Abstract
Objectives
To identify disease-causing mutations in two Chinese families with autosomal dominant retinitis pigmentosa (adRP).
Design
Prospective analysis.
Patients
Two Chinese adRP families underwent genetic diagnosis. A specific hereditary eye disease enrichment panel (HEDEP) based on targeted exome capture technology was used to collect the protein coding regions of targeted 371 hereditary eye disease genes; high throughput sequencing was done with the Illumina HiSeq 2000 platform. The identified variants were confirmed with Sanger sequencing.
Setting
All experiments were performed in a large laboratory specialising in genetic studies in the Department of Ophthalmology, Peking University Third Hospital.
Results
Two novel mutations, including one splice site mutation (Int10 c.1074-2 A>T; p.Y359SfsX29) and one insertion (c.824_825insA; p.Y275X) of PRPF31 were identified in the two families. The two mutations segregated with the disease phenotype in their respective families.
Conclusions
Our findings broaden the spectrum of PRPF31 mutations causing adRP and the phenotypic spectrum of the disease in Chinese patients. The HEDEP based on targeted exome capture technology is an efficient method for molecular diagnosis in adRP patients.
Strengths and limitations of this study.
The HEDEP based on targeted exome capture technology is an efficient method for molecular diagnosis in adRP patients.
Both mutations result in premature termination codons before the last exon, thus insufficient functioning due to haploinsufficiency instead of aberrant function of the mutated proteins seems to be the most probably reason in these two families. However no experiment was done to prove it in this study.
Introduction
Retinitis pigmentosa (RP) is an inherited retinal degeneration that affects approximately one in 3500 individuals, with an estimated total of 1.5 million patients worldwide.1 2 Typically patients affected by RP first suffer from night blindness, most often during adolescence. Rod and cone photoreceptor cells start to degenerate from the mid periphery to the far periphery and the centre of the retina, resulting in the so-called tunnel vision. Later in life, central vision is also lost, leading to legal or complete blindness.3 Clinical hallmarks of RP include bone spicule deposits, attenuated retinal blood vessels, optic disc pallor, visual field loss, and abnormal, diminished or non-recordable electroretinographic responses.
Autosomal dominant RP (adRP) accounts for 20–25% of all RP patients.4 To date, more than 24 genes (Best 1, C1QTNF5, CA4, CRX, FSCN2, GUCA1B, IMPDH1, KLHL7, NR2E3, NRL, PRPF3, PRPF31, PRPF6, PRPF8, PRPH2, RDH12, RHO, ROM1, RP1, RP9, RPE65, SEMA4A, SNRP200 and TOPORS) have been reported to be associated with adRP (https://sph.uth.edu/retnet/disease.htm). Traditionally, patients from RP families are studied with linkage analysis, or gene-by-gene screening,5 6 which is costly, requires substantial human resources, and is time-consuming, thus making molecular diagnosis difficult and complex. This has led to the development of mutation screens based on arrayed primer extension technology, which enable the simultaneous detection of multiple mutations from one individual.7 However, this approach is limited by the fact that it can only detect sequence variants previously reported, with novel mutations unreported.8 It would be advantageous to screen for all the variants in the protein coding regions of targeted genes. One approach to achieve this is to develop a specific hereditary eye disease enrichment panel (HEDEP) based on exome capture technology to re-sequence simultaneously all the exons from targeted genes.
The aim of this study is to describe the development of a specific HEDEP and its application in molecular diagnosis of two Chinese families with adRP, and to characterise the phenotypic manifestation associated with the mutation.
