Abstract
Objectives: This study aimed to elucidate the relative involvement of drug resistance gene copy number and overexpression in fluconazole resistance in clinical C. glabrata isolates using a population-based approach.
Methods: Fluconazole resistance levels were quantified using the minimal inhibitory concentration (MIC) via Etest method. Both gene expression levels and gene copy number of CgCDR1, CgPDH1, CgERG11, and CgSNQ2 were assessed via quantitative real-time PCR. The influence of the main effects and first-level interactions of both the expression level and copy number of these genes on fluconazole resistance levels were analyzed using a multivariate statistical model.
Results: Forty-three C. glabrata isolates were collected from 30 patients during in a hospital survey. In the multivariate analysis, C. glabrata fluconazole MICs were independently increased by CgSNQ2 overexpression (p < 10−4) and the interaction between CgPDH1 gene copy number and CgPDH1 expression level (p = 0.038). In contrast, both CgPDH1 overexpression (p = 0.049) and the interaction between CgSNQ2 and CgERG11 expression (p = 0.003) led to a significant decrease in fluconazole MICs.
Conclusion: Fluconazole resistance in C. glabrata involves complex interactions between drug resistance gene expression and/or copy number. The population-based multivariate analysis highlighted the involvement of the CgSNQ2 gene in fluconazole resistance and the complex effect of the other genes such as PDH1 for which overexpression was associated with reduced fluconazole resistance levels, while the interaction between PDH1 overexpression and copy number was associated with increased resistance levels.
Keywords: Candida glabrata, fluconazole, resistance mechanisms, gene expression, gene copy number, quantitative real-time PCR, human infection
Introduction
Candida glabrata is responsible for the increase in cases of systemic and superficial candidiasis in many countries (Diekema et al., 2002; Li et al., 2007; Pfaller and Diekema, 2007). Several reports have revealed that a significant percentage of C. glabrata clinical isolates are resistant to fluconazole. Pfaller et al. have found that between 2001 and 2007, isolation frequencies and fluconazole resistance among C. glabrata blood stream infection isolates was increased compared with the 1992–2001 period (Pfaller and Diekema, 2004; Pfaller et al., 2009). Similarly, antifungal susceptibility surveys have detected 17 and 18% resistance levels in Europe and North America, respectively (Pfaller et al., 2009). Many studies have shown that drug efflux mediated by the overexpression of ATP-binding cassette (ABC) transporter family membrane proteins was the main molecular mechanism associated with C. glabrata fluconazole resistance (Sanglard et al., 1999, 2001; Bouchara et al., 2000; Niimi et al., 2002; Wada et al., 2002; Cernicka and Subik, 2006; Coleman and Mylonakis, 2009). Studies have also shown that among ABC transporters, CgCDR1, and CgPDH1, but not CgSNQ2, play a major role in fluconazole resistance (Sanguinetti et al., 2005; Torelli et al., 2008). Azole resistance in C. albicans was primarily associated with mutation or overexpression of ERG11, which encodes the key azole target protein lanosterol 14-alpha demethylase (Chau et al., 2004; Cernicka and Subik, 2006). In contrast, CgERG11 mutations were marginally involved in C. glabrata fluconazole resistance (Sanguinetti et al., 2005). However, CgERG11 overexpression has been associated with chromosome duplication and aneuploidy due to genome plasticity as well as fluconazole resistance in Cryptococcus neoformans, C. albicans, and C. glabrata (Marichal et al., 1997; Kwon-Chung and Chang, 2012). This study aimed to dissect the relative contribution of drug resistance gene copy number and overexpression on fluconazole resistance in clinical C. glabrata isolates.
Methods
Patients and isolates
The population of Candida glabrata clinical isolates included in the present study were recovered during an epidemiological survey of antifungal resistance conducted at the Habib Bourguiba University Hospital in Sfax from January 2005 to December 2007 as previously described (Abbes et al., 2011, 2012; Amouri et al., 2011; Sellami et al., 2011). The specimens were isolated from 30 patients with urinary tract infection, vaginal infection or systemic infections. Candida glabrata was identified by assessing the isolates using Candiselect medium (bioMérieux, Craponne, France) and the ID32C profile (bioMérieux). Isolate fluconazole minimum inhibitory concentrations (MICs) were then measured using the E-test method (AB Biodisk, Sweden) on RPMI 1640 medium (AES, France) as recommended by the manufacturer. Isolates with MICs >64 mg/l were defined as fluconazole resistant (Clinical and Laboratory Standards Institute, 2008).
RNA and DNA isolation
Total RNA was extracted from yeast cells isolated from RPMI plates during the logarithmic growth phase at 48 h of incubation using an RNAeasy Protect™ mini kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. RNA extracts were treated with RNase-free DNase (Qiagen) to avoid DNA contamination. Genomic DNA was extracted using the NucleoSpin™ Tissue method (Macherey-Nagel, Dueren, Germany) according to the manufacturer's instructions. Genomic DNA (gDNA) extracts were treated with DNase-free RNase (Qiagen) to eliminate RNA contamination.
Quantitative real-time PCR
Quantitative real-time PCR analysis was performed to measure the expression levels and copy number of each of the four target genes: CgCDR1, CgPDH1, CgSNQ2, and CgERG11 (Table 1). Gene transcript levels and copy number values were normalized to URA3, a single copy gene encoding orotidine 5-phosphate decarboxylase.
Figure 1.
