf Technol Biomed 2011;15:640–6 [DOI] [PubMed] [Google Scholar]
- 60.Caicedo JC, González FA, Romero E. Content-based histopathology image retrieval using a kernel-based semantic annotation framework. J Biomed Inform 2011;44:519–28 [DOI] [PubMed] [Google Scholar]
- 61.Yu F, Ip Horace HS. Semantic content analysis and annotation of histological images. Comput Biol Med 2008;38:635–49 [DOI] [PubMed] [Google Scholar]
- 62.Tang HL, Hanka R, Ip HHS. Histological image retrieval based on semantic content analysis. IEEE Trans Inf Technol Biomed 2003;7:26–36 [DOI] [PubMed] [Google Scholar]
- 63.Gutiérrez R, Gómez F, Roa-Peña L, et al. A supervised visual model for finding regions of interest in basal cell carcinoma images. Diagn Pathol 2011;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romo D, Romero E, González F. Learning regions of interest from low level maps in virtual microscopy. Diagn Pathol 2011;6(Suppl. 1):S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DiFranco MD, O'Hurley G, Kay EW, et al. Ensemble based system for whole-slide prostate cancer probability mapping using color texture features. Comput Med Imaging Graph 2011;35:629–45 [DOI] [PubMed] [Google Scholar]
- 66.Thomas K, Sottile M, Salafia C. Unsupervised segmentation for inflammation detection in histopathology images. In: Elmoataz A, Lezoray O, Nouboud F, Mammass D, Meunier J, eds. Image and signal processing. Berlin/Heidelberg: Springer, 2010:541–9 [Google Scholar]
- 67.Roullier V, Lézoray O, Ta VT, et al. Multi-resolution graph-based analysis of histopathological whole slide images: Application to mitotic cell extraction and visualization. Comput Med Imaging Graph 2011;35:603–15 [DOI] [PubMed] [Google Scholar]
- 68.Chang H, Fontenay GV, Han J, et al. Morphometic analysis of tcga glioblastoma multiforme. BMC Bioinformatics 2011;12:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samsi S, Krishnamurthy AK, Gurcan MN. An efficient computational framework for the analysis of whole slide images: Application to follicular lymphoma immunohistochemistry. J Comput Sci 2012;3:269–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang CH, Veillard A, Roux L, et al. Time-efficient sparse analysis of histopathological whole slide images. Comput Med Imaging Graph 2011;35:579–91 [DOI] [PubMed] [Google Scholar]
- 71.Herold J, Loyek C, Nattkemper Tim W. Multivariate image mining. WIREs Data Mining Knowl Discov 2011;1:2 [Google Scholar]
- 72.Liu CL, Prapong W, Natkunam Y, et al. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol 2002;161:1557–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cooper LAD, Jun K, Fusheng W, et al. Morphological signatures and genomic correlates in glioblastoma. Proceedings of the IEEE International Symposium on Biomedical Imaging. 2011;1624–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lobenhofer EK, Boorman GA, Phillips KL, et al. Application of visualization tools to the analysis of histopathological data enhances biological insight and interpretation. Toxicol Pathol 2006;34:921–8 [DOI] [PubMed] [Google Scholar]
- 75.Lessmann B, Nattkemper TW, Hans VH, et al. A method for linking computed image features to histological semantics in neuropathology. J Biomed Inform 2007;40:631–41 [DOI] [PubMed] [Google Scholar]
- 76.Iglesias-Rozas JR, Hopf N. Histological heterogeneity of human glioblastomas investigated with an unsupervised neural network (som). Histol Histopathol 2005;20:351–6 [DOI] [PubMed] [Google Scholar]
- 77.Stephanakis I, Anastassopoulos G, Iliadis L. Color segmentation using self-organizing feature maps (sofms) defined upon color and spatial image space. In: Diamantaras K, Duch W, Iliadis L, eds. Artificial neural networks—icann. Berlin/Heidelberg: Springer, 2010: 500–10 [Google Scholar]