Materials and methods
Study subjects and clinical evaluation
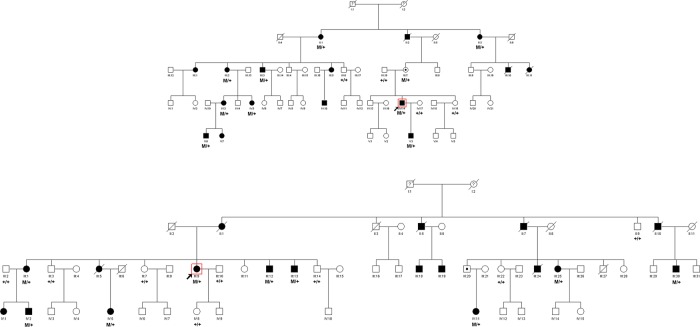
Two Chinese families of Han ethnicity with adRP were identified in the Anhui and Hubei provinces, respectively; there was no history of other ocular or systemic abnormalities in the families. The family adRP-19 has 16 affected individuals in four generations; 14 individuals (nine affected and five unaffected) participated in the study (figure 1A). The family adRP-61 has 18 affected individuals in three generations; 17 individuals (nine affected and eight unaffected) participated (figure 1B). Medical and ophthalmic histories were obtained, and ophthalmological examination was carried out. One hundred sporadic RP patients recruited in the Department of Ophthalmology, Peking University Third Hospital were used for RPPF31 gene mutation screening. One hundred healthy individuals from the Chinese Han ethnic population were recruited to serve as controls. All procedures used in this study conformed to the tenets of the Declaration of Helsinki. All experiments involving DNA and RNA of the patients and their relatives were approved by the Peking University Third Hospital Medical Ethics Committee. Informed consent was obtained from all participants.
Figure 1.
Pedigrees of two Chinese autosomal dominant retinitis pigmentosa (adRP) families with PRPF31 mutations and co-segregation in available family members. Filled symbols represent affected members, unfilled symbols represent unaffected members, and dotted symbols represent asymptomatic carriers. Question marks indicate that it is not clear whether the individual is affected or not. Squares signify male, and circles female members. Arrows mark the index patients. M refers to the mutant allele, and + indicates a normal allele.
Screening for mutations
Blood samples were collected and genomic DNA was extracted by standard protocols (D2492 Blood DNA Maxi Kit, Omega Bio-Tek, Norcross, Georgia, USA). A specific HEDEP based on targeted exome capture technology was used to collect the protein coding regions of targeted genes. This HEDEP was able to capture 371 hereditary eye disease genes, which cover 24 adRP associated genes (Best 1, C1QTNF5, CA4, CRX, FSCN2, GUCA1B, IMPDH1, KLHL7, NR2E3, NRL, PRPF3, PRPF31, PRPF6, PRPF8, PRPH2, RDH12, RHO, ROM1, RP1, RP9, RPE65, SEMA4A, SNRP200 and TOPORS). Genomic DNA (50 μg) from two patients (III3 and IV14), one carrier (III7) and one control (III18) of family adRP-19, and three patients (III13, III30 and IV2) and one control (III2) of family adRP-61 were used for targeted exome capture. The exon-enriched DNA libraries were then prepared for high throughput sequencing with the Illumina HiSeq 2000 platform. The obtained mean exome coverage was more than 98%, with variants accuracy at more than 99%. In these two families, we only analysed mutations that occurred in 24 adRP related genes. The shared changes in the affected individuals but not in the normal control were identified. The changes were filtered against exome data from ethnic Han Chinese Beijing available in the 1000 Genomes Project (fttp://www.1000genome.org), and against the Han Chinese Beijing SNPs in the dbSNP131. Sanger sequencing was then used to validate the identified potential disease-causing variants. Splice-site variants were analysed using the prediction program AUGUSTUS (http://bioinf.uni-greifswald.de/augustus/submission).
Mutation validation
The shared variants in the affected individuals but not in the normal control were then confirmed by direct PCR product sequencing using Bigdye terminator V3.1 cycle sequencing kits (Applied Biosystems, Foster City, California, USA) and analysed on an ABI 3130XL genetic analyzer. Sanger sequencing was used to determine whether the variant co-segregated with the disease phenotype in these two families. Primer pairs for individual exons were designed using the primer program (http://www.yeastgenome.org/cgi-bin/web-primer) (DNA reference number NG_009759). PCR primers, annealing temperatures, and amplimer-specific details are listed in table 1.