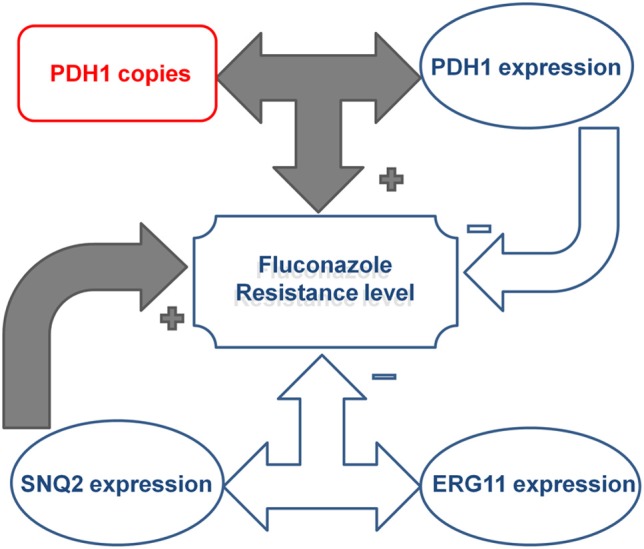
Illustration of the effect of the complex interactions between CgPDH1, CgSNQ2, and CgERG11 drug resistance gene copy numbers and expression levels on fluconazole resistance in Candida glabrata clinical isolates. CgPDH1 gene copy number and expression were correlated, and their interaction led to an increase in fluconazole MIC levels. In contrast, the main effect of CgPDH1 overexpression independently led to a decrease in MIC levels. The main effect of CgSNQ2 overexpression independently promoted an increase in fluconazole MIC levels, while its interaction with CgERG11 expression led to a decrease in MIC levels.
Reverse transcriptase PCR (RT-PCR)
First, the RNA extract of each isolate was transcribed into cDNA using reverse transcriptase (RT), which was performed with a 25-μl reaction volume containing 2.5 μl MuLva buffer (Biology, France), 2 U of MuLva RT (Biology), 5 μl RNA sample and 0.5 μM random primer and polyA mixture (Eurogentec, France). The samples were subjected to reverse transcription at 37°C for 60 min.
Quantitative real-time PCR (qPCR)
Two μl of either cDNA or gDNA template was used to separately assess expression levels and gene copy number of the four genes: CgCDR1, CgPDH1, CgSNQ2, and CgERG11, as previously described (Sanguinetti et al., 2005). The primers utilized are detailed in Table 1. Briefly, 200 pM of each primer, 200 pM hydrolysis probe and 25 μl TaqMan mix (Applied Biosystems, Courtaboeuf, France) were included in the PCR assay. Copy number PCR cycling conditions consisted of an initial step at 94°C for 10 min and then 40 cycles of 15 s at 94°C, 30 s at 55°C and 20 s at 72°C. Fluorescence data were collected during the extension step and analyzed with a MX 4000 real-time PCR system (Stratagene, France). Each reaction was performed in duplicate. Copy number efficiency was determined for each gene. The normalized relative quantities of the gene-specific product or copy number for each sample were then calculated, i.e., the quantity of the samples of interest was compared with a calibrator (TU10, a fluconazole susceptible control, MIC = 0.125 mg/l). The quantity of both the calibrator and the samples of interest were normalized to URA3.
Table 1.
Primers and probes used to amplify CgCDR1, CgPDH1 (formerly denoted CDR2), CgSNQ2, CgERG11, and CgURA3 in separate quantitative Real-Time PCR reactions as described in Wada et al. (2002).
| Gene (genbank accession no) | Primer or probe | Sequence | Annealing site (5′3′) |
|---|---|---|---|
| CgCDR1 (AF109723) | CDR1a | TAGCACATCAACTACACGAACGT | 4500–4522 |
| CDR1b | AGAGTGAACATTAAGGATGCCATG | 4647–4670 | |
| CDR1pr | 6FAM-TGCTGCTGCTTCTGCCACCTGGTT-TAMRA | 4621–4644 | |
| CgPDH1 (AF251023) | CDR2a | GTGCTTTATGAAGGCTACCAGATT | 164–187 |
| CDR2b | TCTTAGGACAGAAGTAACCCATCT | 251–274 | |
| CDR2pr | 6FAM-TACCTTTGCGTGCTGGGCGTCACC-TAMRA | 217–240 | |
| CgSNQ2 (AF251022) | SNQ2a | ACCATGTGTTCTGAATCAATCAAT | 360–383 |
| SNQ2b | TCGACATCATTACAATACCAGAAA | 462–485 | |
| SNQ2pr | 6FAM-AACTAATCGCCGCAGGTTGTGACA-TAMRA | 394–317 | |
| CgERG11 (L40389) | ERGa | ATTGGTGTCTTGATGGGTGGTC | 928–949 |
| ERGb | TCTTCTTGGACATCTGGTCTTTCA | 1019–1042 | |
| ERGpr | 6FAM-ACTTCCGCTGCTACCTCCGCTTGG-TAMRA | 955–978 | |
| CgURA3 (L13661) | URAa | GAAAACCAATCTTTGTGCTTCTCT | 168–191 |
| URAb | CATGAGTCTTAAGCAAGCAAATGT | 268–291 | |
| URApr | VIC-ACGTCACCACCACCAGCGAATTGT-TAMRA | 194–217 |
C. glabrata genotyping
The C. glabrata isolates were genotyped using six markers, five microsatellites (RPM2, MTI, GLM4, GLM4, and GLM6) and irregular patterns in the ERG3 gene as previously described (Abbes et al., 2011, 2012).
