Abstract
Background and objectives
Advances in MRI hardware and sequences are continually increasing the amount and complexity of data such as those generated in high-resolution three-dimensional (3D) scanning of the spine. Efficient informatics tools offer considerable opportunities for research and clinically based analyses of magnetic resonance studies. In this work, we present and validate a suite of informatics tools for automated detection of degenerative changes in lumbar intervertebral discs (IVD) from both 3D isotropic and routine two-dimensional (2D) clinical T2-weighted MRI.
Materials and methods
An automated segmentation approach was used to extract morphological (traditional 2D radiological measures and novel 3D shape descriptors) and signal appearance (extracted from signal intensity histograms) features. The features were validated against manual reference, compared between 2D and 3D MRI scans and used for quantification and classification of IVD degeneration across magnetic resonance datasets containing IVD with early and advanced stages of degeneration.
Results and conclusions
Combination of the novel 3D-based shape and signal intensity features on 3D (area under receiver operating curve (AUC) 0.984) and 2D (AUC 0.988) magnetic resonance data deliver a significant improvement in automated classification of IVD degeneration, compared to the combination of previously used 2D radiological measurement and signal intensity features (AUC 0.976 and 0.983, respectively). Further work is required regarding the usefulness of 2D and 3D shape data in relation to clinical scores of lower back pain. The results reveal the potential of the proposed informatics system for computer-aided IVD diagnosis from MRI in large-scale research studies and as a possible adjunct for clinical diagnosis.
Keywords: Computer-aided diagnosis, Classification, Intervertebral discs, Disc degeneration, Statistical shape models, Morphology
Introduction
Symptomatic intervertebral disc (IVD)-related disorders account for the largest proportion of musculoskeletal complaints in industrialized countries and may be associated with acute or chronic disability.1 MRI is an important diagnostic tool in clinical decision-making, providing highly detailed patho-anatomical examination of the spine,2 with significant radiological resources and specialist time devoted to the assessment of IVD in developed countries.1 3 Given the burgeoning amount and complexity of MRI data4 (multiple image sequences per patient, increasing resolution, three-dimensional (3D) imaging), informatics tools such as computer-aided diagnosis (CAD) systems have potentially large roles to play in clinical and research trial settings.
Common pathological conditions affecting IVD include disc degeneration, bulging or herniations (protrusion, extrusion or sequester), frequently associated with dysfunction, spinal canal and foraminal stenosis, segmental instability and pain.2 Radiological analyses in MRI aim to relate morphological and signal changes in the IVD to the patient's clinical symptoms.5 Routine clinical MRI scans of lumbar IVD are typically acquired as a set of two-dimensional (2D) slices (eg, slices with >3 mm thickness) of both T1-weighted and T2-weighted contrast in sagittal and axial orientation. Radiological assessment of disc pathology requires synthesis of IVD signal intensity characteristics from T2-weighted scans (eg, nucleus pulposus (NP) signal intensity, annulus fibrosis (AF) tear, relative intensity difference amongst neighboring discs) with disc morphology (eg, disc height, broad based bulging or focal herniations in the case of disc protrusion, extrusion or sequestration). Early diagnosis of IVD degenerative changes and reliable assessment during follow-up may have important implications for clinical decision-making for emerging conservative and surgical interventions.6
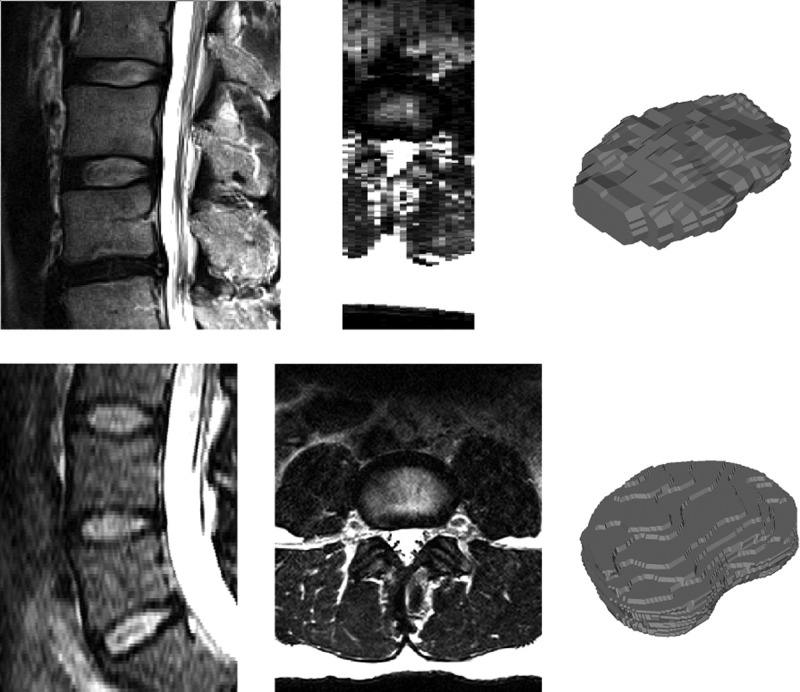
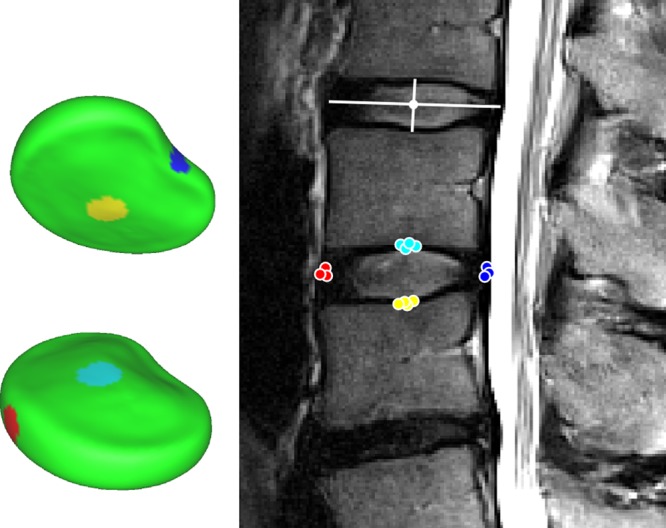
Morphological information extracted from 2D MRI has involved mostly manual delineation and measurements performed on relatively thick non-contiguous slices,7–9 providing planar-based measures such as IVD height, width or interpolated volume. Manual assessment is a tedious and resource-intensive task (expertise, time, equipment) prone to intra and inter-observer variability, and represents a major hindrance for performing large-scale research studies or for routine implementation in clinical practice. Moreover, recent advances in magnetic resonance hardware, software and pulse sequence design have provided the capacity to acquire 3D volumetric MRI scans of the human spine with nearly isotropic resolution under 0.5 mm3 (eg, the SPACE sequence).10 The volumetric scans can be used to acquire detailed 3D morphological information about the IVD shapes (figure 1) for research studies investigating relations between morphological and structural IVD alterations and clinical symptoms.5 However, the manual delineation of volumetric MRI scans for large-scale clinical and research environments is impractical and automatic computer approaches are required for fast, cost-effective and objective analysis.