Table 1.
PCR primers used for PRPF31 amplification
| Primer | Forward | Reverse | Product |
|---|---|---|---|
| PRPF31 Exon 2&3 | CTGGGGGAGAATCATCGCTC | AAGGCTCTGGAAAAGGCT | 557 |
| PRPF31 Exon 4 | CGAGAGGGGGTAGGGATTTAGATAC | ACCTCGATCTGAGCTTGGGCTTAG | 252 |
| PRPF31 Exon 5 | AAGAAGGGGACATGGGTGTTA | TCCTCTCCATCGTCTCCAGA | 287 |
| PRPF31 Exon 6–7 | CAAGAGAGGTTCTCGAGCCTT | TTTCCCAAGGTCACAGTGTCA | 589 |
| PRPF31 Exon 8 | AGCCCCCAGGCAGATTTACT | TCCTGAGTGCTACCGTCAGCT | 350 |
| PRPF31 Exon 9 | TAGAGCCCAAGGGTGGAAA | TTGGTAGGACAGTGCTCGCT | 333 |
| PRPF31 Exon 10&11 | GGCAGCATTAGGTGCTGATTT | GTCGCTTTGGGGCTGAAT | 599 |
| PRPF31 Exon 12&13 | CAACTCTGAGCTCACAGAGCA | TCATCCTGGCCTTCTTCACA | 632 |
| PRPF31 Exon 14 | CTGTCTCATGCCCACCAA | TGGACCTCTGTGTCCCTTCA | 295 |
Isolation of total RNA and reverse transcription PCR analysis
Total RNA was extracted from peripheral whole blood samples by standard protocols (R6814 Blood RNA Kit, Omega). Reverse transcription (RT) was performed with oligonucleotide primers using Superscript II reverse transcriptase according to the manufacturer's protocol (Invitrogen Corporation, Grand Island, New York, USA). Primers for RT-PCR were designed to amplify exons 7–12 of PRPF31 mRNA (mRNA reference number NM_015629). The forward primer is 5′-GCCAAGATCATGGGTGTGG-3′, and the reverse primer is 5′-TGCAGCGTCTTGGAGATCCT-3′. The RT-PCR products were further cloned into plasmids for sequence analysis.
Results
Phenotype details
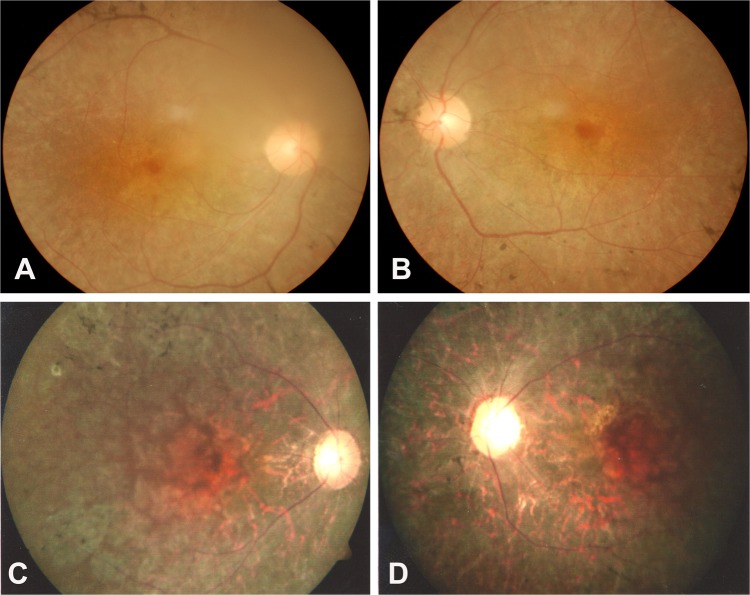
The clinical features of 14 members of family adRP-19 and 17 members of family adRP-61 who participated in this study are shown in table 2. All the tested affected individuals except III7 complained of night blindness and photophobia since childhood. Onset of the disease was noted range from 2 to 12 years of age. Fundus examination in the 38-year-old proband (IV14) of family adRP-19 showed bone spicule like pigmentation in the peripheral part of the retina, retinal arteriolar attenuation and retinal pigment epithelium (RPE) degeneration (figure 2A,B). Fundus examination in the 48-year-old proband (III9) of family adRP-61 showed bone spicule like pigmentation in the mid-periphery retina, bilateral attenuation of retinal vessels, RPE degeneration and pale optic disc (figure 2C,D). The 68-year-old asymptomatic carrier (III7) of family adRP-19 did not complain of night blindness, and fundus examination showed no RP changes in both eyes. Visual acuity in the majority of the patients declined quickly after 40 years of age.
