Abstract
Objective
To develop, evaluate, and share: (1) syntactic parsing guidelines for clinical text, with a new approach to handling ill-formed sentences; and (2) a clinical Treebank annotated according to the guidelines. To document the process and findings for readers with similar interest.
Methods
Using random samples from a shared natural language processing challenge dataset, we developed a handbook of domain-customized syntactic parsing guidelines based on iterative annotation and adjudication between two institutions. Special considerations were incorporated into the guidelines for handling ill-formed sentences, which are common in clinical text. Intra- and inter-annotator agreement rates were used to evaluate consistency in following the guidelines. Quantitative and qualitative properties of the annotated Treebank, as well as its use to retrain a statistical parser, were reported.
Results
A supplement to the Penn Treebank II guidelines was developed for annotating clinical sentences. After three iterations of annotation and adjudication on 450 sentences, the annotators reached an F-measure agreement rate of 0.930 (while intra-annotator rate was 0.948) on a final independent set. A total of 1100 sentences from progress notes were annotated that demonstrated domain-specific linguistic features. A statistical parser retrained with combined general English (mainly news text) annotations and our annotations achieved an accuracy of 0.811 (higher than models trained purely with either general or clinical sentences alone). Both the guidelines and syntactic annotations are made available at https://sourceforge.net/projects/medicaltreebank.
Conclusions
We developed guidelines for parsing clinical text and annotated a corpus accordingly. The high intra- and inter-annotator agreement rates showed decent consistency in following the guidelines. The corpus was shown to be useful in retraining a statistical parser that achieved moderate accuracy.
Keywords: natural language processing, syntactic parsing, annotation guidelines, corpus development
Introduction
With the increasing deployment of electronic health record (EHR) systems, many institutions sit on vast amounts of clinical data that can contribute to diverse and valuable uses. Discrete data from EHRs have been widely used in driving healthcare operation, clinical research, and business reporting. On the other hand, free-text data in narrative clinical documents remains relatively under-utilized, because it is more challenging to extract the information from free-text data accurately and efficiently. With advancements in computational power and machine learning algorithms, modern natural language processing (NLP) technologies have benefited many productive applications such as information retrieval/extraction, document classification/summarization, machine translation, and question answering systems.1 As a result, much public attention and effort has been invested in transferring the success of NLP to the biomedical domain.
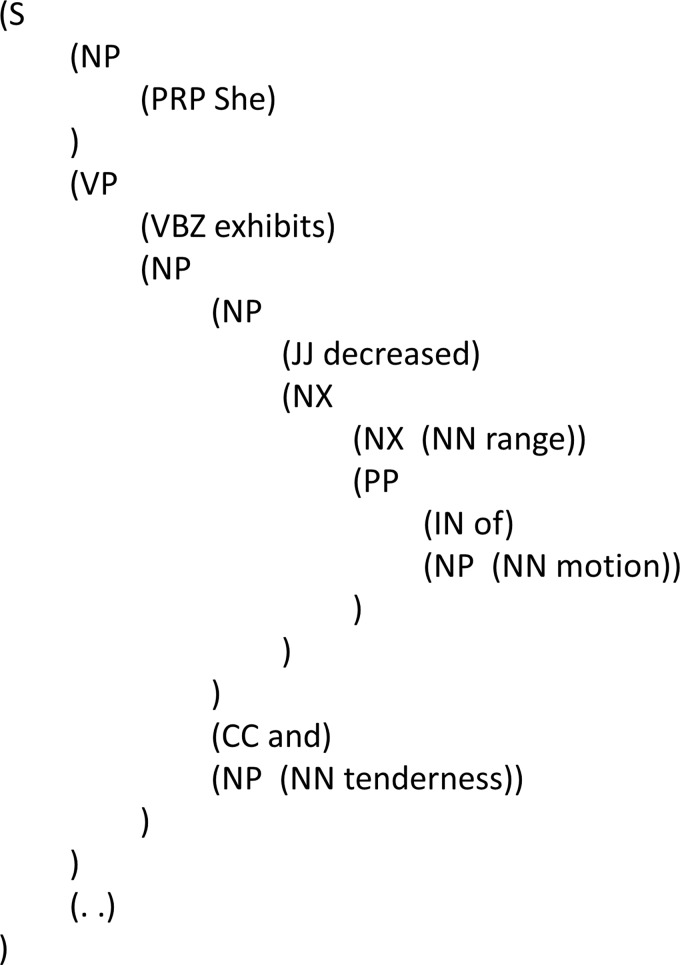
Among the prominent NLP analytics, syntactic parsing, which determines the structure of a sentence, is considered to be one of the most resource-demanding but also potentially high-yield tasks in the general English domain. Syntactic structures have been leveraged in biomedicine to extract coordination constructs (eg, ‘allopurinol 300 mg/day or placebo’) from randomized clinical trial reports,2 extract protein–protein interactions in research literature,3 identify noun phrases in clinical documents,4 and determine negation scope in radiology reports.5 Figure 1 demonstrates a parse tree for a sentence taken from a physical exam: ‘She exhibits decreased range of motion and tenderness’. Syntactically, ‘decreased’ should modify only ‘range of motion’ but not ‘tenderness’, and this reflects the intended semantic structure. A person with domain knowledge would not have trouble in determining the correct syntax, but parsing a clinical sentence like this example is still challenging for machines. A dedicated layer of syntactic representation that is based on common industry standards will be valuable training data especially when used in synergy with other complementary layers of annotation (eg, temporal, semantic, and discourse). It is expected that if we can make computers reproduce parses of comparable quality to humans, we will be able to develop numerous innovative and high-impact applications through the effective processing of unstructured big data in EHRs.
Figure 1.
Syntactic parse tree of an example clinical sentence. S, sentence; NP, noun phrase; PRP, personal pronoun; VP, verb phrase; VBZ, 3rd person singular present verb; JJ, adjective; NX, head constituent in complex NP; NN, singular or mass noun; PP, prepositional phrase; IN, preposition or subordinating conjunction; CC, coordinating conjunction.
Clinical NLP has emerged as a discipline with considerable history (see reviews by Spyns,6 Meystre et al,7 and Nadkarni et al8). However, there is still a dearth of studies that focus on syntactic parsing. This is likely due to the resource-intensive nature in establishing an infrastructure to support development of clinical text parsers—according to Albright et al's9 estimation, nominally ∼$100 000 is needed for annotating 13 091 sentences.
The limited availability of raw and annotated clinical texts, and lack of standards for annotations,10 contribute to the cost of efficiently developing syntactic parsers for the clinical domain and limit the capability of producing comparable, consistent results between systems. In contrast, general NLP parsers leverage large collaborative repositories of syntactically annotated sentences and well defined guidelines. This allows the parsers to produce accuracies in the low 90s.11 12 For example, the Penn Treebank project13 syntactically annotated sentences in the over one million word Wall Street Journal (WSJ) corpus and released the annotation guidelines to the public. Both the corpus and the guidelines have subsequently served as the basis for an enormous amount of research in parser development as well as annotation of new corpora. The clinical NLP community, while aware of the benefits of high-accuracy parsers, has focused primarily on semantic or even higher level annotation for specific applications.14–16 Initial attempts within the clinical community to create syntactic annotation guidelines and corpora have been limited. A recent noteworthy achievement by Mayo Clinic and its collaborating institutions was the development of the MiPACQ (Multi-source Integrated Platform for Answering Clinical Questions) clinical corpus with layered annotations and shared guidelines.9 17 However, there does not appear to be any existing approach that has adequately addressed the challenges in syntactic representation for ill-formed sentences, which are especially common in clinical narratives.