Figure 1.
Sagittal (left) and axial (middle) views of an example 2D turbo spin echo (top, 3.3 mm slice spacing) and 3D SPACE (bottom, 1 mm slice spacing) MRI scans with manually segmented IVD shapes (right). The high resolution 3D image provides information about the anatomical shape that is not available in the sparser 2D scan. IVD, intervertebral disc; 2D, two dimensional; 3D, three dimensional.
In our research we aim to construct a suite of automated informatics tools for analysis of IVD degenerative changes apparent in high-resolution 3D MRI as well as in 2D scans routinely acquired in a clinical setting. Advanced image preprocessing techniques are implemented to reduce imaging artifacts and to adjust for varying image resolutions. In our recent work,11 an automated segmentation algorithm was developed to extract 3D shapes of the IVD from high-resolution 3D SPACE MRI scans. The extracted 3D parameters were used in a preliminary analysis of IVD shape characteristics in asymptomatic subjects presenting with no and early signs of disc abnormalities.11 The high accuracy of this initial proof of concept motivated further study to test the 3D morphological parameters for automated IVD classification. The aim of the current paper is to extract and quantify magnetic resonance-based information of likely relevance for the detection of degenerative IVD changes. In particular, the segmentation algorithm is tested on both high-resolution 3D and routine clinical 2D MRI scans for comparison of the extracted IVD morphological characteristics between the two sequences. A set of morphological and signal intensity features is proposed and evaluated in an IVD classification task to test whether morphological features based on novel 3D segmentations can improve the detection of IVD degeneration when compared to previously used morphological features extracted from a single 2D slice. Similar automated informatics tools have the potential to benefit large-scale studies investigating disc degeneration, trials of pharmaceutical or surgical interventions and to provide novel CAD tools for clinical practice. The developed algorithm circumvents resource-intensive manual segmentation procedures and enables automated 3D analysis of the IVD.
Background
Intervertebral disc degeneration
MRI has become a key investigative tool in radiological assessment of IVD degeneration stemming from its excellent soft tissue contrast and multiple scanning sequences for depicting various patho-anatomical states.12 13 An IVD consists of two main structural elements—the central gel-like NP, surrounded by the peripheral fibrocartilagenous AF. In T2-weighted MRI, a healthy NP appears as a bright elliptical structure, while the AF is imaged as a hypointense region bordering the NP.14 The characteristics used to describe degenerative disc changes during a typical magnetic resonance radiological assessment include: structural cues (height of IVD, IVD space narrowing, bulging or herniation), signal intensity cues (hypointense/hyperintense NP to cerebrospinal fluid, NP homogeneity and distribution) and the level distinction between the NP and AF (AF tear with prolapse of NP or intact AF surrounding the NP).15 A successful CAD classification algorithm would likely include both morphological and signal intensity features.
Automated detection of disc abnormalities
Several automated approaches for MRI-based classification of IVD degenerative changes have been presented in the literature. Alomari et al16 detected desiccated IVD in 2D MRI in the lumbar spine by using signal intensity features (median intensity level of pixel neighborhood) extracted from single mid-sagittal T2-weighted slices. A Gaussian model of IVD appearance was combined with a Bayesian model to describe the relationship between signal intensities of neighboring IVD. In a subsequent study, Alomari et al17 detected disc abnormalities (desiccation, degeneration) by a probabilistic modeling of three energy terms: appearance (several magnetic resonance contrasts), location and context (describing spatial neighboring relations) from a mid-sagittal MRI slice. In a further study based on MRI, disc herniation was detected by Alomari et al18 with a Bayesian classifier based on two shape features: major and minor axis of the segmented IVD. Ghosh et al19 used the mean signal intensities of eight rectangular regions covering the IVD in a mid-sagittal slice together with a shape feature (height to width ratio of IVD bounding box) and statistical texture features to detect herniated IVD from 2D MRI of the lumbar spine. Several advanced intensity and texture features extracted from a mid-sagittal MRI slice were also compared in Ghosh et al.20 First and second order texture features extracted from a segmented 2D MRI were evaluated by Michopoulou et al21 to quantify the degree of disc degeneration. Recently, Hao et al22 modeled the anatomical shape space of 2D IVD contours in T2-weighted images of the lumbar spine and defined a geodesic metric that is used for an automated classification.
To the best of our knowledge, all previous MRI classification approaches have been based on features extracted from 2D slices. Many signal intensity features have been proposed varying from raw intensity values to higher-order statistical texture characteristics, while elementary planar (2D) shape features such as disc height, width or their ratio have been presented. However, the degeneration affects the IVD morphology in 3D and precise quantification of the volumetric changes can provide novel tools to study and describe the degenerative processes.
Current study
In this paper, we extended on our previous work11 that involved automated analysis of 3D T2-weighted SPACE MRI scans of the lumbar IVD in asymptomatic individuals. The aims of the current work are: (1) to compare and validate manual and automated quantification of IVD morphology in a sample of subjects imaged with both 3D T2-weighted SPACE and standard clinical 2D T2-weighted turbo spin echo (TSE) sequences; (2) to evaluate our previously presented automated spine segmentation algorithm on routine clinical scans of patients with varying degree of IVD degeneration; and (3) to introduce and evaluate morphological (2D and 3D) and signal intensity classification features for automated detection of IVD abnormalities from T2-weighted MRI scans. The overreaching aim of this research is to develop an automated processing pipeline for analyses of IVD in MRI examinations with easily interpretable classification features applicable for wider use in clinical practice.4
Methods
Imaging database
The classification performance of proposed features was evaluated on two MRI datasets. The first dataset consisted of MRI scans from 28 apparently healthy subjects acquired using the T2-weighted 3D SPACE pulse sequence (details in table 1). Although all subjects self-reported as being asymptomatic at the time of imaging, signs of early degenerative IVD changes were identified by an experienced radiologist (DW) in 15 cases (17/140 lumbar IVD). A subset of 14 cases (69 IVD) was manually segmented (by AN, under supervision of CE) and used to build a statistical shape model (SSM) of lumbar IVD. Incidental radiological findings in these cases included magnetic resonance observed degenerative changes in seven IVD, Schmorl's nodes (vertebral endplate fractures with focal disc protrusion) and vertebral hemangiomas.