Statistical analysis
Normalized gene copy numbers and expression levels are indicated using range, median, and interquartile range (IQR). The correlation between fluconazole MICs and the relative expression level and copy number of each gene was analyzed using the Spearman rank correlation coefficient. For qualitative analysis, we selected a threshold value of 2.5 normalized gene expression or normalized gene copy number to define gene overexpression or an increase in gene copy number, respectively. Regarding quantitative analysis, fluconazole MIC values were log-transformed, standardized and centered. The effect on fluconazole MICs due to the gene copy number and expression levels of each of the four genes studied was modeled using the Genmod procedure. All first-level interactions (the product of the two main effect terms) between each covariate were tested. When an interaction was statistically significant, each of the two main effects was retained in the model, regardless of the level of statistical significance. The most parsimonious best-fitting model was selected using the log-likelihood ratio test. After selection of the model, the generalized estimating equation option was used to account for the non-independence of isolates sampled from a same patient. Statistical analyses were performed with SAS 9.2 statistical software (Cary, NC, USA). All statistical tests were two-sided, and p < 0.05 was considered significant.
Results
The characteristics of the 43 C. glabrata clinical isolates collected during a hospital survey from 30 patients (9 patients had ≥2 episodes) at the Habib Bourguiba University hospital in Sfax (Tunisia) are detailed in Table 2. Briefly, most isolates were grown from urine samples, and 15 isolates were involved in invasive infections. Fluconazole MICs ranged from 0.016 to 256 mg/l; MIC50 and MIC90 were 4 mg/l and 256 mg/l, respectively. Nine isolates were classified as resistant to fluconazole with a MIC >256 mg/l. Microsatellite analysis displayed 17 distinct genotypes. Twenty-two isolates were sequentially collected (7–220 days lag in the same patient). In patients 1, 2, 3, and 4 treated with fluconazole, MICs were significantly increased in sequential C. glabrata isolates, whereas the microsatellite-based genotypes remained stable (Tables 3, 4). These patients were exposed to 150, 30, 220, or 120 days of fluconazole therapy with a cumulative dose of 4, 5.6, 8, and 12 g fluconazole, respectively (Table 3). In patients 6 and 11, the fluconazole MICs were increased in sequential isolates with a distinct microsatellite genotype, thereby indicating that they probably acquired resistant C. glabrata strains upon exposure to fluconazole (Table 2). Finally, both microsatellite genotypes and fluconazole MICs remained stable in the sequential isolates derived from patients 19, 21, and 26 (Table 2).
Table 2.
Relative gene expression levels and copy numbers of CgCDR1, CgPDH1, CgERG11, and CgSNQ2 in fluconazole sensitive and resistant Candida glabrata isolates.
| Patients | Isolates | Sample site | Microsatellite type | Fluconazole MIC (μg/ml) | Relative increase in | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDR1 expression | CDR1 copies | PDH1 expression | PDH1 copies | ERG11 expression | ERG11 copies | SNQ2 expression | SNQ2 copies | |||||
| 1 | 1.1 | Urine | 3 | 1.5 | 3.09 | 4.59 | 6.62 | 1.99 | 0.4 | 0.74 | 1.11 | 0.18 |
| 1 | 1.2 | Urine | 3 | 256 | 5.64 | 4.7 | 20.04 | 4.13 | 0.54 | 1549.94 | 0.48 | 364.12 |
| 2 | 2.1 | Urine | 5 | 16 | ND | 2.01 | ND | 1.85 | ND | 1.55 | ND | 2.13 |
| 2 | 2.2 | Urine | 5 | 256 | 92.61 | 1.83 | 169.56 | 1.37 | 0.12 | 1.6 | 5.41 | 0.94 |
| 3 | 3.1 | Vaginal | 9 | 2 | 0.12 | 2.16 | 0.44 | 1.61 | 0.24 | 2.3 | 0.07 | 1.58 |
| 3 | 3.2 | Vaginal | 9 | 256 | 0.29 | 2.18 | 0.57 | 1.27 | 1.16 | 1.41 | 0.19 | 1.65 |