Table 2.
Clinical data of the family members participating in the study
| Family | ID | Sex | Age (y) | Disease status | Onset age (y) | Visual acuity unaided (corrected) |
Fundus features | |
|---|---|---|---|---|---|---|---|---|
| R eye | L eye | |||||||
| adRP-19 | II:1 | F | 82 | Yes | Childhood | LP | LP | Cataract, can't see the fundus |
| II:3 | F | 80 | Yes | 8 | LP | LP | Cataract, can't see the fundus | |
| III:2 | F | 58 | Yes | 3 | LP | LP | Cataract, can't see the fundus | |
| III:3 | M | 54 | Yes | 3 | 0.1 | 0.1 | Bilateral attenuation of retinal vessels, bone spicule pigments throughout the fundus, RPE degeneration, pale optic disc | |
| III:6 | M | 46 | No | – | 1.5 | 1.5 | No RP changes in both eyes | |
| III:7 | F | 65 | No | – | 0.9 | 0.8 | No RP changes in both eyes | |
| III:18 | M | 68 | No | – | 1.2 | 1.2 | No RP changes in both eyes | |
| IV:3 | F | 29 | Yes | Childhood | 1.0 | 1.0 | Bone spicule pigments in peripheral part of the fundus | |
| IV:5 | F | 27 | Yes | Childhood | 1.0 | 1.0 | Bone spicule pigments in peripheral part of the fundus | |
| IV:14 | M | 38 | Yes | 3 | 0.3 | 0.5 | Bilateral attenuation of retinal vessels, bone spicule pigments in peripheral part of the fundus, RPE degeneration | |
| IV:15 | F | 34 | No | – | 1.5 | 1.5 | No RP changes in both eyes | |
| IV:17 | F | 37 | No | – | 1.5 | 1.2 | No RP changes in both eyes | |
| V:3 | M | 16 | Yes | 2 | 1.2 | 1.2 | Bone spicule pigments in peripheral part of the fundus | |
| V:6 | M | 3 | Yes | 2 | 1.2 | 1.2 | Bone spicule pigments in peripheral part of the fundus | |
| adRP-61 | II:9 | M | 80 | No | – | 1.0 | 1.0 | No RP changes in both eyes |
| III:1 | F | 63 | Yes | 12 | 0.4 | 0.5 | Bone spicule pigments in peripheral part of the fundus | |
| III:2 | M | 63 | No | – | 0.8 | 0.8 | No RP changes in both eyes | |
| III:3 | M | 61 | No | – | 1.0 | 1.0 | No RP changes in both eyes | |
| III:7 | F | 49 | No | – | 1.2 | 1.0 | No RP changes in both eyes | |
| III:9 | F | 48 | Yes | Childhood | 0.06 | 0.06 | Bilateral attenuation of retinal vessels, bone spicule pigments throughout the fundus, RPE degeneration, pale optic disc | |
| III:10 | M | 50 | No | – | 1.2 | 1.2 | No RP changes in both eyes | |
| III:12 | M | 40 | Yes | Childhood | 0.5 | 0.6 | Bone spicule pigments in peripheral part of the fundus | |
| III:13 | M | 38 | Yes | Childhood | 0.7 | 0.6 | Bone spicule pigments in peripheral part of the fundus | |