Table 1.
MRI acquisition parameters for the 3D SPACE and 2D TSE images
| Parameter | 3D T2-weighted SPACE | 2D T2-weighted TSE |
|---|---|---|
| Slices | 176 | 11–21 |
| Image matrix | 640×640 | 448×448 |
| Pixel spacing (mm) | 0.3437×0.3437* | 0.7142×0.7142 |
| Slice thickness (mm) | 1 | 3 |
| Slice spacing (mm) | – | 3.3 |
| TR (ms) | 1500 | 2790–4620 |
| TE (ms) | 131 | 105–113 |
| Echo train length | 167 | 19–25 |
*The image matrix is interpolated in the scanner from the acquisition of 0.7×0.7 mm.
Subjects at the University of Queensland were scanned using Siemens Trio 3 T system, patients in Heidelberg with Siemens Verio 3 T scanner.
2D, two dimensional; 3D, three dimensional; TSE, turbo spin echo.
The second dataset contained 16 2D T2-weighted TSE scans acquired in the sagittal plane (details in table 1). Eleven cases were patients presenting to the University Hospital of Heidelberg, Germany, for MRI investigation of symptomatic conditions of the lumbar spine. There was a variety of radiological findings (performed by DW) with respect to the IVD, including severe degeneration, IVD bulges and herniations (including cord impingement). The remaining five cases were follow-up scans of volunteers from the first dataset with previously identified IVD degenerative changes. Degenerative changes were identified (by DW) altogether in 53/93 IVD from the combined set of 16 scans.
The five follow-up scans (12 months) were used for validation of the proposed features and for comparisons between 2D SPACE and 3D TSE sequences. Four cases were apparently healthy asymptomatic individuals and one case (becoming symptomatic during the 12 months) had been diagnosed with posterior L4/L5 bulge. Incidental MRI findings by an experienced radiologist (DW) revealed at least one lumbar IVD with degenerative changes in every subject, with the total number equaling to 10 IVD (out of 29). The set of 29 lumbar IVD (T12/L1–L5/S1) were manually segmented in a mid-sagittal slice in both 3D and 2D scans independently by two experienced raters (Dr Mark W Strudwick and CE) and the manual ground truths were used to evaluate the inter-rater variability and the accuracy of the automated segmentation.
Image preprocessing and segmentation
All images were preprocessed using a customized intensity adjustment procedure based on the N4 bias field correction algorithm,23 which was applied to the region of the lumbar spinal column. Smoothing by anisotropic diffusion (15 iterations with a time step 0.01 and conductance 1.0) was applied next to reduce the acquisition noise. Image histogram normalization was then performed to standardize the intensity values by matching the histogram (extracted from the spinal column) to the histogram of an atlas image. The 2D clinical scans were reformatted using B-Spline image interpolation to an isotropic resolution (0.7142 mm).
An active shape model (ASM) based segmentation approach11 was used to extract the IVD shapes. The algorithm was previously applied to the SPACE images in dataset 1 and the segmentation accuracy was reported.11 Furthermore, the 3D SPACE and 2D TSE of five volunteers were manually segmented in the mid-sagittal slice by two experienced raters and the inter-rater variability and segmentation accuracy was evaluated using the Dice similarity coefficient24 (DSC) to evaluate the applicability of the method to 2D TSE scans:
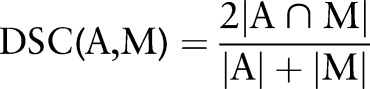 |
The DSC measures the overlap between automatic (A) and manual (M) segmentation masks.
Feature extraction
Two sets of morphological features were extracted from the automatically segmented IVD: planar (2D) measurements, similar to those previously reported,19 20 and 3D shape/volume characteristics (see below). Signal appearance features were extracted from histograms of signal intensities within the segmented IVD. The 2D morphological features were quantitatively evaluated against manual reference and both sets of morphological features were compared between 2D and 3D MRI scans. Finally, the features were analyzed for the ability to detect IVD degenerative changes automatically.
Planar measurements
There were three morphological features extracted from the planar IVD measurements: mid-sagittal IVD middle height (inferior–superior), mid-sagittal IVD middle width (anterior–posterior) and the height–width ratio (figure 2). The measures were extracted using triangular mesh of the automatically segmented IVD, in which four areas of interest were marked: anterior, posterior, inferior and superior central areas (figure 2). Mesh points from all areas were projected to the mid-sagittal slice and the IVD height was computed as the minimal distance between the superior–inferior points (similarly for the width). The computed values were compared (on five cases with 29 IVD) against manually extracted measurements (by AN under supervision of CE) using two-way analysis of variance (the factors are IVD level T12/L1–L5/S1 and the measurement method) and Spearman's rank correlation coefficient r was computed. Correlation of the features between 2D and 3D MRI scans was analyzed by Spearman's rank correlation coefficient r and two-way analysis of variance was used to investigate effects of MRI acquisition sequence (2D, 3D) and parameters (inter-slice gap, slice thickness, resolution) that influence the anatomy visualization, and consequently the extracted features.
Figure 2.
Automated location and measurement of the IVD height and width was done using the statistical shape model with four areas of interest. Points from the areas of the segmented IVD are projected to the mid-sagittal slice and the minimal distance between superior–inferior and anterior–posterior clouds are used as IVD height and width, respectively. Example manual measurements are illustrated by white lines. IVD, intervertebral disc. Access the article online to view this figure in colour.