- 78.Datar M, Padfield D, Cline H. Color and texture based segmentation of molecular pathology images using hsoms. Proceedings of the IEEE International Symposium on Biomedical Imaging. 2008;292–5 [Google Scholar]
- 79.Rabinovich A, Krajewski S, Krajewska M, et al. Framework for parsing, visualizing and scoring tissue microarray images. IEEE Trans Inf Technol Biomed 2006;10:209–19 [DOI] [PubMed] [Google Scholar]
- 80.Yang L, Chen W, Meer P, et al. Virtual microscopy and grid-enabled decision support for large-scale analysis of imaged pathology specimens. IEEE Trans Inf Technol Biomed 2009;13:636–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marchevsky AM, Dulbandzhyan R, Seely K, et al. Storage and distribution of pathology digital images using integrated web-based viewing systems. Arch Pathol Lab Med 2002;126:533–9 [DOI] [PubMed] [Google Scholar]
- 82.Mayerich D, Abbott L, McCormick B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J Microsc 2008;231:134–43 [DOI] [PubMed] [Google Scholar]
- 83.Won-Ki J, Schneider J, Turney SG, et al. Interactive histology of large-scale biomedical image stacks. IEEE Trans Vis Comput Graph 2010;16:1386–95 [DOI] [PubMed] [Google Scholar]
- 84.Zwönitzer R, Kalinski T, Hofmann H, et al. Digital pathology: Dicom-conform draft, testbed, and first results. Comput Methods Programs Biomed 2007;87:181–8 [DOI] [PubMed] [Google Scholar]
- 85.Triola MM, Holloway WJ. Enhanced virtual microscopy for collaborative education. BMC Med Educ 2011;11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nock R. Fast and reliable color region merging inspired by decision tree pruning. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2001;1:271–6 [Google Scholar]
- 87.Monaco JP, Tomaszewski JE, Feldman MD, et al. High-throughput detection of prostate cancer in histological sections using probabilistic pairwise markov models. Med Image Anal 2010;14:617–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol 2005;3:185–205 [DOI] [PubMed] [Google Scholar]
- 89.Sidiropoulos K, Glotsos D, Kostopoulos S, et al. Real time decision support system for diagnosis of rare cancers, trained in parallel, on a graphics processing unit. Comput Biol Med 2012;42:376–86 [DOI] [PubMed] [Google Scholar]
- 90.Phan JH, Quo CF, Cheng C, et al. Multiscale integration of -omic, imaging, and clinical data in biomedical informatics. IEEE Rev Biomed Eng 2012;5:74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Srivastava S, Rodríguez JJ, Rouse AR, et al. Computer-aided identification of ovarian cancer in confocal microendoscope images. J Biomed Opt 2008;13:024021. [DOI] [PubMed] [Google Scholar]
- 92.Lei Z, Wetzel AW, Gilbertson J, et al. Design and analysis of a content-based pathology image retrieval system. IEEE Trans Inf Technol Biomed 2003;7:249–55 [DOI] [PubMed] [Google Scholar]
- 93.Foran DJ, Yang L, Chen W, et al. Imageminer: a software system for comparative analysis of tissue microarrays using content-based image retrieval, high-performance computing, and grid technology. J Am Med Inform Assoc 2011;18:403–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doyle S, Monaco J, Feldman M, et al. An active learning based classification strategy for the minority class problem: application to histopathology annotation. BMC Bioinformatics 2011;12:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cosatto E, Miller M, Graf HP, et al. Grading nuclear pleomorphism on histological micrographs. Proceedings of the International Conference on Pattern Recognition. 2008:1–4 [Google Scholar]
- 96.Begelman G, Pechuk M, Rivlin E, et al. A microscopic telepathology system for multiresolution computer-aided diagnostics. J Multimed 2006;1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.