| III:14 | M | 45 | No | – | 1.0 | 1.0 | No RP changes in both eyes | |
| III:22 | F | 64 | No | – | 1.2 | 1.2 | No RP changes in both eyes | |
| III:25 | M | 50 | Yes | Childhood | 0.2 | 0.1 | Bilateral attenuation of retinal vessels, bone spicule pigments throughout the fundus, RPE degeneration, pale optic disc | |
| III:30 | M | 49 | Yes | Childhood | 0.1 | 0.06 | Bilateral attenuation of retinal vessels, bone spicule pigments throughout the fundus, RPE degeneration, pale optic disc | |
| IV:2 | M | 32 | Yes | Childhood | 0.7 | 0.6 | Bone spicule pigments in peripheral part of the fundus | |
| IV:5 | F | 35 | Yes | Childhood | 0.9 | 0.7 | Bone spicule pigments in peripheral part of the fundus | |
| IV:8 | F | 28 | No | – | 1.5 | 1.5 | No RP changes in both eyes | |
| IV:11 | F | 40 | Yes | Childhood | 0.5 | 0.6 | Bone spicule pigments in peripheral part of the fundus | |
RP, retinitis pigmentosa; RPE, retinal pigment epithelium.
Figure 2.
Fundus photographs of two probands with mutations in the PRPF31 gene. (A, B) Proband IV14 is from family autosomal dominant retinitis pigmentosa (adRP)-19. (C, D) Proband III9 is from family adRP-61. Typical RP changes can be seen.
Identification of mutations in PRPF31
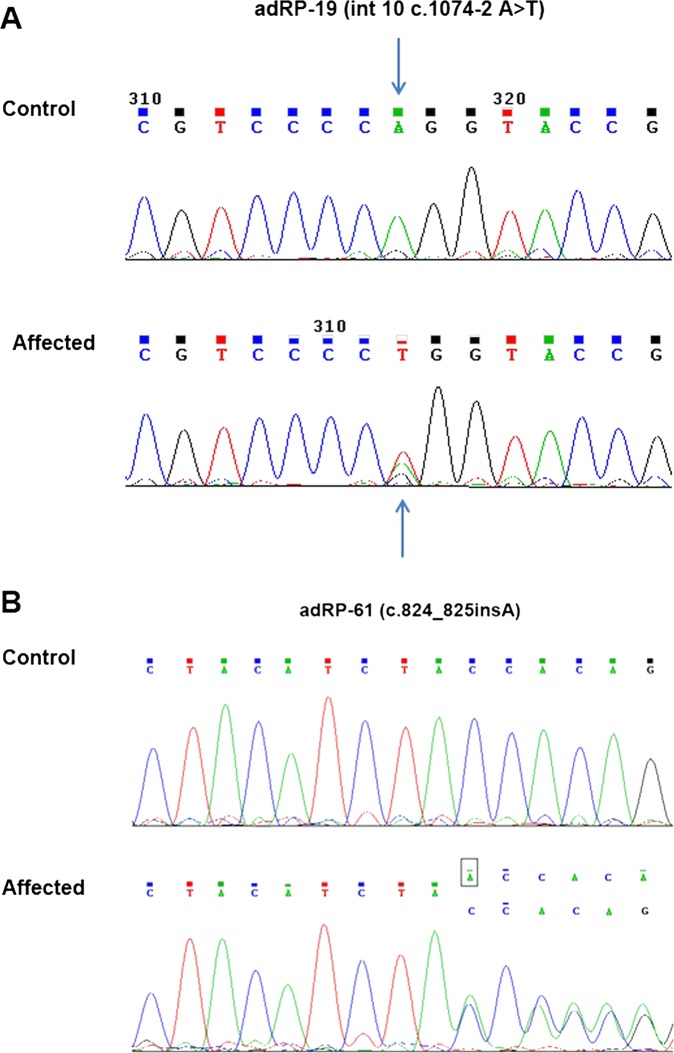
We selected four individuals in each family for targeted exome capture. We generated an average of 0.77 Gb of sequence with 228× average coverage for each individual with paired 100 bp reads. The generated sequence covered an average of 99.2% of the targeted bases with the accuracy of a variant call more than 99%, which is sufficient to pass the thresholds for calling SNPs and short insertions or deletions (indels). We filtered all the detected variants and found the potential disease causing mutations. Then we compared the shared variants in affected individuals with the ethnic Han Chinese Beijing available in the 1000 Genomes Project (fttp://www.1000genome.org), and against the Han Chinese Beijing SNPs in the dbSNP131. This left one splice site variation (Int10 c.1074-2 A>T) in family adRP-19 and one insertion (c.824_825insA; p.Y275X) in family adRP-61, which was shared among affected individuals but not in the normal control in each family. Sanger sequencing validation and segregation analysis was carried out, which demonstrated that these two variants (figure 3) co-segregated with the disease phenotype in each family (figure 1), but was absent in 100 matched normal controls.