3D shape parameters
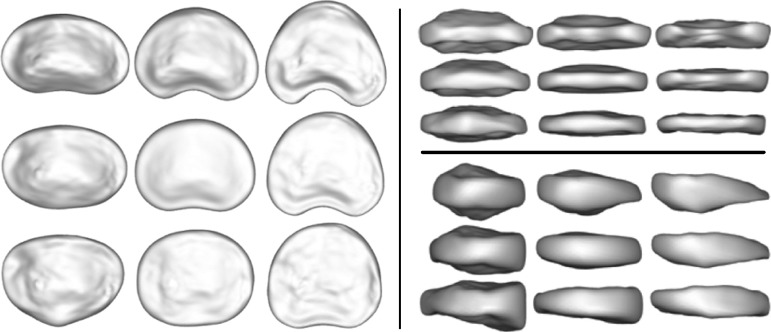
In our previous work, we used a dedicated algorithm incorporating a 3D SSM approach for automatic segmentation of the individual lumbar IVD.11 In brief, SSM describe statistical variations of objects in a training database that are represented by corresponding landmarks (mesh point coordinates in this case). Point distributions of the landmarks are analyzed, and modes of variation are computed by principal component analysis. A segmented IVD shape is projected on the SSM and weights of the first three principal modes of variation (b1, b2 and b3 also called shape parameters) are used as 3D shape features (figures 3 and 4A):
Figure 3.
The statistical shape model of lumbar IVD extracted from the 3D SPACE MRI scans. The mean shape (middle of each panel) and shapes generated at ± 3 SD are shown (left, superior view; top right, posterior view; bottom right, side view) for the first (abscissa axis) and second (ordinate axis) modes of variation. Both modes are associated with relative disc thinning, the second mode includes further information on anterior–posterior wedging of the disc. IVD, intervertebral disc; 3D. three dimensional.
Figure 4.
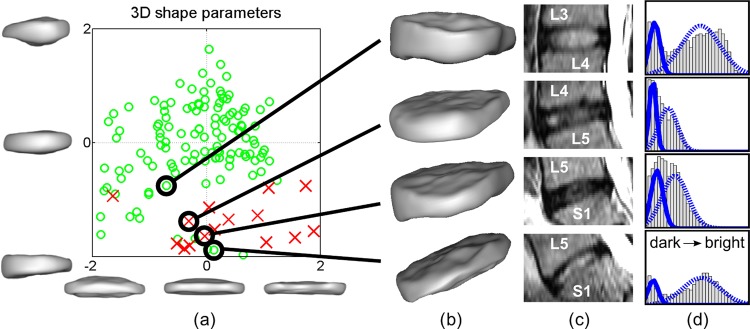
Projection of dataset 1 onto the first two modes of variation (±2 SD) is shown in (A). Most of the abnormal discs (red crosses) are located in the bottom right area, describing IVD narrowing (as can be seen in figure 3). Four individual example IVD automatic segmentations are shown in (B), corresponding MRI in panel (C) and the corresponding histograms with fitted distributions in (D). A progressive IVD space narrowing can be observed in (B) from top to bottom, which is reflected by the spatial relationship of the corresponding points in (A). The nucleus pulposus signal intensity is lower in the middle two IVD, as seen in (C), resulting in a compressed ‘single’ intensity peak in the histograms (D) demonstrating two close and overlapping Gaussian distributions. IVD, intervertebral disc; 3D, three dimensional. Access the article online to view this figure in colour.
where 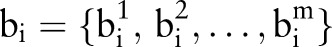 are the shape parameters of shape xi, x is the mean shape and P is the matrix of principal modes of variation. The SSM of lumbar IVD (T12/L1−L4/L5) and the first two modes of variation are shown in figure 3. Figure 4 shows the use of the SSM and the 3D shape parameters to quantify 3D IVD shape information and relative IVD shape changes automatically.
are the shape parameters of shape xi, x is the mean shape and P is the matrix of principal modes of variation. The SSM of lumbar IVD (T12/L1−L4/L5) and the first two modes of variation are shown in figure 3. Figure 4 shows the use of the SSM and the 3D shape parameters to quantify 3D IVD shape information and relative IVD shape changes automatically.
Signal intensity features
Disc degeneration is commonly associated with NP desiccation, observed as signal intensity attenuation in T2-weighted MRI. The degree of attenuation is a recognized sign of the degenerative change and an indicative marker within diagnostic procedures. Changes in magnetic resonance signal intensities can be well observed from image histograms. A histogram of a healthy disc is typically characterized by two distinctive intensity peaks, corresponding to the hypointense AF and brighter NP on T2-weighted MRI. In IVD with degenerative changes on MRI, the peak of brighter signal intensities will decrease and shift towards the lower peak (darker T2-weighted signal). To quantify the appearance of the discs automatically, we model the disc signal intensity histogram by a Gaussian mixture model of two normal distributions, corresponding to the signal intensities of the AF and NP. This model can fit and separate two distinct peaks in the histogram, and will result in two largely overlapping distributions if only one peak is present (figure 4D). Consequently, the degree of degeneration can be described by parameters of the two Gaussians (figure 4).
Histograms were extracted using IVD segmentation masks, eroded by 1 mm to eliminate extraneous surrounding tissue, and the Gaussian mixture model was fitted using the expectation maximization algorithm. Five appearance features were used in the experiments, corresponding to the two Gaussian means and variations and a Gaussian weight (of either the first or second peak).
Classification
In total, 11 features from three feature sets (planar measurements, 3D shape parameters, signal intensity features) were defined. Every feature was first evaluated separately on both datasets using two established classification techniques—linear discriminant analysis (LDA) and support vector machine (SVM) with linear kernel. The LDA classifier was chosen to reveal how linearly separable the data are, while the SVM is a significantly more advanced technique that allows a better estimate of performance and comparison among different features. Repeated (1000 times) stratified twofold cross-validation was performed by splitting both imaging datasets into a training (60%) and a cross-validation set (40%) while maintaining the ratio of healthy and degenerative discs in each. Next, all three feature sets were evaluated using the same strategy and the backward search feature selection with the area under the receiver operating curve (area under the curve; AUC) as the performance metric. Subsequently, the 2D and 3D morphological features were combined with signal intensity features (one at a time and both together). The performance among different feature sets was evaluated for statistical significance using the unpaired Student's t test. In all statistical tests, an effect was considered to be significant if the p value was 0.05 or less. The p values were not adjusted for multiple testing and interpretation of p values was explorative.