| 4 | 4.1 | Urine | 1 | 4 | ND | 1.48 | 0.11 | 1.14 | 0.53 | 3.35 | 0.04 | 0.99 |
| 4 | 4.2 | Urine | 1 | 256 | 0.1 | 2.32 | 9.42 | 0.83 | 0.3 | 3.14 | 1.19 | 1.44 |
| 5 | 5 | Urine | 1 | 256 | 1.84 | 1.11 | 7.64 | 1.16 | 0.56 | 0.91 | 0.97 | 1.22 |
| 6 | 6.1 | Urine | 8 | 32 | 0.1 | 1.02 | 0.49 | 0.9 | 0.94 | 0.93 | 0.56 | 0.82 |
| 6 | 6.2 | Urine | 2 | 256 | 2.54 | 4.04 | 375.57 | 5.7 | 4.34 | 12.7 | 73.61 | 0.56 |
| 7 | 7 | Blood culture | 7 | 256 | 0.39 | 1.89 | 5.6 | 2.71 | 0.42 | 2.01 | 5.77 | 1.2 |
| 8 | 8 | Urine | 9 | 256 | 12.06 | 3.64 | 0.31 | 3.21 | 0.23 | 3.74 | 1.17 | 3.24 |
| 9 | 9 | Blood culture | 7 | 256 | 12.14 | 1.27 | 0.59 | 0.89 | 0.87 | 1.01 | 2.17 | 1.03 |
| 10 | 10 | Blood culture | 7 | 32 | 1.42 | 1.25 | 0.27 | 1.01 | 0.25 | 1.84 | 0.26 | 1.56 |
| 11 | 11.1 | Urine | 12 | 1 | 0.34 | ND | 15.08 | ND | 1.11 | ND | 12.93 | ND |
| 11 | 11.2 | Urine | 4 | 32 | 0.46 | 1.21 | 4.86 | 1.38 | 0.57 | 0.79 | 0.45 | 1.04 |
| 11 | 11.3 | Urine | 10 | 32 | 0.65 | 1.25 | 0.87 | 0.86 | 0.54 | 1.84 | 1.72 | ND |
| 12 | 12 | Blood culture | 10 | 1 | 0.43 | 1.41 | 0.46 | 2.05 | 0.09 | 1.45 | 0.01 | 1.28 |
| 13 | 13 | Blood culture | 6 | 2 | ND | 1.73 | 0.04 | 1.21 | 0.27 | 1.29 | 0.22 | 0.85 |
| 14 | 14 | Blood culture | 10 | 2 | 0.3 | 1.68 | 0.26 | 1.17 | 0.16 | 1.68 | 0.37 | 1.39 |
| 15 | 15 | Kidney abscess | 13 | 3 | ND | 1.59 | 0.76 | 1.93 | 0.42 | 1.6 | 0.99 | 0.7 |
| 16 | 16 | Lung abscess | 14 | 2 | 0.23 | 1.24 | 0.35 | 0.79 | 0.31 | 1.07 | 0.47 | 0.95 |
| 17 | 17 | Urine | 14 | 2 | 0.18 | 1.75 | 0.46 | 1.52 | 0.26 | 3.86 | 0.42 | 0.77 |
| 18 | 18 | Blood culture | 15 | 2 | 0.09 | 1.74 | 0.25 | 0 | 0.26 | 2.1 | 0.3 | 1.12 |
| 19 | 19.1 | Urine | 7 | 0.064 | 0.019 | 1.26 | 0.94 | 1.32 | 1.23 | 1.19 | 0.49 | 1.31 |
| 19 | 19.2 | Urine | 7 | 0.125 | 0.04 | 1.2 | 0.79 | 1.14 | 0.38 | 0.99 | 0.52 | 0.69 |
| 20 | 20 | Blood culture | 7 | 4 | 1.3 | 1.47 | 17.05 | 1.67 | 1.78 | 1.32 | 0.48 | 1.83 |
| 21 | 21.1 | Urine | 4 | 6 | 6.71 | 1.16 | 3.59 | 1.25 | 2.34 | 4.03 | 3.56 | 0.41 |
| 21 | 21.2 | Urine | 4 | 4 | 0.86 | 2.76 | 0.74 | 2.57 | 0.71 | 2.57 | 0.69 | 2.82 |
| 21 | 21.3 | Urine | 4 | 4 | 0.82 | 1.4 | 0.96 | 1.51 | 1.32 | 1.38 | 0.79 | 1.67 |
| 21 | 21.4 | Urine | 4 | 8 | 0.01 | 2.16 | 0.57 | 1.61 | 1.89 | 2.3 | 1.72 | 1.75 |
| 21 | 21.5 | Urine | 4 | 4 | 2.63 | 2.31 | 2.9 | 1.96 | 1.05 | 1.76 | 2.14 | 1.6 |
| 22 | 22 | Blood culture | 3 | 4 | 1.59 | 1.71 | 1.13 | 1.75 | 0.74 | 1.34 | 1.18 | 1.43 |
| 23 | 23 | Urine | 4 | 4 | 1.81 | 1.62 | 2.79 | 1.48 | 0.52 | 1.46 | 1.79 | 1.61 |
| 24 | 24 | Urine | 16 | 4 | 0.12 | 1.71 | 0.29 | 1.18 | 0.43 | 1.1 | 0.25 | 1.28 |
| 25 | 25 | Urine | 10 | 0.5 | 0.03 | ND | 1.61 | 2.61 | 0.09 | 1.36 | 0.1 | 48.88 |
| 26 | 26.1 | Urine | 9 | 0.016 | ND | 4.19 | 2.76 | 2.23 | 1 | 1.15 | 0.35 | 0.37 |
| 26 | 26.2 | Urine | 9 | 6 | 0.4 | ND | 0.88 | ND | 0.84 | ND | 0.59 | ND |
| 27 | 27 | Blood culture | 11 | 8 | ND | ND | 0.76 | ND | 0.7 | ND | 0.15 | ND |
| 28 | 28 | Blood culture | 3 | 1 | 0.2 | 4.27 | 1.16 | 2.41 | 2.09 | 1.59 | 0.34 | 0.66 |
| 29 | 29 | Blood culture | 9 | 4 | ND | 1.73 | 1.26 | 0.64 | 0 | 0.55 | 0.69 | 0.77 |
| 30 | 30 | Blood culture | 17 | 2 | 0.01 | 1.18 | 1.04 | 1.09 | 0.98 | 0.78 | 2.08 | 0.75 |
Table 3.
Characteristics and fluconazole treatment features in six patients infected with C. glabrata that acquired in vitro resistance under fluconazole treatment.
| Patient no. | Age (yr)/sex | Underling disease | No. of positive culture (sampling times in days) | Fluconazole treatment dosage (administration days) |
|---|---|---|---|---|
| 1 | 43/M | Nephrolithiasis | 3 (0, 60, 150) | 200 mg (60–80) |
| 2 | 42/F | Diabetes | 2 (0, 30) | 200 mg (0–28) |
| 3 | 53/F | Asthma (corticotherapy) | 5 (0, 150, 220, 250, 280) | 150 mg (120–135), 100 mg (150–165), 400 mg (225–232), 200 mg (250–257) |
| 4 | 72/F | Nephrolithiasis and diabetes | 3 (0, 130, 446) | 400 mg (130–160) |
Table 4.