Figure 3.
Sequencing results of the PRPF31 mutations in the two families. (A) Family autosomal dominant retinitis pigmentosa (adRP)-19 carried the mutation Int10 c.1074-2 A>T; p.Y359SfsX29. (B) Family adRP-61 carried the mutation c.824_825insA; p.Y275X.
We further carried out direct PCR sequencing of the PRPF31 exons in an additional 100 unrelated sporadic RP patients. All of these patients showed typical RP fundus features, including bilateral attenuation of retinal vessels, bone spicule pigments throughout the fundus, RPE degeneration and pale optic disc. No disease causing mutations were identified in these 100 sporadic patients.
Functional characterisation of the Int10 c.1074-2 A>T mutation using RT-PCR
To determine whether the Int10 c.1074-2 A>T splicing mutation in family adRP-19 has any effect on mRNA splicing, we performed RT-PCR for PRPF31 using total RNA samples isolated from peripheral blood samples from two patients (IV3 and V3), one unaffected family member (IV15) and two normal controls not related to the family. The RT-PCR products were further cloned into plasmids for sequence analysis. It yielded a 590 bp product from normal control samples as expected with normal splicing. One 590 bp fragment and another 517 bp product were amplified from the sample of affected individuals. Sequence analysis showed that the 517 bp product skipped exon11, resulting in frameshift—ie, p.Y359SfsX29 leading to premature termination with 28 new amino acids downstream. The sequence comparison details between wild type and mutated are listed in online supplementary figure S1.
Discussion
In this study we reported the identification of two novel mutations—one splice site mutation (Int10 c.1074-2 A>T; p.Y359SfsX29) and one insertion (c.824_825insA; p.Y275X) of PRPF31 in two Chinese adRP families. The identified splice site mutation (Int10 c.1074-2 A>T; p.Y359SfsX29) in family adRP-19 segregated in all nine affected patients and one asymptomatic carrier, the mother of an affected patient who manifested the disease from childhood. The identified insertion mutation (c.824_825insA; p.Y275X) in family adRP-61 segregated in all the nine affected patients, including one woman who inherited the mutation from her 65-year-old asymptomatic father. This is consistent with incomplete penetrance of the PRPF31 mutation.