Results
Comparison against manual reference
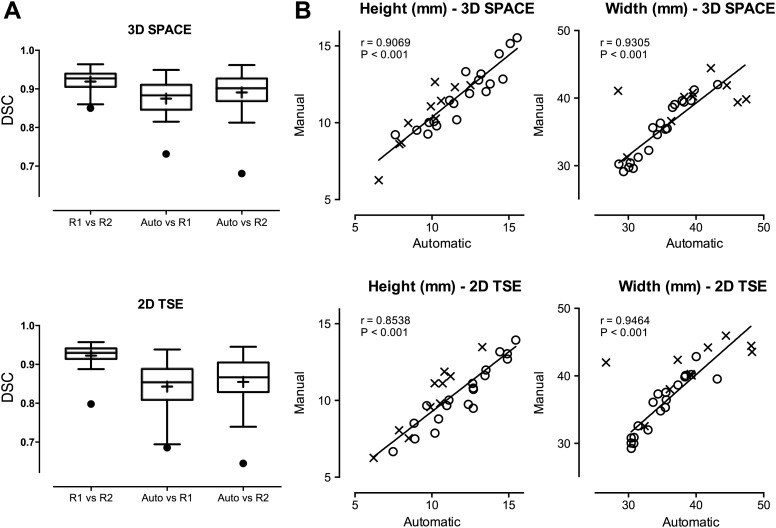
Manual and automatic segmentations for the 3D SPACE and 2D TSE obtained on five cases (29 IVD) are compared in figure 5. The inter-rater DSC is 0.916±0.031 and 0.923±0.029 (for 3D SPACE and 2D TSE, respectively), the DSC for manual versus automatic segmentations is 0.873±0.044 and 0.843±0.064 for rater 1, and 0.889±0.055 and 0.854±0.067 for rater 2 (3D SPACE and 2D TSE, respectively). Figure 5 presents a comparison of the automatically extracted IVD widths and heights against manual measurements. The main effect of the measurement method and interaction (IVD level and method), respectively, does not reach statistical significance for IVD heights in 3D SPACE images (p=0.7669, p=0.9929) but a statistically significant bias (1.01 mm) for the automated method was observed for IVD heights in 2D TSE (p<0.05). No statistically significant effect of the method or interaction was observed for IVD widths in 3D SPACE (p=0.6966, p=0.9561) or 2D TSE (p=0.4151, p=0.8984).
Figure 5.
DSC scores for inter-rater variability and accuracy of the automated segmentation algorithm evaluated on 29 IVD (R1, rater 1; R2, rater 2; Auto, automatic segmentation) are presented in (A). (B) Presents correlations of extracted IVD heights and widths (a healthy IVD is marked with a circle, abnormal with a cross). One outlier in IVD width assessment (top left cross) was removed from linear regression fitting and Spearman's r computation for IVD widths. This case presented particular anterior bulge and posterior herniation combined with adjacent vertebral body degeneration and challenged both manual and automatic assessment (the outlier marked in DSC box plots). DSC, dice similarity coefficient; IVD, intervertebral disc; 2D, two dimensional; 3D, three dimensional; TSE, turbo spin echo.
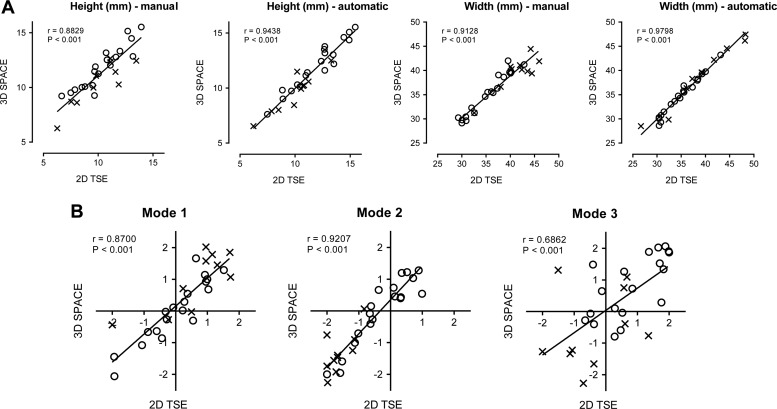
Reproducibility of all manual and automatic morphological features between both image types is presented in figure 6. There is a statistically significant effect of the magnetic resonance sequence on manually extracted IVD heights (p<0.05) with a bias of −1.07 mm. Effects of the imaging sequence or interaction do not reach statistical significance for IVD heights automatically extracted with our method (p=0.8494, p=0.9907). No significant effect was observed on IVD widths with either the manual (p=0.5331, p=0.9976) or automatic (p=0.9068, p>0.9999) methods. Finally, no statistically significant effect was observed on the three modes of SSM variation (p=0.4782, p=0.9850 for mode 1, p=0.2926, p=0.9981 for mode 2, p=0.7797, p=0.5928 for mode 3).
Figure 6.
Reproducibility of the morphological features (2D measurements in (A), 3D modes of variation in (B) between 2D and 3D MRI scans (a healthy IVD is marked with a circle, abnormal with a cross) with Spearman's rank correlation coefficient r. IVD, intervertebral disc; 2D, two dimensional; 3D, three dimensional; TSE, turbo spin echol.
Classification results
Reported results reveal very similar performance for both classification techniques (LDA and SVM) in all experiments. The performance of individual features is reported in table 2. The second mode of SSM variation (IVD thinning and anterio-posterior wedging, figure 3) is the best predictive feature for the classification of IVD abnormalities on the first dataset (AUC 0.95), followed by the mean and variance of the second (corresponding to NP signal) Gaussian (AUC both 0.93). The width to height ratio feature is next (AUC 0.86). The same four features perform best on the second dataset with the Gaussian mean of the NP signal first (AUC 0.90), followed by variance and the second mode of variation (AUC 0.88) and then the width to height ratio (AUC 0.86).
Table 2.