Details of the relative expression levels and copy numbers of CDR1, PDH1, ERG11, and SNQ2 genes in four patients' genetically stable Candida glabrata sequential isolates that displayed distinct fluconazole MIC levels.
| Patient | Isolate | Sampling times in days | Fluconazole MIC (mg/l) | Microsatellite markers alleles | Relative increase in gene | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RPM2 | MTI | ERG3 | GLM4 | GLM5 | GLM6 | CDR1 expression | CDR1 copies | PDH1 expression | PDH1 copies | ERG11 expression | ERG11 copies | SNQ2 expression | SNQ2 copies | ||||
| 1 | 1.1 | 0 | 1.5 | 127 | 238 | 197 | 273 | 298 | 295 | 3.09 | 4.59 | 6.62 | 1.99 | 0.4 | 0.74 | 1.11 | 0.18 |
| 1.2 | 150 | 256 | 127 | 238 | 197 | 273 | 298 | 295 | 5.64 | 4.70 | 20.04 | 4.13 | 0.54 | 1549.94 | 0.48 | 364.12 | |
| 2 | 2.1 | 0 | 16 | 139 | 227 | 228 | 276 | 262 | 298 | ND | 2.01 | ND | 1.85 | ND | 1.55 | ND | 2.13 |
| 2.2 | 30 | 256 | 139 | 227 | 228 | 276 | 262 | 298 | 92.61 | 1.83 | 169.56 | 1.37 | 0.12 | 1.6 | 5.41 | 0.94 | |
| 3 | 3.1 | 0 | 2 | 127 | 238 | 197 | 288 | 271 | 295 | 0.12 | 2.16 | 0.44 | 1.61 | 0.24 | 2.3 | 0.07 | 1.58 |
| 3.2 | 60 | 256 | 127 | 238 | 197 | 288 | 271 | 295 | 0.29 | 0.57 | 0.57 | 1.27 | 1.16 | 1.41 | 0.19 | 1.65 | |
| 4 | 4.1 | 0 | 4 | 127 | 238 | 197 | 270 | 271 | 289 | ND | 1.48 | 0.11 | 1.14 | 0.53 | 3.35 | 0.04 | 0.99 |
| 4.2 | 120 | 256 | 127 | 238 | 197 | 270 | 271 | 289 | 0.1 | 2.32 | 9.42 | 0.83 | 0.30 | 3.14 | 1.19 | 1.44 | |
CgCDR1, CgPDH1, CgSNQ2, and CgERG11 expression
To investigate the mechanism of fluconazole resistance in C. glabrata we analyzed the gene expression levels of CgCDR1, CgPDH1, CgERG11, and CgSNQ2. In the 43 C. glabrata isolates, the normalized gene expression levels were as follows: CDR1 levels ranged from 0.010 to 92.61, median 0.415 IQR [0.120–1.825]; CDR2 levels ranged from 0.04 to 375.57, median 0.91 IQR [0.46–3.59]; SNQ2 levels ranged from 0.010 to 73.61, median 0.575 IQR [0.34–1.72] and ERG11 levels ranged from 0 to 4.34, median 0.54 IQR [0.27–1.00] (Table 2).
When considering the nine fluconazole resistant isolates, CgCDR1 was overexpressed in five isolates, CgPDH1 was overexpressed in six isolates, CgSNQ2 was overexpressed in three isolates and CgERG11 was overexpressed in one isolate (Table 2). Overall, the expression of the efflux pump genes CgCDR1, CgPDH1, and CgSNQ2 was increased in two resistant isolates compared with the susceptible initial isolates. One resistant isolate displayed simultaneous overexpression of CgCDR1, CgPDH1 and CgERG11, while another isolate displayed overexpression of both CgPDH1 and CgSNQ2. CgCDR1 and CgPDH1 were each overexpressed in two resistant isolates. In contrast, at least one gene was overexpressed in nine fluconazole susceptible isolates. Although overexpression of CgERG11 was never observed, CgPDH1, CgCDR1 and CgSNQ2 were overexpressed in seven, three and two fluconazole-susceptible isolates, respectively (Table 2). In four of these isolates, CgPDH1 overexpression was observed in parallel with either CgSNQ2 or CgCDR1 overexpression. When considering sequential isolates derived from the same patients, the acquisition of fluconazole resistance was associated with increased expression of CgCDR1 and CgPDH1 in patient 1 and increased expression of CgPDH1 in patient 4 (Table 4).
CgCDR1, CgPDH1, CgSNQ2, and CgERG11 copy number
To investigate one possible mechanism of C. glabrata fluconazole resistance we analyzed gene copy number of CgCDR1, CgPDH1, CgERG11, and CgSNQ2. The normalized gene copy number values were as follows: CDR1 copy numbers ranged from 1.02 to 4.70, median 1.71 IQR [1.2–2.18]; CDR2 copy numbers ranged from 0.00 to 5.70, median 1.43 IQR [1.14–1.98]; SNQ2 copy numbers ranged from 0.18 to 364.12, median 1.22 IQR [0.77–1.61] and ERG11 copy numbers ranged from 0.55 to 1549.94, median 1.51 IQR [1.13–2.20]. In some isolates, either overexpression or increased gene copy number was observed for CgCDR1 and CgPDH1 (Table 2). Nevertheless, three out of five resistant isolates that displayed CgCDR1 overexpression also displayed an increase in CgCDR1 gene copy number (4.7, 4.04 and 3.64 copies for isolates 1.2, 6.2, and 8, respectively) (Table 2). Three of the six resistant isolates that displayed CgPDH1 overexpression also exhibited an increase in CgPDH1 gene copy number (2.71, 4.13, and 5.7 copies in isolates 7, 1.2, and 6.2, respectively). Similarly, resistant isolates displaying CgERG11 overexpression also exhibited an increase in CgERG11 gene copy number (12.7 times more than TU10). For CgSNQ2, gene overexpression did not correlate with gene copy number (Table 2). Among the fluconazole susceptible isolates, the copy number of at least one gene was increased in seven isolates. More precisely, copy number of the four genes was increased in isolate 21.2, while only one gene copy number was increased in the remaining isolates as follows: CgERG11 copy number was increased in four isolates and the gene copy number of CgSNQ2 and CgCDR1 was increased, each in one isolate.