Another contributor to the difficulty in creating a clinical syntactic parser is the intrinsic properties of medical sublanguage.18 19 Similar to other domain-specific sublanguages, many clinical sentences are telegraphic, with missing subjects and/or verbs that usually result from the assumption that omission of self-evident information would not hinder understanding in context. Such sentences make interpretation difficult due to the abundant ambiguities involved and pose a challenge to existing parsers that are modeled over well-formed sentences. For example, the expression ‘completed acyclovir and rash better but still a stinging pain’ is actually a reduced form of ‘the patient completed acyclovir, and her rash got better, but she still has a stinging pain’. Filling in the gaps in such cases requires proper domain knowledge, without which the process of analyzing the syntactic structure and interpreting the meaning would be highly error-prone. This example also represents technical obstacles in applying generic guidelines that target mainly well-formed sentences to annotating fragments like ‘rash better’, where only the noun and the comparative adjective cannot be annotated as a grammatical sentence without accounting for the missing verb. These obstacles have discouraged people from pursuing the potentially fruitful track of parsing clinical text, but they nevertheless deserve the community's persistent investigation for a solution.
Despite the challenges summarized above, the benefit of developing/sharing syntactically annotated corpora for the clinical domain and pursuing linguistically sound annotation guidelines is clear. Ideally the annotations/guidelines should be compatible with commonly accepted style of syntactic annotation for general English (eg, that of the Penn Treebank) so that a great number of existing methods/tools can be reused. Our long-term objective is to facilitate research and development in clinical NLP, with the corpus constantly attracting exploration of questions and solutions. As an initial step the current study focuses on:
Adapting the Penn Treebank guidelines for parsing clinical text. While managing to be compatible with the Penn Treebank, the adaptation included special instructions for dealing with ill-formed sentences and provided real examples. The guidelines were designed to be self-contained and easy-to-follow for annotators with moderate linguistic/clinical background.
Annotating a clinical corpus by following the newly developed guidelines. Part of the annotation process was to support iterative refinements of the guidelines. Over several rounds, we evaluated the progressive inter-annotator agreement and addressed human feedback on the consistency and readability of the guidelines.
We report our experience/findings within a cross-institutional setting for guideline development and corpus annotation (using de-identified clinical notes from the 2010 i2b2/VA Clinical NLP Challenge16). Specifically, our iterative guideline development resulted in an inter-annotator agreement rate of 0.930 between Kaiser Permanente and Vanderbilt University. In addition to releasing this clinical Treebank (currently containing 1100 sentences), our work is the first one that applies Foster's computationally verified approach20 to annotating ungrammatical clinical sentences. To verify the usefulness of the corpus, we also used it to train a statistical parser and achieved an accuracy of 0.811 with 10-fold cross validation, demonstrating a significant improvement over the same parser trained on only non-clinical sentences. The guidelines and annotations are available for download, with instructions on how to load and visualize the syntactic parses. Through sharing it is hoped that more constructive inputs will be received and further collaborations will be formed to continue improving the resources.
Background
Syntactic parsing for natural language
Parsing, or formal syntactic analysis, has been a fundamental subject in studying natural language. As an essential step to truly understanding sentential meaning, parsing is performed to represent the structure of a sentence by annotating the relations among its components. There are two major paradigms in syntactic analysis: constituency (phrase structure) parsing and dependency parsing. The paradigms are basically complementary in terms of expressiveness, and automated methods are available for conversion from one to the other.21 The constituency paradigm is relatively popular in English, with a huge amount of annotations accumulated over more than a decade (eg, the Penn Treebank). An example parse in the Treebank style is illustrated in figure 1, following its latest guidelines22: the innermost layer of brackets denote the part-of-speech (POS) tags, which are lexical classes of the tokens and are usually obtained from a separate preprocessor before parsing. On top of the POS tags are the actual phrase-level constituent labels, which represent syntactic roles and imply the semantic scope of each role via the nested bracketing. The rich information in such sentential structure empowers NLP to perform various useful relation-based information extraction and inference tasks.
Importance of sharing annotated corpora: the Penn Treebank model
The influence of the Penn Treebank has lasted for almost two decades because it provides abundant value-added annotations to facilitate data-driven linguistics. One could argue that corpus linguistics already existed (eg, work by Harris23) long before modern extensive digitization and that computational linguistics would still deliver significant impact by using raw corpora (eg, through unsupervised learning24). However, it was not until the availability of large-scale, computable, annotated corpora that we started to benefit immensely from:
Quantifying our qualitative linguistic knowledge in more systematic and efficient ways.
Manipulating the behavior of computers more effectively in making them reproduce our interpretations (annotations).
The first benefit may inevitably come with certain implicit subjectivity in any annotation due to the targeted usage, but such subjectivity should be considered acceptably realistic in data science and does not dwarf the actual benefits. The second benefit has been strengthened by advancements in (symbolic and statistical) machine-learning techniques and hardware performance over the past decades.
We can use the number of scholarly citations to the Penn Treebank as a rough estimate of its influence in facilitating NLP research and development. According to Google Scholar25 there have been 4036 publications (as of September 21, 2012) directly citing the seminal article by Marcus et al13 in 1993, which on average equals about 200 publications per year. Topics of these citing publications are diverse, including automated sentence parsing, semantic role labeling, question answering, discourse processing, and measuring syntactic complexity in psycholinguistics. Another example is Treebanking of the GENIA corpus,26 which contains annotated sentences from biomedical literature and has been cited 72 times (as of September 21, 2012) since 2005. Apparently we see that once the linguistically annotated corpora have been released to the public, over time they steadily attract users searching for solutions and/or questions from the data. Both the Penn Treebank and the GENIA corpus have indeed pushed forward the NLP state-of-the-art in their respective as well as related domains. Our previous study on sharing clinical NLP resources also suggested that collaborating institutions can benefit more from developing and sharing corpora based on a common and coherent set of annotation guidelines, rather than from sharing exclusively pre-packaged programs or pre-trained models.27
Current status and challenges in parsing clinical text
Full sentence parsing of clinical text is generally considered implausible and impractical, primarily due to the prevalence of syntactically ill-formed expressions in such text. To elude the challenge, many existing clinical parsers rely on semantic grammars (or even simpler templates)28–30 that are often manually crafted by domain experts and only perform shallow parsing (ie, obtaining phrasal units). Despite its success, one issue with the semantics based approach is that it usually involves proprietary class/role/relation labels, which are not always universal and therefore can limit interoperability across systems. As a result, those proprietary solutions have not addressed the missing piece in establishing a data-driven, sustainable model exemplified by Penn Treebank, especially in terms of reusable common corpora and unified symbolic representation/guidelines.