Evaluation (AUC) of individual features on both datasets (data 1, data 2)
| AUC | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Data 1 LDA | 0.762 | 0.716 | 0.866 | 0.668 | 0.950 | 0.646 | 0.437 | 0.479 | 0.604 | 0.933 | 0.931 |
| Data 1 SVM | 0.761 | 0.713 | 0.864 | 0.672 | 0.950 | 0.641 | 0.435 | 0.484 | 0.604 | 0.930 | 0.934 |
| Data 2 LDA | 0.795 | 0.650 | 0.862 | 0.758 | 0.879 | 0.686 | 0.783 | 0.538 | 0.499 | 0.907 | 0.880 |
| Data 2 SVM | 0.792 | 0.650 | 0.859 | 0.753 | 0.882 | 0.686 | 0.785 | 0.538 | 0.498 | 0.907 | 0.883 |
1, IVD height; 2, IVD width; 3, IVD width/height; 4, SSM mode 1; 5, SSM mode 2; 6, SSM mode 3; 7, Gaussian weight; 8, mean 1 (AF); 9, variance 1 (AF); 10, mean 2 (NP); 11, variance 2 (NP).
AF, annulus fibrosis; AUC, area under the curve; LDA, linear discriminant analysis; NP, nucleus pulposus; SSM, statistical shape model; SVM, support vector machine.
Classification results of three feature sets (2D and 3D morphological and signal intensity features) are summarized in tables 3 and 4 and include lists of selected features for each scenario. In the first dataset with 3D SPACE data (table 3), both the 3D-based signal intensity data and shape parameters deliver good classification results and clearly outperform the planar (2D) measurement features. The combination of 3D morphological features and signal intensity features results in a statistically significant improvement against the combination of signal intensity and 2D morphological features (LDA AUC 0.984 to 0.976 with p<0.0001, SVM AUC 0.978 to 0.964 with p<0.0001) or when the intensity features are used alone (LDA AUC 0.984 to 0.977 with p<0.0001, SVM AUC 0.978 to 0.964 with p<0.0001). Combination of all 11 features did not reach a statistically significant improvement over the combination of 3D morphological and signal intensity features (p=0.9843 for LDA, p=0.6800 for SVM).
Table 3.
Classification results on dataset 1 (AUC/accuracy/sensitivity/specificity)
| Features | Linear discriminant analysis | Support vector machine |
|---|---|---|
| A: Planar measurement | 0.870/0.811/0.678/0.829: {1, 2} | 0.860/0.798/0.668/0.815: {3} |
| B: 3D shape | 0.954/0.919/0.941/0.916: {4, 5} | 0.952/0.920/0.927/0.919: {4, 5} |
| C: Intensity | 0.977/0.887/0.942/0.879: {9, 10, 11} | 0.964/0.898/0.840/0.906: {9, 10} |
| A + C | 0.976/0.883/0.937/0.876: {9, 10, 11} | 0.964/0.919/0.906/0.922: {1, 2, 7, 10} |
| B + C | 0.984/0.945/0.969/0.942: {4, 5, 9, 10, 11} | 0.978/0.940/0.879/0.949: {4, 5, 7, 10} |
| A+B+C | 0.984/0.942/0.975/0.937: {2, 4, 5, 9, 10, 11} | 0.977/0.936/0.911/0.941: {1, 5, 6, 9, 10} |
Selected features are noted in the braces (feature index corresponds to table 2), the best results are highlighted in bold.
AUC, area under the curve; 3D, three dimensional.
Table 4.
Classification results on dataset 2 (AUC/accuracy/sensitivity/specificity)
| Features | Linear discriminant analysis | Support vector machine |
|---|---|---|
| A: Planar measurement | 0.859/0.773/0.771/0.777: {1, 2} | 0.860/0.767/0.757/0.780: {3} |
| B: 3D shape | 0.922/0.843/0.839/0.849: {5, 6} | 0.923/0.848/0.847/0.849: {5, 6} |
| C: Intensity | 0.978/0.921/0.937/0.900: {7, 8, 9, 10, 11} | 0.961/0.910/0.874/0.959: {8, 10} |
| A+C | 0.983/0.934/0.929/0.941: {1, 7, 8, 10, 11} | 0.972/0.930/0.901/0.968: {1, 2, 8, 10} |
| B+C | 0.988/0.930/0.943/0.913: {5, 7, 8, 10, 11} | 0.981/0.925/0.929/0.920: {5, 6, 8, 10} |
| A+B+C | 0.988/0.930/0.944/0.913: {5, 7, 8, 10, 11} | 0.981/0.923/0.924/0.923: {5, 6, 8, 10} |
Selected features are noted in the braces (feature index corresponds to table 2), the best results are highlighted in bold.
AUC, area under the curve; 3D, three dimensional.
In the second dataset (table 4), the signal intensity features alone deliver good classification results (LDA AUC 0.978, SVM AUC 0.961). The 3D shape parameters outperform the planar (2D) measurement features when used alone (LDA AUC 0.954 to 0.870 with p<0.0001, SVM AUC 0.952 to 0.868 with p<0.0001), and deliver a small but statistically significant improvement in combination with the intensity features for the LDA classifier (AUC 0.988 to 0.983 with p<0.05), and the SVM classifier (AUC 0.981 to 0.972 with p<0.0001). None of the 2D morphological features were selected from the combination of all 11 features.
Discussion
Feature evaluation against manual reference
The good accuracy (figure 5) of the automated segmentation (in light of the inter-rater variability involving manual segmentations) provides a promising basis for the use of the algorithm11 on both 2D and 3D MRI sequences. The higher variance of the automated method is partly due to the presence of one outlier and, in the case of the 2D TSE scans, to the slightly lower accuracy on some cases. The overall better results on 3D SPACE images are expected as the ASM was trained on these high-resolution data with a larger field of view, leading to potential segmentation imprecisions towards the margins of the field of view in 2D TSE scans. However, the morphological features extracted from these segmentations are strongly correlated with manual measurements, which provides a sound argument for further development of the automated approach. There was a bias between automatic and manual measurements for IVD heights in 2D TSE. This is likely to be a consequence of ASM training on SPACE data that, according to our experiment, bias the visualization and measurement of manually measured IVD heights. A strong correlation between 2D and 3D MRI scans was, however, found for all morphological features, with the exception of the third mode of variation. Investigations on larger datasets will be needed to assess further the relationship of morphological features between the acquisitions for both healthy and degenerative IVD. Possible sources of variation can include local morphological degenerative changes not captured by the 2D TSE scans with inter-slice gap, or bias of the segmentation algorithm trained on 3D SPACE images.