Population-based gene interaction study
Univariate correlation analysis indicated a complex interaction between gene copy number and expression levels (Table 5). A correlation between CgPDH1, CgCDR1, and CgSNQ2 gene copy numbers was observed, while a correlation between the expression levels of CgPDH1, CgERG11, and CgSNQ2 was also found. Fluconazole MIC values positively correlated with both CgCDR1 and CgSNQ2 expression levels. A positive correlation between gene copy number and expression level was observed only for the CgPDH1 gene. Figure 1 summarizes the significant combined main effects and first-level interactions between gene copy number and gene expression levels influencing fluconazole MIC levels in C. glabrata.
Table 5.
Spearman rank correlation coefficients between the fluconazole MICs and the normalized expression level and copy number of CDR1, PDH1, ERG11, and SNQ2 genes.
| Fluconazole | Gene expression | Gene copy number | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC | CDR1 | PDH1 | ERG11 | SNQ2 | CDR1 | CDR2 | ERG11 | ||
| GENE EXPRESSION | |||||||||
| CDR1 | 0.50551 | ||||||||
| PDH1 | 0.21498 | 0.38320 | |||||||
| ERG11 | 0.11168 | 0.04558 | 0.34565 | ||||||
| SNQ2 | 0.38022 | 0.41744 | 0.58012 | 0.33921 | |||||
| GENE COPY NUMBER | |||||||||
| CDR1 | 0.04854 | 0.19972 | 0.18646 | −0.04077 | −0.00235 | ||||
| PDH1 | −0.04227 | 0.29146 | 0.36136 | 0.18184 | 0.06981 | 0.62306 | |||
| ERG11 | 0.30065 | 0.18173 | −0.05340 | −0.00061 | 0.09112 | 0.43376 | 0.32972 | ||
| SNQ2 | 0.29037 | 0.01279 | −0.07420 | −0.07782 | −0.13971 | 0.17851 | 0.24155 | 0.29510 | |
The Spearman rank correlation coefficient values >0.3 (and p < 0.05) are bolded.
The best-fitting multivariate model of the factors influencing fluconazole MIC values included (i) the main effects of the expression levels of CgSNQ2, CgERG11, and CgPDH1 and the gene copy number of CgPDH1 and (ii) the interaction between CgSNQ2 and CgERG11 gene expression and the interaction between CgPDH1 copy number and expression. In this model, C. glabrata fluconazole MICs were significantly increased due to both CgSNQ2 overexpression (p < 10−4) and the interaction between CgPDH1 copy number and expression (p = 0.038). In contrast, both CgPDH1 overexpression (p = 0.049) and the interaction between CgSNQ2 and CgERG11 expression (p = 0.003) led to a statistically significant decreased in fluconazole MICs. Neither CgERG11 expression (p = 0.823) nor CgPDH1 gene copy number (p = 0.985) independently displayed a statistically significant influence on fluconazole MICs. These multivariate analysis findings are illustrated in Figure 1.
Discussion
This population-based approach allowed us to confirm the finding that ABC transporter family membrane gene overexpression is a pivotal fluconazole resistance mechanism in clinical C. glabrata isolates, which had been previously evidenced primarily using laboratory strains. Moreover, this study enabled us to further elucidate the main effects and complex interactions between gene expression and copy number associated with in vitro fluconazole resistance. The population-based analysis revealed that CgPDH1 and CgERG11 overexpression indirectly influence fluconazole MIC levels by increasing CDR1 and SNQ2 expression levels. Although CgERG11, CgSNQ2, and CgPDH1 expression and, to a lesser extent, copy number levels were significantly associated with fluconazole resistance, our findings highlight the complex interaction between these genes and suggest the involvement of unmeasured confounding effects. Indeed, mutations in CgPDR1, CgERG11 or others genes that might impact C. glabrata fluconazole resistance were not analyzed in this study. Univariate analysis (Table 5) suggested that CgCDR1 overexpression plays a major role in the regulation of MIC levels. However, its effect did not remain significant in the multivariate analysis, which highlighted the major independent effect of CgSNQ2 overexpression. Both univariate and multivariate analyses showed that copy number and overexpression were markedly correlated for CgPDH1 but not for the other genes. In patient 1, for whom sequential C. glabrata isolates displayed a stable genotype, switching to fluconazole resistance was associated with both overexpression and increased gene copy number of CgPDH1 (Table 4). A high correlation between the expression levels of all membrane transporter genes was detected, which is probably because they are controlled by the same transcription factor CgPDR1 (Ferrari et al., 2009; Paul et al., 2011). This transcription factor has been associated with both resistance to azole antifungals via the overexpression of ABC transporter genes and enhanced virulence in C. glabrata due to gain-of-function mutations (Ferrari et al., 2009).
CgCDR1 and CgPDH1 overexpression-driven drug efflux is the most frequent fluconazole resistance mechanism observed in C. glabrata (Miyazaki et al., 1998; Sanglard et al., 1999, 2001; Wada et al., 2002; Coleman and Mylonakis, 2009). Numerous reports have documented acquired resistance upon exposure to long-term fluconazole therapy (Coste et al., 2007; Shin et al., 2007), as observed in the four patients who acquired resistant isolates (Table 3). Although CgCDR1 and CgPDH1 expression could not be determined in the fluconazole susceptible isolate, overexpression of both genes was associated with fluconazole resistance in the isolate derived from patient 2. Acquisition of both CgPDH1 overexpression and increased gene copy number was observed in isolates derived from patient 1 (Table 4).