The human PRPF31 gene contains 14 exons and encodes a 61 kDa protein of 499 amino acids, the core component of the U4/U6•U5 tri-snRNP complex which constitutes a major building block of the pre-mRNA processing spliceosome.9 Although PRPF31 is ubiquitously expressed, patients with mutant PRPF31 alleles only show symptoms in the retina and not in other organs.10 At present, up to 50 mutations (including 15 splice defects, 10 missenses, 23 deletions and 2 insertions) in PRPF31 have been reported to be linked with adRP and sporadic RP cases.5 11 12 Splice defect have been reported in Int1,13 Int2,14 Int4,14 Int5,15 Int6,16 Int8,14 Int10,17 Int11 and Int13.10 18 Insertions have been reported in exon 7 and exon 8.19 All the above splice defect and insertion mutations resulted in frameshift, leading to premature termination. A previous study from Rio Frio et al20 demonstrated that most PRPF31 mutations bearing premature termination codons (PTCs) before the last exon behave as null allele, resulting in haploinsufficiency as their corresponding mRNA is degraded by nonsense-mediated mRNA decay (NMD). However, if the PTC produced by the mutation occurs in the last exon of a given gene, the mutant mRNA is insensitive to NMD and is thought to be translated into a truncated protein.21 The splice site mutation (Int10 c.1074-2 A>T) in the present study caused the skipping of exon 11, resulting in frameshift, leading to premature termination with 28 new amino acids downstream (ie, p.Y359SfsX29). The insertion mutation (c.824_825insA) in the present study happened in exon 8, resulting in the following amino acid changing from Thr to stop codon (p.Y275X). Since both mutations result in PTC before the last exon, insufficient functioning due to haploinsufficiency instead of aberrant function of the mutated proteins seems to be the most probable cause in these two families.
Several studies have been reported on the use of genotyping microarray for genetic diagnosis of retinal disease, such as Stargardt disease,22 Leber congenital amaurosis,23 Usher syndrome,24 autosomal recessive RP25 and adRP.7 Different from arrayed primer extension technology used in most of previous microarray studies, the HEDEP in this study was based on target exon capture technology. HEDEP has been able to capture, for example, 371 hereditary eye disease genes, which cover 53 RP associated genes, 7 Stargardt associated genes, 19 Leber congenital amaurosis associated genes, 11 Usher syndrome associated genes, 33 cone and rod dystrophy associated genes, 13 chorioretinal atrophy associated genes, 35 microphthalmia associated genes, 46 congenital cataract associated genes, 4 glaucoma associated genes, 6 familial exudative vitreoretinopathy associated genes and 35 RP related syndrome associated genes. It is applicable to genetic study of broader inherited eye disease. Sanger sequencing in this study confirmed the mutation previously detected with the HEDEP, which proved the sensitivity and specificity of the HEDEP for application in molecular diagnosis of inherited eye diseases.
We presented a successful genetic diagnosis with a specific HEDEP, and identified two novel mutations— one splice site mutation (Int10 c.1074-2 A>T; p.Y359SfsX29) and one insertion (c.824_825insA; p.Y275X) of PRPF31—in two Chinese adRP families with incomplete penetrance. Both mutations resulted in PTC before the last exon, thus insufficient functioning due to haploinsufficiency was the most probable cause in these two families. Our findings broaden the spectrum of PRPF31 mutations causing adRP and the phenotypic spectrum of the disease in Chinese patients, which will be helpful for genetic consultation and genetic diagnosis in the future.
Supplementary Material
Acknowledgments
The authors are grateful to all family members for their participation in this study.
Footnotes
Contributors: Conceived and designed the experiment: LY, HZ, GL and ZM. Performed the experiments: LY, XY, LW and NC. Analysed the data: LY, LW and NC. Wrote the paper: LY and ZM.
Funding: This study was supported by the National Natural Science Foundation of China (Grant 81170877).
Competing interests: None.
Patient consent: Obtained.
Ethics approval: This study was conducted with the approval of Peking University Third Hospital Medical Ethics Committee (No. 2012093).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The original sequencing results are available on request.