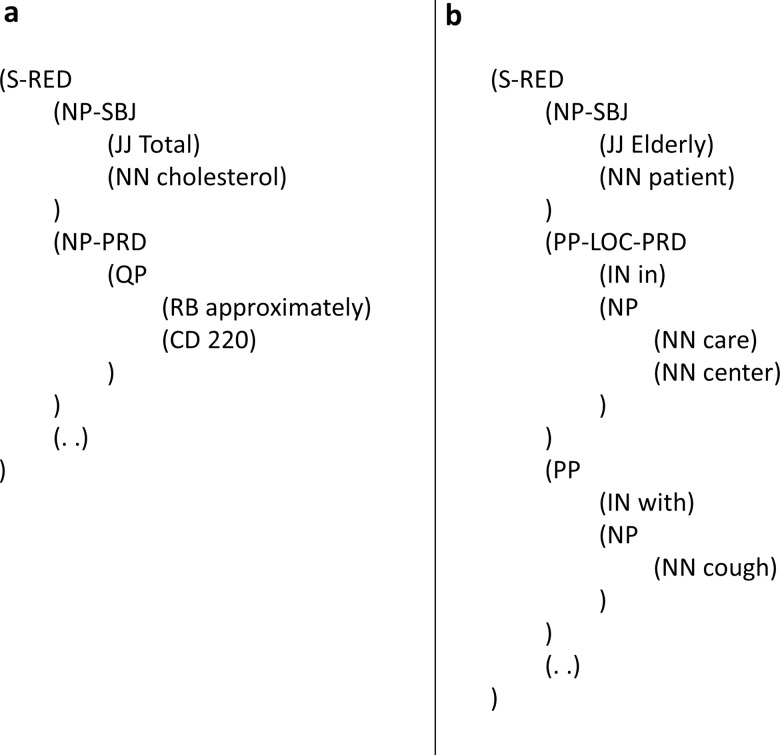
There is nevertheless a general awareness of this data gap in the clinical NLP community, and there has been increasing effort to create annotated corpora for the clinical domain. Among the most well-organized endeavors are the SHARP projects31 by Mayo Clinic and its collaborators, whose NLP team has recently released a set of linguistic annotations/guidelines for clinical text (the MiPACQ corpus).9 Their guidelines specifically include a well-thought clinical addendum17 to the original Penn Treebank document, with an inter-annotator agreement rate of 0.926. The addendum's main solution to handling ungrammatical sentences is the introduction of a new function tag RED (REDuced) to denote that a sentence has missing elements and requires marking phrase-level function tags such as SBJ (subject) and PRD (non-verbal predicate) to represent the implied syntactic relations. Using examples from the addendum, figure 2 shows how sentences with a missing verb can be annotated by function tagging. In figure 2A the PRD tag helps indicate the second noun phrase ‘approximately 200’ as the subject complement linked by a missing copular verb ‘is’; similarly in figure 2B the PRD marks the first prepositional phrase ‘in care center’ as a complement linked by the missing ‘is’. However, there are potential limitations with the specific function tagging approach:
Although a parse like figure 2A appears syntactically correct with the additionally annotated grammatical roles, the annotation yields an undesired grammar rule at the constituent (phrase structure) level, whereby a sentence is formed with just two consecutive noun phrases (without any verb) and a period. An implication of having only noun phrases form a sentence is the increased ambiguity in parsing, since bare noun phrases are generally parsed as part of a larger noun phrase (not a sentence). Thus, more complicated models would be needed for learning how to disambiguate (namely, between an NP and an S parse) and automatically assign the function tags.
Role-based function tagging falls short when defining effective phrase scope in structures such as parallel prepositional attachments. In figure 2B (with the reading ‘Elderly patient in care center with cough’) apparently only one predicate is allowed per sentence, and the first prepositional phrase ‘in care center’ has occupied the PRD tag as associated with the predicate role. The annotation, however, remains inaccurate because it does not distribute the predicate's scope over the second prepositional phrase, leaving ‘Elderly patient is with cough’ unrepresented.
While the function tagging guidelines specify how missing elements like subject noun phrases, copulas, and auxiliaries are handled, they are not clear about how other frequently missing elements (eg, prepositions) are to be handled in general.
Figure 2.
Sentences with missing elements, parsed by following (and excerpted from) Warner et al.'s guidelines.17 S, sentence; RED, function tag for a sentence with missing element(s); NP, noun phrase; SBJ, function tag for subject of a sentence; JJ, adjective; NN, singular or mass noun; PRD, function tag for a non-verbal predicate in sentence; QP, quantifier phrase; RB, adverb; CD, cardinal number; PP, prepositional phrase; LOC, function tag for locative expression; IN, preposition or subordinating conjunction.
The MiPACQ corpus annotated with the function tags was used to retrain a dependency parser and achieved a highest labeled attachment score of 0.836. Although using a dependency model has the advantage of eluding limitations in the constituency annotations, it does not resolve core issues such as 2 above—inaccurate phrase scoping (ie, the model cannot rectify what has been inadequately annotated). In summary, while clinical NLP is moving promisingly towards sharing syntactically annotated text, there still remain fundamental knowledge representation challenges that are not addressed and deserve continuing pursuit of linguistically sound guidelines.
Alternative solutions to handling ungrammatical sentences
Ungrammatical sentences as a linguistic phenomenon are not restricted to clinical texts. Therefore, it is reasonable to look for existing work in general or specialized NLP domains that already addresses the challenge or at least offers insight into addressing it. Originally Treebank had the FRAG constituent for annotating any clause that lacks essential elements and is therefore deemed unparsable.22 Since FRAG annotates a fragment ‘as is’ without attempting to restore grammatical structure, we actually want to minimize its use and search for a more constructive annotation approach that can represent the implicit syntax of a fragment. A closely related language phenomenon is disfluency, which refers to filled pause, repetition, speaker self-correction, etc. in spontaneous speech.32 However, the existing guidelines33 for annotating disfluent expressions are not suitable to our goal, as (1) many ungrammatical disfluencies result from dialog interruptions and are self-corrected by the speaker subsequently, which exhibit different nature from most clinical narratives, and (2) the special tags help mark up comprehensive types of disfluency but are not meant to restore grammatical structures that are useful to post-processing programs. For German language, guidelines were developed and applied to create the SINBAD corpus, a collection of syntactically annotated ungrammatical sentences.34 The applicability of the SINBAD guidelines to English has not been reported, and it is not clear if computer parsers could be trained from the proposed annotation schema. There have also been principles and algorithms proposed to deal with ill-formed sentences when parsing.35 36 Instead of creating offline gold annotations with properly restored sentence structures, they focus on enhancing a parser's run-time robustness by enabling extraction of correct semantic representation regardless of syntactic deformity or by preventing failure on extra-grammatical (outside of grammar) sentences. The run-time strategies are on a different track from addressing the representation issues in creating gold standard annotation for ill-formed sentences.
The most pertinent solution we came across in our survey was the work by Foster,20 which involved: (a) systematically infusing ungrammatical elements into Penn Treebank parses; (b) creating corresponding annotation guidelines and gold standard annotations; and (c) training/testing state-of-the-art statistical parsers with the combined annotations (ie, both grammatical and ungrammatical). In other words, the study was meant to develop error-handling guidelines and assess the learnability of the annotations by computer parsers. The proposed strategy in handling common ungrammatical elements includes the following:
Respect the intended meaning of misspelled words. An example from Foster's paper: the ‘in’ should be treated as a verb for parsing ‘A romance in coming your way’ to avoid an incorrect syntactic structure for the original sentence.
Absorb any redundant word into a nearby phrase that minimizes distortion of the original intended syntactic structure. For example, ‘A the romance is coming your way’ should be parsed with ‘A the romance’ grouped as a single noun phrase.