An overproduction of lanosterol-demethylase in C. glabrata has initially been described by Marichal et al. who found that duplication of the entire chromosome E (Marichal et al., 1997), the chromosome on which ERG11 is located, was associated with a four-fold increase in ERG11 copy number in fluconazole resistant compared with susceptible isolates. A recent study has also shown that lanosterol-demethylase overproduction, for which the mechanism is unknown, was involved in fluconazole resistance (Redding et al., 2003). In our study, an increase in CgERG11 gene copy number was associated with CgERG11 overexpression in only one isolate. Furthermore, CgCDR1, CgPDH1, and/or CgERG11 gene copy number was inconsistently associated with overexpression, thereby suggesting that an increase in gene copy number might sometime result in gene silencing via either heterochromatin formation or DNA methylation (Merrick and Duraisingh, 2006; Mishra et al., 2011).
A strong correlation between CgCDR1, CgPDH1, and CgERG11 gene copy number was observed (Table 4), although these genes are located on chromosomes M, F, and E, respectively. Further genome hybridization experiments may shed light on the underlying mechanisms and enable the localization of these amplified genes. Many gene duplication mechanisms have been described in various regions of the C. glabrata genome. Poláková et al. have suggested that duplication occurs in chromosomal segments that encode for some ABC transporter genes (Poláková et al., 2009). Similarly, Mûller et al. have shown that the mechanisms of microevolution of this asexual species include recombination within tandem arrays of repeated genes, which often encode cell wall proteins, thereby suggesting a possible adaptive function (Mûller et al., 2009). The fact that some genotypically identical isolates (genotype 3 and 7, Table 2) displayed different fluconazole resistance mechanisms support the hypothesis that fluconazole resistance is chiefly associated with the overexpression of normally present genes within the C. glabrata genome rather than gene mutation.
In conclusion, this population-based approach provides further evidence that overexpression of membrane transporter genes is a pivotal fluconazole resistance mechanism in C. glabrata clinical isolates. A high correlation between the expressions of these genes, which are controlled by the same transcription factor, was observed. However, the multivariate analysis revealed, for the first time, that CgSNQ2 exerts a major independent effect on fluconazole resistance. Regarding the other genes assessed, interactions played a greater role than the main effects in fluconazole resistance. Notably, CgPDH1 overexpression was significantly associated with increased gene copy number, which was not observed with the other genes analyzed. Furthermore, a correlation between gene copy number and fluconazole resistance was only observed for CgERG11, thereby suggesting that an increase in copy number of C. glabrata membrane transporter genes might result in gene silencing. This study provides only partial insight into C. glabrata fluconazole resistance mechanisms, as many other mechanisms have not been analyzed, including those targeting other membrane components. Nevertheless, our findings highlight the important role that complex gene copy number and overexpression interactions play in fluconazole resistance, which should been taken into account in further studies.
Author contributions
Salma Abbes carried out the collection of the clinical isolates, the molecular genetic studies and drafted the manuscript. Charles Mary participated in the design of the molecular genetic studies and drafted the manuscript. Hayet Sellami participated in the study design and isolates and clinical data collection. Annie Michel-Nguyen participated in C. glabrata isolates' identification and in vitro susceptibility assays analysis. Stéphane Ranque performed the statistical analysis. Ali Ayadi and Stéphane Ranque conceived the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Salma Abbes received travel grants from Aix Marseille University. We gratefully acknowledge Sandra Moore for English editing.
References
- Abbes S., Sellami H., Sellami A., Hadrich I., Amouri I., Mahfoudh N., et al. (2012). Candida glabrata strain relatedness by new microsatellite markers. Eur. J. Clin. Microbiol. Infect. Dis. 31, 83–91 10.1007/s10096-011-1280-4 [DOI] [PubMed] [Google Scholar]
- Abbes S., Sellami H., Sellami A., Makni F., Mahfoudh N., Makni H., et al. (2011). Microsatellite analysis and susceptibility to FCZ of Candida glabrata invasive isolates in Sfax Hospital, Tunisia. Med. Mycol. 49, 10–15 10.3109/13693786.2010.493561 [DOI] [PubMed] [Google Scholar]
- Amouri I., Sellami H., Borji N., Abbes S., Sellami A., Cheikhrouhou F., et al. (2011). Epidemiological survey of vulvovaginal candidosis in Sfax, Tunisia. Mycoses 54, e499–e505 10.1111/j.1439-0507.2010.01965.x [DOI] [PubMed] [Google Scholar]
- Bouchara J. P., Zouhair R., Le Boudouil S., Renier G., Filmon R., Chabasse D., et al. (2000). In-vivo selection of an azole-resistant petite mutant of Candida glabrata. J. Med. Microbiol. 49, 977–984 [DOI] [PubMed] [Google Scholar]
- Cernicka J., Subik J. (2006). Resistance mechanisms in fluconazole-resistant Candida albicans isolates from vaginal candidiasis. Int. J. Antimicrob. Agents 27, 403–408 10.1016/j.ijantimicag.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Chau A. S., Mendrick C. A., Sabatelli F. J., Loebenberg D., McNicholas P. M. (2004). Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob. Agents Chemother. 48, 2124–2131 10.1128/AAC.48.6.2124-2131.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. (2008). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved standard CLSI document M27-A3. Wayne, PA: Clinical and Laboratory Standards Institute [Google Scholar]