Classification
The more elaborate SVM technique delivers very similar results to LDA, suggesting that the classification boundary is well defined and the data reasonably separable with used features. Classification results show that both 3D-based morphological and signal intensity features contain important information for the correct detection of IVD degenerative signs. The results indicate that the second mode of SSM variation (feature 5) encodes core morphological information for classification on both datasets. This is highlighted by single feature classification analyses in table 2 and confirmed by feature selection procedures reported in tables 3 and 4. Similarly, the mean and variation of the second (NP) histogram Gaussian (features 10 and 11) are key signal intensity features for IVD degeneration classification. The combination of 2D morphological and signal intensity features never outperforms the 3D morphological and signal intensity features, and on the second dataset, all height, width and their ratio were excluded from the classification when all features were combined together. Confirmed by the explorative tests of statistical significance, the SSM-based 3D morphological features offer a clear advantage over simple planar measurements for IVD classification.
To our knowledge, this is the first time that a 3D shape has been used in the classification of IVD degeneration. Although it is difficult to compare classification experiments performed on different datasets with varying acquisition parameters or disease type and severity, our classification accuracy for the combined 3D shape and signal intensity features (0.945 for LDA and 0.940 for SVM on dataset 1, 0.930 for LDA and 0.925 for SVM on dataset 2) compares favorably to classification results presented by Alomari et al,17 who reported the accuracy of 0.913. Michopoulou et al25 achieved comparable accuracy of 0.94 in detecting degeneration of cervical IVD. The values are comparable to the accuracy of 0.949 reported by Ghosh et al20 who, however, focused solely on the detection of IVD herniation.
Our experiments suggest that the proposed automated 3D features relate well to the radiological (magnetic resonance) findings of IVD degenerative changes and achieve comparable accuracy as the state-of-the-art techniques. They are straightforward to interpret and offer novel insight for assessment of the IVD in 3D. This may provide a useful and clinically feasible approach for an efficient CAD for large-scale research and clinical studies.
Study limitations and future work
While the classification results obtained with the automated analysis are promising, an extended validation on a larger dataset and the correlation to clinical symptoms would be beneficial. A logical extension would involve analyses of a fuller dataset containing annotated cases with concomitant registration of lower back pain scores and quality of life scores, acquired with both 2D and 3D MRI sequences, which will allow distinguishing features suitable for the detection of early or advanced stages of IVD degeneration, correlations to clinical symptoms and comparison between 2D and 3D MRI scans on cases with different stages of degeneration. A bigger dataset with increased case numbers and pathological presentations will also allow opportunities for greater comparisons of larger feature sets previously proposed in the literature.
An interesting direction for ongoing investigations could be the combination of morphological features with signal intensity information from multi-modal MRI contrasts and biochemical MRI. Several biochemical magnetic resonance characteristics have recently been studied in the context of IVD degeneration (T2 or T1ρ relaxation times (collagen network and water content),6 26 27 diffusion MRI (extracellular space volume)28 or sodium MRI (glycosaminoglycan content)),29 which might provide new insights into the pathogenesis of the disease.
Conclusion
High-resolution 3D MRI of the spine with automated segmentations of IVD provide novel possibilities for CAD assessment of disc-related pathologies. In this paper, we have introduced an informatics system for automatic extraction and quantification of 3D MRI data of the IVD in the lumbar spine. The 3D morphological and signal intensity features, based on statistical shape analyses and intensity histogram modeling of lumbar IVD, were successfully applied to detect IVD abnormalities automatically. The results suggest that the 3D shape features contain salient information for the detection of degenerative changes, compared to the traditionally used planar morphological features. The results highlight the potential benefit of the presented method for studies concerned with automated quantification of IVD changes, and a straightforward interpretability of the proposed features supports their implementation within CAD systems for clinical practice to aid the processing of ever increasing amounts of radiological data. An automated quantification of IVD changes has the potential of a more accurate and reproducible assessment of IVD changes during follow-up and in relation to clinical symptoms, because, although spine pathology is ubiquitous and is encountered by nearly all medical specialties, the language used by radiologists to describe pathology may be complex and is not directly correlated to clinical symptoms.30 31
Acknowledgments
The authors would like to thank Aiman Al-Najjar from the Centre for Advanced Imaging, University of Queensland, Brisbane, Australia, for his assistance with magnetic resonance data acquisition and Dr Mark W Strudwick for performing manual segmentations.
Footnotes
Contributors: AN designed the study, implemented the software, participated in interpreting the results and drafted the manuscript. JF designed the study, contributed to the software implementation and acquisition of imaging data, participated in interpreting the results and critically revised and edited the manuscript. CE designed the study, participated in acquisition and interpretation of the imaging data, provided manual segmentation of the imaging data, participated in interpreting the results and critically revised and edited the manuscript. DW interpreted the imaging data, contributed to the literature review and participated in interpretation of the results. MAW contributed to the literature review, provided clinical data and critically revised and edited the manuscript. RS contributed to the study design and participated in acquisition and interpretation of the imaging data. SC designed the study, participated in interpreting the results and critically revised and edited the manuscript.
Funding: This research was supported under the Australian Research Council's linkage projects funding scheme LP100200422.
Competing interests: None.