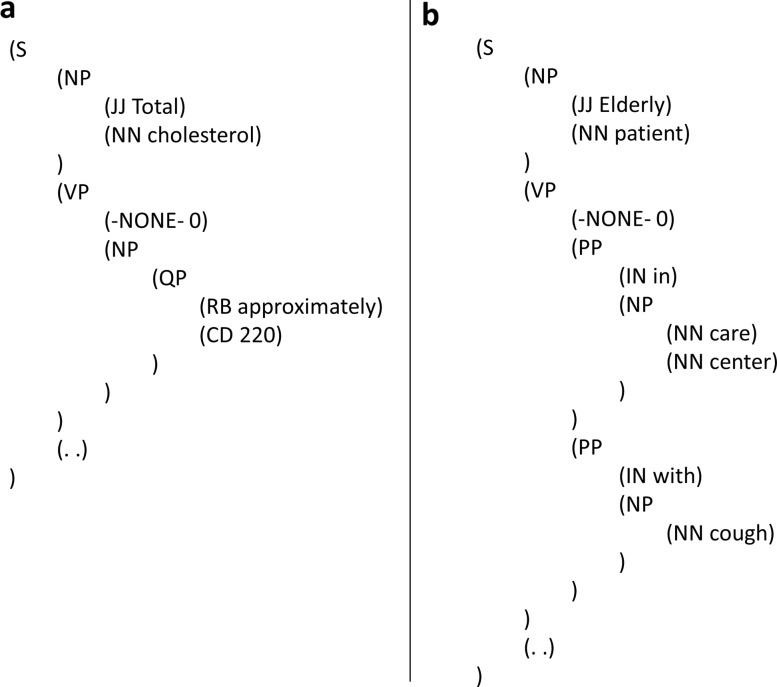
Insert a null element 0-NONE- for an inferred missing word to reconstruct the intended syntactic structure. For example, applying this approach on the sentences in figure 2, we obtained the parses illustrated in figure 3. It can be seen that denoting the missing verb as 0-NONE- not only helps reconstruct the intended grammatical structure with a noun phrase and a verb phrase (both in figure 3A and B) but also helps represent the scope of prepositional phrases properly within a single verb phrase predicate when needed (as in figure 3B).
Figure 3.
Sentences with missing elements, parsed by following Foster's approach.20 S, sentence; NP, noun phrase; JJ, adjective; NN, singular or mass noun; VP, verb phrase; -NONE-0, null element for missing word; QP, quantifier phrase; RB, adverb; CD, cardinal number; PP, prepositional phrase; IN, preposition or subordinating conjunction.
Foster's approach appears intuitive and linguistically sound to us for parsing ungrammatical sentences into constituency structure, which is fundamental to Treebank-style annotations and associated applications. Additionally, Foster reported an accuracy of 0.881 in F-measure achieved by the best-performing parser model37 retrained with a mixture of grammatical and ungrammatical sentences, constituting a small but statistically significant improvement over the parser trained only on grammatical sentences. Therefore, while we believe there is room for improvement in this approach, we considered it to be promising for annotating clinical sentences and incorporated it into our annotation guidelines.
Methods
Dataset selection and preprocess
We used progress notes from the University of Pittsburgh Medical Center (UPMC) distributed in the 2010 i2b2/VA Clinical NLP Challenge.16 A physician (RML) manually reviewed the progress notes and selected 25 of them by identifying those with a specialty of general medicine and precluding those that apparently contained copy-pasted redundant information. To eliminate confounding errors in steps prior to parsing, a linguist (EWY) performed manual tokenization and POS tagging for the 25 notes, with consulting a physician on cases that required domain knowledge. A noteworthy benefit of our manual tokenization is grouping of the special de-identification labels such as ‘**AGE[in 80s]-year-old’ to avoid artificial fragmentation that would hurt POS tagging and parsing. Based on the gold standard tokenization, we automatically sampled sentences with at least three tokens of length for this study. Before distributing the notes for manual parsing, we pre-parsed them by using the Stanford Parser38 (V.2011-09-14) with a general PCFG model (trained from WSJ and English of some other genres) that came in the official package.
Annotation guideline and corpus development
We planned and executed four major stages in developing our annotation guidelines along with iteratively refining the deliverable corpus (see figure 4 for flow chart):
Creating the guidelines: Based on manual analysis of institution-specific notes other than the 25 notes selected for this study, two linguists (RP and EWY) drafted a preliminary set of clinical parsing guidelines adapted from the Penn Treebank.22 We applied the error handling strategy introduced by Foster20 and found, specifically, inserting null elements for ignored (or missing) words to be helpful in restoring the proper syntax of ungrammatical clinical sentences. The guideline creation was not only knowledge-driven but also involved annotating real clinical text. We used the application WordFreak39 for performing our manual annotation. In the initial batch, both the Kaiser Permanente and Vanderbilt teams annotated 6 of the 25 notes (with physician consultation for domain-specific interpretation) and discussed/adjudicated the disagreements. After each round the draft guidelines were revised accordingly to address the discovered issues.
Fine-tuning the guidelines and training the annotators: In the second stage, we performed another three rounds of annotation to train two human annotators and to fine-tune the guidelines. For each round we randomly sampled 150 sentences from the remaining 19 notes (not used in the first stage) and had both the linguist and a computer scientist (MJ) annotate them by following the guidelines and consulting physicians whenever needed. The Evalb program (the version included with the Stanford Parser) was used to compute the inter-annotator agreement rate based on F-measure.40 The annotators met to adjudicate differences and discuss applicability of the guidelines. These three rounds with 450 sentences were meant to facilitate convergence among the annotators and the guidelines (by revising any instructions that were considered problematic).
Testing the guidelines: To test the learnability/stability of our guidelines, we randomly sampled another 216 sentences (intended to round up the deliverable corpus as 1100 sentences) from the 19 notes and had both annotators annotate them. An agreement rate was computed to evaluate whether the annotators could generate consistent annotations by following the latest guidelines. Additionally, to investigate the potential bias introduced by pre-parsing with the Stanford Parser, we had the linguist annotate two of the six stage-one notes (interrupted with a wash-out period of a half year) directly from the POS-tagged sentences. The linguist's self-agreement rate (between her annotations using the pre-parsed versus those using only the POS-tagged) was computed to evaluate whether the consistency holds with/without the factor of pre-parsing.
Finalizing the deliverable corpus: After the agreement rate was assessed to be satisfactory in the last round, the annotators met to discuss/resolve any remaining disagreements and then the linguist applied the latest guidelines in preparing a final version of the deliverable corpus.
Figure 4.
Flow diagram of the guideline and corpus development process.
Analysis of the annotated corpus
Quantitatively, we computed descriptive statistics with regard to distribution of the different types of syntactic constituents in the parsed corpus. We also extracted from our annotations the most prevalent grammar rules that involved insertion of null elements to restore incomplete sentence structures. Qualitatively, we analyzed and summarized the features of the annotations (guidelines), with emphasis on our domain-specific adaptations. Operationally, we retrained the Stanford Parser with our annotated corpus and reported parsing accuracy based on 10-fold cross validation.
Results
Annotation consistency in following the guidelines
Inter-annotator agreement rates were computed to measure the consistency between the institutions in following the guidelines. Table 1 shows the progressive rates from the three iterations of guideline tuning to the final independent testing. We can see the agreement rates climbed steadily over the tuning iterations and culminated to 0.930 in the final testing. The final agreement rate was high and close to the 0.926 reported by Albright et al9 in Treebanking the MiPACQ corpus. The intra-annotator agreement rate of the linguist in annotating from Stanford Parser pre-parsed sentences versus from POS-tagged sentences only was 0.948, with 0.791 perfectly self-agreed parse trees. These results indicate that the annotators were able to perform syntactic parsing with decent consistency by following the guidelines.