- Coleman J. J., Mylonakis E. (2009). Efflux in fungi: la pièce de résistance. PLoS. Pathog. 5:e1000486 10.1371/journal.ppat.1000486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A., Selmecki A., Forche A., Diogo D., Bougnoux M.-E., d' Enfert C., et al. (2007). Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell. 6, 1889–1904 10.1128/EC.00151-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekema D. J., Messer S. A., Brueggemann A. B., Coffman S. L., Doern G. V., Herwaldt L. A., et al. (2002). Epidemiology of candidemia: 3-year results from the emerging infections and the epidemiology of Iowa organisms study. J. Clin. Microbiol. 40, 1298–1302 10.1128/JCM.40.4.1298-1302.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Ischer F., Calabrese D., Posteraro B., Sanguinetti M., Fadda G., et al. (2009). Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS. Pathog. 5:e1000268 10.1371/journal.ppat.1000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Chang Y. C. (2012). Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 8:e1003022 10.1371/journal.ppat.1003022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Redding S., Dongari-Bagtzoglou A. (2007). Candida glabrata: an emerging oral opportunistic pathogen. J. Dent. Res. 86, 204–215 10.1177/154405910708600304 [DOI] [PubMed] [Google Scholar]
- Marichal P., Vanden Bossche H., Odds F. C., Nobels G., Warnock D. W., Timmerman V., et al. (1997). Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41, 2229–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick C. J., Duraisingh M. T. (2006). Heterochromatin-mediated control of virulence gene expression. Mol. Microbiol. 62, 612–620 10.1111/j.1365-2958.2006.05397.x [DOI] [PubMed] [Google Scholar]
- Mishra P. K., Baum M., Carbon J. (2011). DNA methylation regulates phenotype-dependent transcriptional activity in Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 108, 11965–11970 10.1073/pnas.1109631108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H., Miyazaki Y., Geber A., Parkinson T., Hitchcock C., Falconer D. J., et al. (1998). Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents. Chemother. 42, 1695–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mûller H., Thierry A., Coppée J.-Y., Gouyette C., Hennequin C., Sismeiro O., et al. (2009). Genomic polymorphism in the population of Candida glabrata: gene copy-number variation and chromosomal translocations. Fungal. Genet. Biol. 46, 264–276 10.1016/j.fgb.2008.11.006 [DOI] [PubMed] [Google Scholar]
- Niimi M., Nagai Y., Niimi K., Wada S. I., Cannon R. D., Uehara Y., et al. (2002). Identification of two proteins induced by exposure of the pathogenic fungus Candida glabrata to fluconazole. J. Chromatogr. B. Analyt. Technol. Biomed. Life. Sci. 782, 245–252 10.1016/S1570-0232(02)00668-2 [DOI] [PubMed] [Google Scholar]
- Paul S., Schmidt J. A., Moye-Rowley W. S. (2011). Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata. Eukaryot. Cell. 10, 187–197 10.1128/EC.00277-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Diekema D. J. (2004). Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 10(Suppl. 1), 11–23 10.1111/j.1470-9465.2004.t01-1-00844.x [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Diekema D. J. (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Messer S. A., Hollis R. J., Boyken L., Tendolkar S., Kroeger J., et al. (2009). Variation in susceptibility of bloodstream isolates of Candida glabrata to fluconazole according to patient age and geographic location in the United States in 2001 to 2007. J. Clin. Microbiol. 47, 3185–3190 10.1128/JCM.00946-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poláková S., Blume C., Zárate J. A., Mentel M., Jørck-Ramberg D., Stenderup J., et al. (2009). Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. U.S.A. 106, 2688–2693 10.1073/pnas.0809793106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding S. W., Kirkpatrick W. R., Saville S., Coco B. J., White W., Fothergill A., et al. (2003). Multiple patterns of resistance to fluconazole in Candida glabrata isolates from a patient with oropharyngeal candidiasis receiving head and neck radiation. J. Clin. Microbiol. 41, 619–622 10.1128/JCM.41.2.619-622.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Bille J. (2001). Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45, 1174–1183 10.1128/AAC.45.4.1174-1183.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Calabrese D., Majcherczyk P. A., Bille J. (1999). The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43, 2753–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M., Posteraro B., Fiori B., Ranno S., Torelli R., Fadda G. (2005). Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 49, 668–679 10.1128/AAC.49.2.668-679.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellami A., Sellami H., Néji S., Makni F., Abbes S., Cheikhrouhou F., et al. (2011). Antifungal susceptibility of bloodstream Candida isolates in Sfax hospital: Tunisia. Mycopathologia 171, 417–422 10.1007/s11046-010-9388-0 [DOI] [PubMed] [Google Scholar]
- Shin J. H., Chae M. J., Song J. W., Jung S.-I., Cho D., Kee S. J., et al. (2007). Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J. Clin. Microbiol. 45, 2385–2391 10.1128/JCM.00381-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli R., Posteraro B., Ferrari S., La Sorda M., Fadda G., Sanglard D., et al. (2008). The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol. Microbiol. 68, 186–201 10.1111/j.1365-2958.2008.06143.x [DOI] [PubMed] [Google Scholar]
- Wada S. I., Niimi M., Niimi K., Holmes A. R., Monk B. C., Cannon R. D., et al. (2002). Candida glabrata ATP-binding cassette transporters CDR1p and PDH1p expressed in a Saccharomyces cerevisiae strain deficient in membrane transporters show phosphorylation-dependent pumping properties. J. Biol. Chem. 277, 46809–46821 10.1074/jbc.M207817200 [DOI] [PubMed] [Google Scholar]