Ethics approval: The Medical Research Ethics Committee of the University of Queensland approved the study. Datasets from Heidelberg University Hospital were fully pseudonymized and all identifying parameters were omitted.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Woolf AD, Pfleger B. Burden of major musculoskeletal conditions. Bull WHO 2003;81:646–56 [PMC free article] [PubMed] [Google Scholar]
- 2.Cousins JP, Haughton VM. Magnetic resonance imaging of the spine. J Am Acad Orthop Surg 2009;17:22–30 [DOI] [PubMed] [Google Scholar]
- 3.National Institute of Neurological Disorder and Stroke (NINDS) Low back pain fact sheet, 2008
- 4.Van Ginneken B, Schaefer-Prokop CM, Prokop M. Computer-aided diagnosis: how to move from the laboratory to the clinic. Radiology 2011;261:719–32 [DOI] [PubMed] [Google Scholar]
- 5.Modic MT, Ross JS. Lumbar degenerative disk disease. Radiology 2007;245:43–61 [DOI] [PubMed] [Google Scholar]
- 6.Auerbach JD, Johannessen W, Borthakur A, et al. In vivo quantification of human lumbar disc degeneration using T(1rho)-weighted magnetic resonance imaging. Eur Spine J 2006;15(Suppl. 3):S338–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts N, Gratin C, Whitehouse GH. MRI analysis of lumbar intervertebral disc height in young and older populations. J Magn Reson Imaging 1997;7:880–6 [DOI] [PubMed] [Google Scholar]
- 8.Pfirrmann CW, Metzdorf A, Elfering A, et al. Effect of aging and degeneration on disc volume and shape: a quantitative study in asymptomatic volunteers. J Orthop Res 2006;24:1086–94 [DOI] [PubMed] [Google Scholar]
- 9.Luoma K, Vehmas T, Riihimäki H, et al. Disc height and signal intensity of the nucleus pulposus on magnetic resonance imaging as indicators of lumbar disc degeneration. Spine 2001;26:680–6 [DOI] [PubMed] [Google Scholar]
- 10.Lichy MP, Wietek BM, Mugler JP, et al. Magnetic resonance imaging of the body trunk using a single-slab, 3-dimensional, T2-weighted turbo-spin-echo sequence with high sampling efficiency (SPACE) for high spatial resolution imaging: initial clinical experiences. Invest Radiol 2005;40:754–60 [DOI] [PubMed] [Google Scholar]
- 11.Neubert A, Fripp J, Engstrom C, et al. Automated detection, 3D segmentation and analysis of high resolution spine MR images using statistical shape models. Phys Med Biol 2012;57:8357–76 [DOI] [PubMed] [Google Scholar]
- 12.Emch TM, Modic MT. Imaging of lumbar degenerative disk disease: history and current state. Skeletal Radiol 2011;40:1175–89 [DOI] [PubMed] [Google Scholar]
- 13.Fries P, Runge VM, Kirchin MA, et al. Magnetic resonance imaging of the spine at 3 Tesla. Semin Musculoskelet Radiol 2008;12:238–52 [DOI] [PubMed] [Google Scholar]
- 14.Grenier J-M, Scordilis PJ, Wessely MA. Lumbar MRI Part 1: Normal imaging appearance of the lumbar spine. Clinical Chiropr 2005;8:205–15 [Google Scholar]
- 15.Pfirrmann CW, Metzdorf A, Zanetti M, et al. Magnetic Resonance classification of lumbar intervertebral disc degeneration. Spine 2001;26:1873–8 [DOI] [PubMed] [Google Scholar]
- 16.Alomari RS, Corso JJ, Chaudhary V, et al. Desiccation diagnosis in lumbar discs from clinical MRI with a probabilistic model. Int Symp Biomed Imaging 2009:546–9 [Google Scholar]
- 17.Alomari RS, Corso JJ, Chaudhary V, et al. Computer-aided diagnosis of lumbar disc pathology from clinical lower spine MRI. Int J Comput Assist Radiol Surg 2010;5:287–93 [DOI] [PubMed] [Google Scholar]
- 18.Alomari RS, Corso JJ, Chaudhary V, et al. Toward a clinical lumbar CAD: herniation diagnosis. Int J Comput Assist Radiol Surg 2011;6:119–26 [DOI] [PubMed] [Google Scholar]
- 19.Ghosh S, Alomari RS, Chaudhary V, et al. Composite features for automatic diagnosis of intervertebral disc herniation from lumbar MRI. Conf Proc IEEE Eng Med Biol Soc 2011:5068–71 [DOI] [PubMed] [Google Scholar]
- 20.Ghosh S, Alomari RS, Chaudhary V, et al. Computer-aided diagnosis for lumbar MRI using heterogeneous classifiers. 8th IEEE International Symposium on Biomedical Imaging 2011:1179–82 [Google Scholar]
- 21.Michopoulou SK, Costaridou L, Vlychou M, et al. Texture-based quantification of lumbar intervertebral disc degeneration from conventional T2-weighted MRI. Acta Radiologica 2011;52:91–8 [DOI] [PubMed] [Google Scholar]
- 22.Hao S, Jiang J, Guo Y, et al. Intervertebral disc shape analysis with geodesic metric in shape space. Int Conf Image Graph 2011:320–4 [Google Scholar]
- 23.Tustison NJ, Gee JC. N4ITK: Nick's N3 ITK implementation for MRI bias field correction. Insight J 2009:1–8 [Google Scholar]
- 24.Dice L. Measures of the amount of ecologic association between species. Ecology 1945;26:297–302 [Google Scholar]
- 25.Michopoulou SK, Boniatis I, Costaridou L, et al. Computer assisted characterization of cervical intervertebral disc degeneration in MRI. J Instrum 2009;4:P05022 [Google Scholar]
- 26.Mayerhoefer ME, Stelzeneder D, Bachbauer W, et al. Quantitative analysis of lumbar intervertebral disc abnormalities at 3.0 Tesla: value of T2 texture features and geometric parameters. NMR Biomed 2012;25:866–72 [DOI] [PubMed] [Google Scholar]
- 27.Berger R, Fenty M, Fry B, et al. Improving predictability of painful discs by using T1ρ MRI and disc height. Proceedings of International Society for Magnetic Resonance in Medicine (ISMRM) Scientific Meeting, 2011;420 [Google Scholar]
- 28.Beattie PF, Morgan PS, Peters D. Diffusion-weighted magnetic resonance imaging of normal and degenerative lumbar intervertebral discs: a new method to potentially quantify the physiologic effect of physical therapy intervention. J Orthop Sports Phys Ther 2008;38:42–9 [DOI] [PubMed] [Google Scholar]
- 29.Noebauer-Huhmann IM, Juras V, Pfirrmann CWA, et al. Sodium MR imaging of the lumbar intervertebral disk at 7 T: correlation with T2 mapping and modified Pfirrmann score at 3T – preliminary results. Radiology 2012;265:555–64 [DOI] [PubMed] [Google Scholar]
- 30.Fardon DF, Milette PC. Nomenclature and classification of lumbar disc pathology. Recommendations of the Combined Task Forces of the North American Spine Society, American Society of Spine Radiology, and American Society of Neuroradiology. Spine 2001;26:E93–113 [DOI] [PubMed] [Google Scholar]
- 31.Costello RF, Beall DP. Nomenclature and standard reporting terminology of intervertebral disk herniation. Magn Reson Imaging Clin N Am 2007;15:167–74; v–vi. [DOI] [PubMed] [Google Scholar]