Table 1.
Inter-annotator agreement rates in guideline tuning and final testing
| Tuning 1 | Tuning 2 | Tuning 3 | Final testing | |
|---|---|---|---|---|
| Number of sentences | 150 | 150 | 150 | 216 |
| Agreement rate | 0.872 | 0.887 | 0.903 | 0.930 |
| Proportion of perfectly agreed parse trees | 0.633 | 0.660 | 0.693 | 0.713 |
Features of the developed guidelines
Our annotation guidelines are based on the original Bracketing Guidelines for Treebank II Style,22 with modifications to accommodate the properties of medical sublanguage. Several noteworthy adaptations are summarized in the following. For further details, the reader is referred to our guideline's content (see online supplement 1).
Handling missing elements: As described in the Background section, we adopted Foster's approach20 in annotating sentences with omitted words. We insert a null element 0-NONE- for the inferred missing word to restore the intended syntactic structure. A null element can represent any lexical category (eg, verb, noun, preposition) that is grammatically essential or required for deriving the structure of the sentence. The null element is further made to project to the phrasal level with the appropriate constituent label, such as NP (noun phrase), VP (verb phrase), and PP (prepositional phrase), that would have been assigned for the corresponding word, if explicit. For example, in figure 3A the 0-NONE- is inserted to represent the missing verb ‘is’, so that this sentence is restored with a VP and reanalyzed as grammatical. However, grammatically allowed omissions are not considered as missing elements, such as the omission of the relative pronoun in a relative clause (eg, ‘The patient I saw yesterday’ is still grammatical without the relative pronoun ‘who’). In general, we found that this approach was conceptually intuitive and provided flexibility in reconstructing fragments as complete sentences. Note that the guidelines still allow using FRAG when the attempt of reanalyzing any expression into a natural-sounding grammatical sentence fails. This policy respects expressions that are originally not meant to be sentences (eg, section header such as ‘Physical Exam:’) and avoids aggressive re-authoring of fragments that have too many missing elements (eg, the policy does not restore ‘Fe 53 μg/dL Low’ fully into ‘Fe was 53 μg/dL, which was Low’).
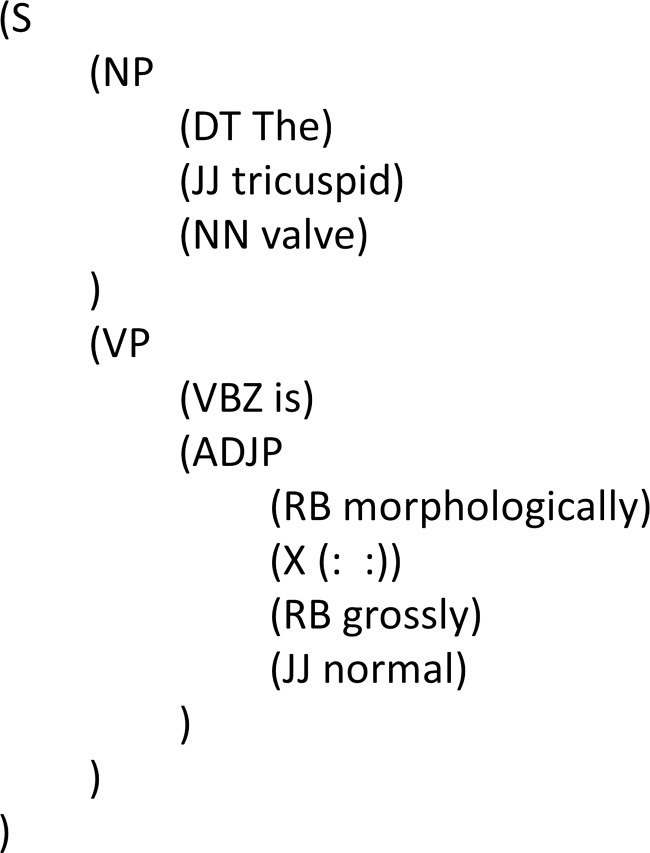
Superfluous and redundant elements: Superfluous and redundant elements that cannot be accommodated in a sentence or phrase structure are annotated with the constituent X, which could be anything such as punctuation marks, symbols, and words. Figure 5 shows a colon ‘:’ as being redundant and bracketed with X inside an ADJP (adjective phrase). This approach aligns with Foster's suggestion on avoiding interruption to the intended syntactic structure.
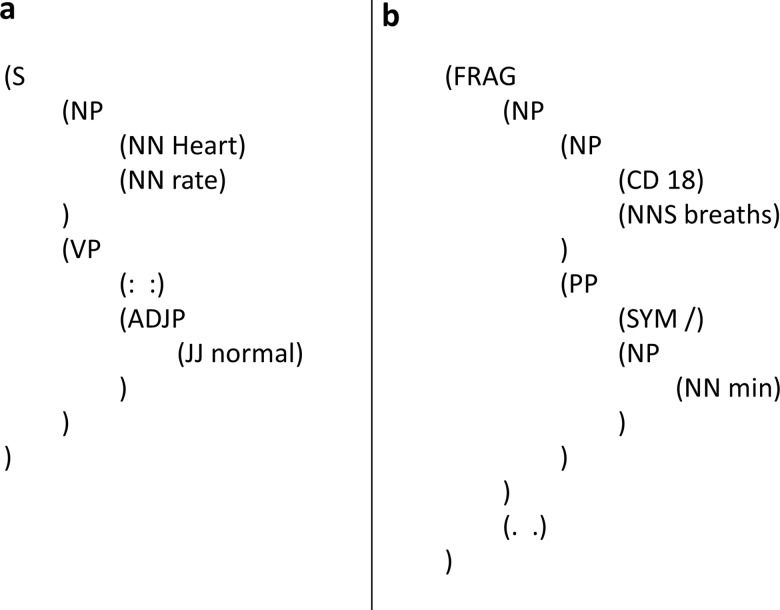
Symbols with inferable syntactic roles: ‘Header: value’ expressions occur frequently in clinical notes. Some of them can be legitimately annotated as complete sentences if the colon is interpreted as a verb. In such cases, the colon is allowed to project a verb phrase while its POS tag should still remain ‘:’. Figure 6A shows such an expression parsed into a sentence. Figure 6B demonstrates a similar example, where ‘/’ is POS-tagged as SYM (symbol) but interpreted as the word ‘per’, therefore projecting a PP (prepositional phrase) in the parse.