References
- 1.Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl 2002;233:1–34 [DOI] [PubMed] [Google Scholar]
- 2.Churchill JD, Bowne SJ, Sullivan LS, et al. Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 2013;54:1411–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis 2006;1:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrari S, Di Iorio E, Barbaro V, et al. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics 2011;12:238–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saini S, Robinson PN, Singh JR, et al. A novel 7 bp deletion in PRPF31 associated with autosomal dominant retinitis pigmentosa with incomplete penetrance in an Indian family. Exp Eye Res 2012;104:82–8 [DOI] [PubMed] [Google Scholar]
- 6.Naz S, Ali S, Riazuddin SA, et al. Mutations in RLBP1 associated with fundus albipunctatus in consanguineous Pakistani families. Br J Ophthalmol 2011;95:1019–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco-Kelly F, García-Hoyos M, Cortón M, et al. Genotyping microarray: mutation screening in Spanish families with autosomal dominant retinitis pigmentosa. Mol Vis 2012;18:1478–83 [PMC free article] [PubMed] [Google Scholar]
- 8.Clark GR, Crowe P, Muszynska D, et al. Development of a diagnostic genetic test for simplex and autosomal recessive retinitis pigmentosa. Ophthalmology 2010;117:2169–77 [DOI] [PubMed] [Google Scholar]
- 9.Linder B, Dill H, Hirmer A, et al. Systemic splicing factor deficiency causes tissue-specific defects: a zebrafish model for retinitis pigmentosa. Hum Mol Genet 2011;20:368–77 [DOI] [PubMed] [Google Scholar]
- 10.Waseem NH, Vaclavik V, Webster A, et al. Mutations in the gene coding for the pre-mRNA splicing factor, PRPF31, in patients with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 2007;48:1330–4 [DOI] [PubMed] [Google Scholar]
- 11.Audo I, Bujakowska K, Mohand-Saïd S, et al. Prevalence and novelty of PRPF31 mutations in French autosomal dominant rod-cone dystrophy patients and a review of published reports. BMC Med Genet 2010;11:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu F, Sui R, Liang X, et al. Novel PRPF31 mutations associated with Chinese autosomal dominant retinitis pigmentosa patients. Mol Vis 2012;18:3021–. [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JY, Dai X, Sheng J, et al. Identification and functional characterization of a novel splicing mutation in RP gene PRPF31. Biochem Biophys Res Commun 2008;367:420–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivolta C, McGee TL, Rio Frio T, et al. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat 2006;27:644–53 [DOI] [PubMed] [Google Scholar]
- 15.Xia K, Zheng D, Pan Q, et al. A novel PRPF31 splice-site mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol Vis 2004;10:361–5 [PubMed] [Google Scholar]
- 16.Chakarova CF, Cherninkova S, Tournev I, et al. Molecular genetics of retinitis pigmentosa in two Romani (Gypsy) families. Mol Vis 2006;12:909–14 [PubMed] [Google Scholar]
- 17.Sullivan LS, Bowne SJ, Birch DG, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci 2006;47:3052–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rio Frio T, McGee TL, Wade NM, et al. A single-base substitution within an intronic repetitive element causes dominant retinitis pigmentosa with reduced penetrance. Hum Mutat 2009; 30:1340–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vithana EN, Abu-Safieh L, Allen MJ, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 2001;8:375–81 [DOI] [PubMed] [Google Scholar]
- 20.Rio Frio T, Wade NM, Ransijn A, et al. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest 2008;118:1519–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 2007;76:51–74 [DOI] [PubMed] [Google Scholar]
- 22.Valverde D, Riveiro-Alvarez R, Bernal S, et al. Microarray-based mutation analysis of the ABCA4 gene in Spanish patients with Stargardt disease: evidence of a prevalent mutated allele. Mol Vis 2006;12:902–8 [PubMed] [Google Scholar]
- 23.Vallespin E, Cantalapiedra D, Riveiro-Alvarez R, et al. Mutation screening of 299 Spanish families with retinal dystrophies by Leber congenital amaurosis genotyping microarray. Invest Ophthalmol Vis Sci 2007;48:5653–61 [DOI] [PubMed] [Google Scholar]
- 24.Jaijo T, Aller E, García-García G, et al. Microarray-based mutation analysis of 183 Spanish families with Usher syndrome. Invest Ophthalmol Vis Sci 2010;51:1311–17 [DOI] [PubMed] [Google Scholar]
- 25.Ávila-Fernández A, Cantalapiedra D, Aller E, et al. Mutation analysis of 272 Spanish families affected by autosomal recessive retinitis pigmentosa using a genotyping microarray. Mol Vis 2010;16:2550–8 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.