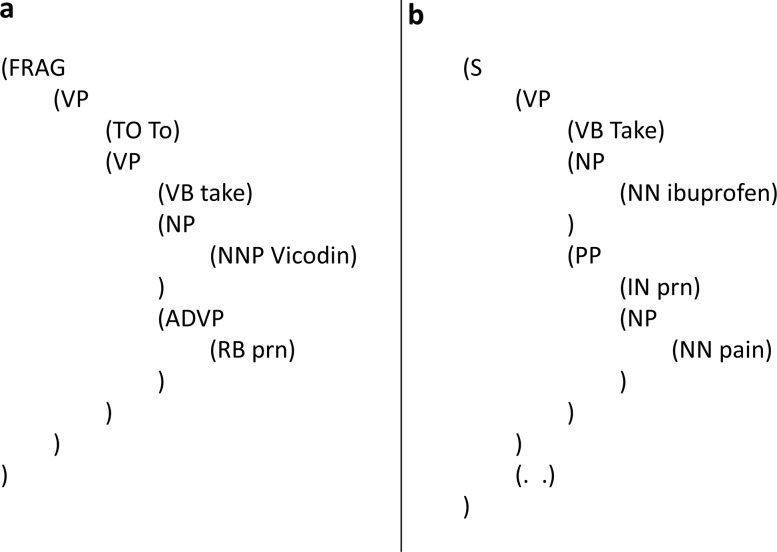
Interpreting syntactic roles of Latin abbreviations: Instead of using FW (foreign word) for Latin abbreviations, we try to infer their functioning POS tags and syntactic roles. Figure 7A shows that ‘prn’ (pro re nata) is interpreted as an adverb meaning ‘when necessary’. Similar adverbial usage includes medication frequency such as ‘qid’ (quarter in die), meaning ‘four times a day’. Figure 7B shows a different context where ‘prn’ is interpreted as a preposition and stands for ‘required if in condition of’. Such an approach is based on the motivation of infusing proper lexical semantics in deciding the syntactic structure, rather than forcing any meaningful abbreviation into an indistinguishable FW.
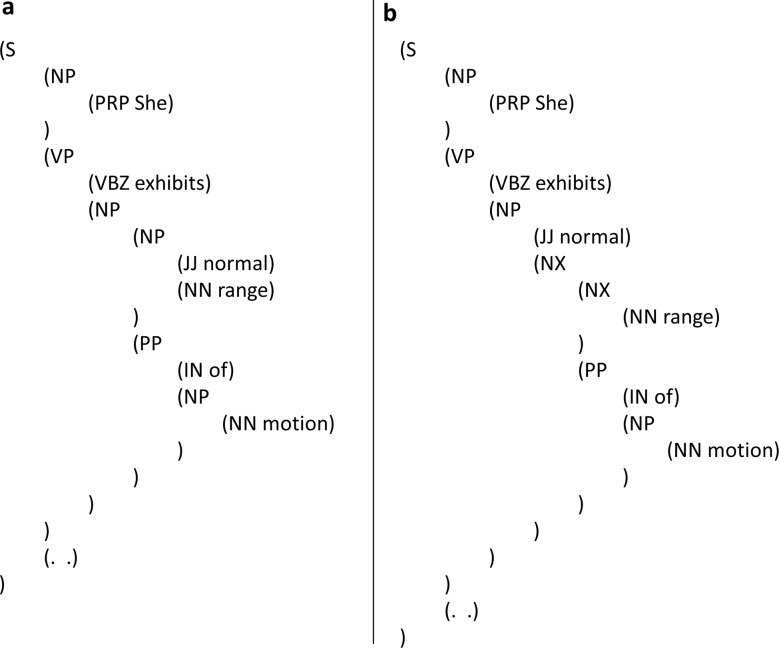
Respect domain-specific semantic structure of complex phrases: We try to accurately represent the internal semantic structure of medical expressions in annotating the constituents. When there are alternative grammatical parses, the one that better captures the intended meaning is preferred. Figure 8A shows a domain-neutral and grammatical parse where ‘range of motion’ is not treated with semantic pre-coordination. On the other hand, figure 8B is a parse that treats ‘range of motion’ as a domain-specific semantic chunk. Corresponding to such pre-coordination, the constituent NX is especially used to mark ‘range’ being the head of the noun phrase ‘range of motion’, which as a consolidated unit, projects into a single NX and is modified by the adjective ‘normal’.
Figure 5.
A sentence with redundant element, parsed by our guidelines. S, sentence; NP, noun phrase; DT, determiner; JJ, adjective; NN, singular or mass noun; VP, verb phrase; VBZ, 3rd person singular present verb; ADJP, adjective phrase; RB, adverb; X, unknown/uncertain element.
Figure 6.
Sentences with role-inferred symbol, parsed by our guidelines. S, sentence; NP, noun phrase; NN, singular or mass noun; VP, verb phrase; ADJP, adjective phrase; JJ, adjective; FRAG, fragment; CD, cardinal number; NNS, plural noun; PP, prepositional phrase; SYM, symbol.
Figure 7.
The Latin abbreviation ‘prn’ parsed with different syntactic roles. FRAG, fragment; VP, verb phrase; TO, the word ‘to’; VB, base form verb; NP, noun phrase; NNP, singular proper noun; ADVP, adverb phrase; RB, adverb; S, sentence; NN, singular or mass noun; PP, prepositional phrase; IN, preposition or subordinating conjunction.
Figure 8.
Parsing ‘range of motion’ with/without considering domain semantics. S, sentence; NP, noun phrase; PRP, personal pronoun; VP, verb phrase; VBZ, 3rd person singular present verb; JJ, adjective; NN, singular or mass noun; PP, prepositional phrase; IN, preposition or subordinating conjunction; NX, special constituent used with certain complex NPs to mark the head.
Descriptive and operational analysis of the annotated corpus
The annotated corpus contains 1100 sentences, with a median length of eight tokens per sentence. Table 2 shows the distribution of syntactic constructs (constituents) in our annotated corpus, aligned side by side with that in an arbitrarily selected WSJ sub-corpus (the 00 section) of 1921 sentences. Constituents marked with an asterisk (*) symbol in the first column indicate statistical significance (p<0.05) of the difference between the two corpora. For example, the 7.65% of FRAG in the clinical genre (with many telegraphic fragments) is understandably higher than the 0.21% in news text, while the more rhetorical SBAR structure is relatively rare in clinical text (1.42% vs 3.93%). We also automatically extracted all the grammar rules from the annotated parse trees, and table 3 shows a list of those rules that involve restoration of missing elements by inserting a 0-NONE- node. For example, we can see the most frequent rule involving element restoration is VP→-NONE- NP, as extracted from a parse like figure 3A (restoring ‘is’ of the verb phrase for the intended meaning ‘is approximately 220’). As summarized in the guideline features above, we still left out some expressions that did not qualify for restoration as sentences. For example, most section headers were annotated as fragments composed of a noun phrase followed by a colon (ie, FRAG→NP:).
Table 2.
Constituent distribution in the annotated clinical corpus with comparison to a newspaper corpus
| Constituent label | Constituent description | % in our annotated clinical corpus | % in a subset of the WSJ corpus |
|---|---|---|---|
| NP* | Noun phrase | 40.00 | 42.37 |
| VP* | Verb phrase | 16.85 | 19.68 |
| S | Sentence | 12.18 | 12.93 |
| PP* | Prepositional phrase | 8.56 | 12.74 |
| FRAG* | Fragment | 7.65 | 0.21 |
| ADJP* | Adjective phrase | 4.51 | 1.66 |
| ADVP | Adverb phrase | 2.87 | 2.57 |
| NX* | Head in complex NP | 2.14 | 0.26 |
| SBAR* | Clause introduced by a subordinating conjunction | 1.42 | 3.93 |
| PRN* | Parenthetical | 1.41 | 0.50 |
| LST* | List marker | 0.78 | 0.03 |
| X* | Unknown, uncertain, or unbracketable constituent | 0.58 | 0.01 |
| WHNP* | Wh-noun phrase | 0.44 | 0.99 |
| PRT* | Particle | 0.21 | 0.35 |
| UCP* | Unlike-coordinated phrase | 0.17 | 0.04 |
| WHADVP | Wh-adverb phrase | 0.14 | 0.25 |
| QP* | Quantifier phrase | 0.06 | 0.91 |
| CONJP | Conjunction phrase | 0.02 | 0.04 |
| WHPP | Wh-prepositional phrase | 0.01 | 0.05 |
| SQ | Inverted yes/no question, or main clause of a wh-question | 0 | 0.04 |
| SINV* | Inverted declarative sentence | 0 | 0.30 |
| SBARQ | Direct question introduced by a wh-word or wh-phrase | 0 | 0.03 |
| RRC | Reduced relative clause | 0 | 0.02 |
| NAC* | Not a constituent | 0 | 0.08 |
| INTJ | Interjection | 0 | 0.01 |
*Indicates significant difference with p<0.05.
WSJ, Wall Street Journal.
Table 3.
Grammar rules involving restoration of missing elements in our annotated corpus
| Frequency | Grammar rule |
|---|---|
| 161 | VP→-NONE- NP |
| 52 | NP→-NONE- |
| 41 | VP→-NONE- VP |
| 25 | PP→-NONE- NP |
| 21 | VP→-NONE- ADJP |
| 6 | VP→-NONE- PP |
| 3 | VP→-NONE- NP PP |
| 2 | NP→JJ -NONE- |
| 1 | VP→-NONE- ADVP VP |
| 1 | VP→-NONE- NP PRN |
| 1 | PP→-NONE- NP ADVP |
| 1 | VP→-NONE- NP NP |
| 1 | ADJP→ADVP -NONE- |
See online supplement 2 for examples.
In order to verify the usefulness of the annotated corpus operationally, we retrained the Stanford Parser by using the corpus and performed 10-fold cross validation with comparisons. In table 4 we see that the general English model (from Stanford Parser package) does not work well consistently on clinical text (with both low accuracy and low standard error (SE)), and the low perfect parse rate (0.154) suggests that sublanguage differences systematically penalized the off-domain parser in many of the sentences. Independent training/testing on 9:1 partitions of the clinical corpus itself yielded accuracy of 0.769, but with a higher SE across the folds. The accuracy was improved to 0.811 by combining the clinical corpus (each training partition) with a fixed number of news sentences, with more perfect parses and lower SE observed. The amount of mixed off-domain sentences were determined by simply incrementing one section of the WSJ corpus at a time, and we found inclusion of the 00 and 01 sections reached a peak accuracy before it dropped by including more sections. The level of accuracy was not comparable to the low 90s achieved by state-of-the-art parsers trained/tested on news text but could still be considered moderate, given the challenging nature of clinical text. The accuracy was interestingly close to the 0.81 reported in a preliminary result of using the MiPACQ corpus for training a constituency parser,41 which also mixed a portion of the Penn Treebank annotations. However, the comparison needs to be interpreted with reservation because the settings in the two studies could not be perfectly aligned.
Table 4.
Stanford Parser performance from training on general English, on our annotated clinical corpus alone, and on our corpus mixed with some parsed news text
| Accuracy | Standard error of accuracy | Proportion of perfect parses | |
|---|---|---|---|
| General English (mainly news) | 0.656 | 0.017 | 0.154 |
| Our corpus alone | 0.769 | 0.028 | 0.553 |
| Our corpus plus news | 0.811 | 0.022 | 0.570 |
Discussion
Contribution of the work
The main purpose of this study is to develop/evaluate/share domain-customized parsing guidelines along with a real clinical corpus annotated accordingly. The promising inter-annotator agreement rate (0.930) indicates reliability of the guidelines, and the accuracy (0.811) of a statistical parser retrained with the corpus demonstrates reasonable usability of the annotations. The structure-restoring annotation should benefit development of NLP applications (eg, information extraction) by providing explicitly represented syntax to facilitate composing unambiguous extraction rules/templates. For example, figure 3B has both PPs properly scoped and enables syntactically distributing ‘with cough’ to ‘elderly patient’ via the restored VP. Conversely, syntax-free methods or those with inadequate syntax-restoring capability would be disadvantaged, as they rely more on heuristics that leverage proximity and/or semantic constraints with less transferability across systems. To our knowledge, the current work is the first one introducing Foster's error-handling approach to ill-formed clinical sentences. We do not claim it to be the most suitable and final solution to annotating ungrammatical clinical sentences, and actually new ideas on further refining Foster's approach have burgeoned along our analysis into the results. In the long run, it is hoped that the community will gradually converge on a common consensus by absorbing advantages of different proposals.
As mentioned in the Background section, the study was partly motivated towards addressing the limited interoperability in medical text parsers that involve proprietary semantic grammar. We believe our sharing of the standard-conforming corpus is critical in a data-driven, sustainable model that attracts constant pursuit of questions/solutions for research and development. However, it should be emphasized that a general syntactically annotated corpus is not contradictory to the value of any existing semantic approaches. If coupled appropriately, syntax and semantics can in fact complement each other in forming a robust parsing solution to clinical narratives.42 Specifically, when superimposed with semantic annotations on the same corpus, the Treebank constituents can facilitate automated derivation of a semantic grammar in flexible ways.
As by-products, the study yielded interesting findings as well as research questions:
The simple comparison of constituent distribution between the annotated corpus and a WSJ subset served as proof of concept that clinical text does differ syntactically from non-clinical English. However, it is an open question whether the progress note sample in this study is representative of any clinical text. In other words, could syntactic composition differ considerably even between different clinical genres?
The number of iterations and amount of training notes provided a hint on the effort required to achieve reasonable annotation consistency. Our results suggest it would take at least three iterations of annotating/adjudicating on more than 500 sentences in total for the annotators to reach a higher than 0.9 agreement rate. However, it definitely needs a larger scale comparative study to verify the generality of our findings.
Combining clinical with a certain amount of Treebank training sentences resulted in the most accurate parser model. A purely general English model achieved an accuracy of only 0.656, but the mixture boosted the purely clinical parser model's accuracy from 0.769 to 0.811. One hypothesis is that the size of our corpus (1100 sentences) is still not sufficient to train a statistically robust parser, and therefore even off-domain annotations can help with a smoothing effect. The research question here is: How large should the clinical corpus be in order to independently train a parser? And before the sufficiency of domain-specific training data is achieved, how can we reliably estimate the optimal ratio to mix the heterogeneous corpora?
Limitations
There was no experiment/evaluation performed to formally assess quality of the gold standard token and POS tag annotations. For institutional development interest, we selected progress notes in this study. We are aware that the results obtained on progress notes cannot represent all possible specialties and genres. Because of limited resources, we could not annotate more sentences and did not have more annotators to independently adjudicate or explore various parameters (eg, homogeneous versus heterogeneous knowledge backgrounds). The implication involves limited quantitative power in interpreting the results and limited qualitative generality based on only the two annotators.
Conclusion
With an iterative approach, we developed syntactic parsing guidelines for clinical text and annotated a set of 1100 sentences in progress notes accordingly. The guidelines are compatible with the standard Penn Treebank syntactic annotation style and include special adaptations to accommodate clinical sublanguage properties. Two annotators (a linguist and a computer scientist) reached an agreement rate of 0.930 in the final independent evaluation, which indicates consistency in following the guidelines. As simple validation of usefulness, retraining a statistical parser with the annotated corpus achieved a best accuracy of 0.811 (by involving also some off-domain training sentences). Both the guidelines and annotations are made available at https://sourceforge.net/projects/medicaltreebank.
Acknowledgments
We thank Dr Rommel Yabut for explaining the clinical content to our annotators. The de-identified clinical records in i2b2/VA NLP Challenge were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr Özlem Uzuner, i2b2, and SUNY.
Footnotes
Contributors: JF, HX, and YH oversaw the design, development, and evaluation of the study and contributed to the writing. RML selected the clinical notes of non-redundant content. EWY and MJ performed the annotations for iterative guideline and corpus development. EWY drafted (with assistance from RP), revised, and finalized the annotation guidelines. JF performed the data processing and analysis. All authors contributed to conceptual development and revision of the manuscript, and approved the final submission.
Funding: This work was partly supported by the National Library of Medicine (NLM) grant number R01-LM007995 and the National Cancer Institute (NCI) grant number R01-CA141307.
Competing interests: The KP team is regularly salaried by Kaiser Permanente, which does not have any financial interest or influence over the study; the Vanderbilt team received grant supports from the NLM and NCI.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Both the guidelines and annotations are made available to the research community and can be downloaded from https://sourceforge.net/projects/medicaltreebank.
References
- 1.Manning C, Schütze H. Foundations of statistical natural language processing. Cambridge, MA: MIT Press, 1999 [Google Scholar]
- 2.Chung GY. Towards identifying intervention arms in randomized controlled trials: extracting coordinating constructions. J Biomed Inform 2009;42:790–800 [DOI] [PubMed] [Google Scholar]
- 3.Miyao Y, Sagae K, Sætre R, et al. Evaluating contributions of natural language parsers to protein–protein interaction extraction. Bioinformatics 2009;25:394–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Lowe HJ, Klein D, et al. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS specialist lexicon. J Am Med Inform Assoc 2005;12:275–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Lowe HJ. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform Assoc 2007;14:304–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med 1996;35:285–301 [PubMed] [Google Scholar]
- 7.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 8.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc 2011;18:544–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc 2013.10.1136/amiajnl-2012-001317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc 2011;18:540–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charniak E. A maximum-entropy-inspired parser. Proceedings of 1st NAACL Conference; Stroudsburg, PA, USA: Association for Computational Linguistics, 2000:132–9 [Google Scholar]
- 12.Petrov S, Klein D. Improved inference for unlexicalized parsing. Proceedings of NAACL HLT 2007; Association for Computational Linguistics, 2007:404–11 [Google Scholar]
- 13.Marcus MP, Santorini B, Marcinkiewicz MA. Building a large annotated corpus of English: the Penn Treebank. Comput Linguist 1993;19:313–30 [Google Scholar]
- 14.Uzuner Ö. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warner C, Lanfranchi A, O'Gorman T, et al. Bracketing biomedical text: an addendum to Penn Treebank II guidelines. Institute of Cognitive Science, University of Colorado at Boulder; http://clear.colorado.edu/compsem/documents/treebank_guidelines.pdf (accessed Jun 4, 2012) [Google Scholar]
- 18.Hirschman L, Sager N. Automatic information formatting of a medical sublanguage. In: Kittredge R, Lehrberger J. eds. Sublanguage: studies of language in restricted semantic domains. Berlin: Walter de Gruyter, 1982:27–80 [Google Scholar]
- 19.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform 2002;35:222–35 [DOI] [PubMed] [Google Scholar]
- 20.Foster J. Treebanks gone bad: parser evaluation and retraining using a Treebank of ungrammatical sentences. Int J Doc Anal Recognit 2007;10:129–45 [Google Scholar]
- 21.De Marneffe M-C, Maccartney B, Manning CD. Generating typed dependency parses from phrase structure parses. Proceedings of 5th International Conference on Language Resources and Evaluation (LREC 2006); Genoa, Italy, 2006:449–54 [Google Scholar]
- 22.Bies A, Ferguson M, Katz K, et al. Bracketing guidelines for Treebank II style. 1995. ftp://ftp.cis.upenn.edu/pub/treebank/doc/manual/root.ps.gz (accessed 25 Mar 2011)
- 23.Harris Z. Methods in structural linguistics. Chicago: University of Chicago Press, 1951 [Google Scholar]
- 24.Solan Z, Horn D, Ruppin E, et al. Unsupervised learning of natural languages. Proc Natl Acad Sci USA 2005;102:11629–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Google Scholar. http://scholar.google.com/ (accessed Sep 21, 2012)
- 26.Tateisi Y, Yakushiji A, Ohta T, et al. Syntax annotation for the GENIA corpus. Proceedings of International Joint Conference on Natural Language Processing; JejuIsland, Republic of Korea, 2005:222–7 [Google Scholar]
- 27.Fan J, Prasad R, Yabut RM, et al. Part-of-speech tagging for clinical text: wall or bridge between institutions? AMIA Annual Symposium Proceedings; Washington, DC, USA, 2011:382–91 [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn U, Romacker M, Schulz S. MEDSYNDIKATE—a natural language system for the extraction of medical information from findings reports. Int J Med Inform 2002;67:63–74 [DOI] [PubMed] [Google Scholar]
- 30.Taira RK, Bashyam V, Kangarloo H. A field theoretical approach to medical natural language processing. IEEE Trans Inf Technol Biomed 2007;11:364–75 [DOI] [PubMed] [Google Scholar]
- 31.Rea S, Pathak J, Savova G, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project. J Biomed Inform 2012;45:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shriberg E. Disfluencies in Switchboard. Proceedings of International Conference on Spoken Language Processing; Philadelphia, PA, USA, 1996:11–14 [Google Scholar]
- 33.Meteer M, Taylor A, MacIntyre R, et al. Dysfluency annotation stylebook for the Switchboard Corpus. 1995. ftp://ftp.cis.upenn.edu/pub/treebank/swbd/doc/DFL-book.ps (accessed 13 May 2013)
- 34.Kepser S, Steiner I, Sternefeld W. Annotating and querying a treebank of suboptimal structures. In: Kübler S, Nivre J, eds. Proceedings of 3rd Workshop on Treebanks and Linguistic Theories; Tübingen, Germany, 2004:63–74 [Google Scholar]
- 35.Carbonell JG, Hayes PJ. Recovery strategies for parsing extragrammatical language. Am J Comput Linguist 1983;9:123–46 [Google Scholar]
- 36.Jensen K, Heidorn GE, Miller LA, et al. Parse fitting and prose fixing: getting a hold on ill-formedness. Am J Comput Linguist 1983;9:147–60 [Google Scholar]
- 37.Charniak E, Johnson M. Coarse-to-fine n-best parsing and MaxEnt discriminative reranking. Proceedings of 43rd Annual Meeting of the Association for Computational Linguistics (ACL'05); 2005;vol 1:173–80 [Google Scholar]
- 38.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of 41st Annual Meeting of the Association for Computational Linguistics (ACL'03); 2003;vol 1:423–30 [Google Scholar]
- 39.Morton T, Lacivita J. WordFreak: an open tool for linguistic annotation. Proceedings of Conference of the North American Chapter of the Association of Computational Linguistics Human Language Technologies; 2003:17–8 [Google Scholar]
- 40.Hripcsak G, Rothschild AS. Agreement, the F-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005;12:296–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng J, Chapman WW, Miller TA, et al. A system for coreference resolution for the clinical narrative. J Am Med Inform Assoc 2012;19:660–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan J, Friedman C. Deriving a probabilistic syntacto-semantic grammar for biomedicine based on domain-specific terminologies. J Biomed Inform 2011;44:805–14 [DOI] [PMC free article] [PubMed] [Google Scholar]