Abstract
CRISPR-Cas systems have been used with single-guide RNAs for accurate gene disruption and conversion in multiple biological systems. Here we report the use of the endonuclease Cas9 to target genomic sequences in the C. elegans germline, utilizing single-guide RNAs that are expressed from a U6 small nuclear RNA promoter. Our results demonstrate that targeted, heritable genetic alterations can be achieved in C. elegans, providing a convenient and effective approach for generating loss-of-function mutants.
Clustered, regularly interspaced, short palindromic repeats (CRISPR) and CRISPR-associated (Cas) systems are adaptive mechanisms evolved by bacteria and archaea to repel invading viruses and plasmids1, 2. CRISPR-Cas systems incorporate foreign DNA sequences into host CRISPR loci to generate short CRISPR RNAs (crRNAs) that direct sequence-specific cleavage of homologous target double-stranded DNA by Cas endonucleases3, 4. Recent work with the S. pyogenes type II CRISPR system, which requires the nuclease Cas9, a targeting crRNA, and an additional trans-activating crRNA (tracrRNA), has shown that fusing the crRNA to the tracrRNA to form a single guide RNA (sgRNA) is sufficient to direct Cas9-mediated target cleavage4. This system has been used for genome engineering in yeast5, Drosophila6, human and mouse cell lines7–10, and in zebrafish and mouse11, 12. Here we configured Cas9 and sgRNAs for targeted gene disruption in the nematode C. elegans.
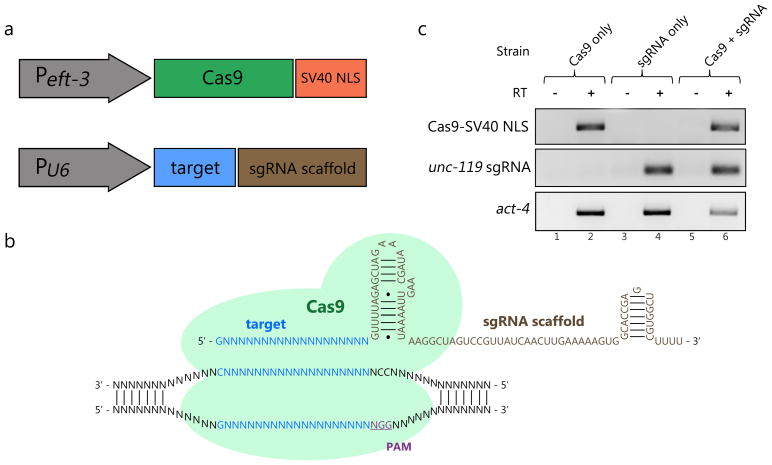
We first generated vectors to express Cas9 and sgRNAs in the germline (Fig. 1a). An SV40 nuclear localization signal (NLS) was added to the 3′ end of the Cas9 open reading frame to ensure the enzyme would be properly localized to the nucleus8, 10. To drive expression of transcripts encoding this Cas9-SV40 NLS fusion protein, we utilized the promoter sequence from the gene eft-3, selected for its effectiveness in driving expression in the germline13. While previous studies have utilized vectors containing RNA polymerase III (pol III) promoters to transcribe small RNAs14 or sgRNAs in mammalian systems, no equivalent vector has been described in C. elegans. Studying conserved upstream and downstream regulatory sequences flanking a U6 snRNA gene in C. elegans, we derived a putative pol III promoter for sgRNA expression (Fig. 1a; Supplementary Fig. 1). It has been suggested that optimal expression from pol III promoters occurs when the first base transcribed is a purine15, 16. Combining this finding with the known sequence requirements of CRISPR-Cas guided cleavage, our sgRNA expression system enables the selection of target sequences of the form G/A(N)19NGG, where the G/A(N)19 represents a 20 nucleotide sequence that will recognize a homologous stretch of double-stranded DNA in the genome, and the 3′ NGG sequence represents the essential protospacer-associated motif (PAM)1 (Fig. 1b).
Figure 1.
A set of vectors that drive expression of Cas9 and sgRNAs in C. elegans. (A) The C. elegans eft-3 promoter drives transcription of Cas9 with a 3′ SV40 nuclear localization sequence. A pol III promoter (derived from a U6 snRNA locus) drives transcription of the sgRNA, which contains a target sequence and a scaffold sequence. (B) A schematic illustration of Cas9 interacting with sgRNA and its genomic target. (C) RT-PCR results demonstrating expression of Cas9 and sgRNA transcripts. Total RNA was tested from strains carrying Cas9 vector alone (lanes 1 and 2), unc-119 sgRNA vector alone (lanes 3 and 4), and both vectors (lanes 5 and 6) with primers specific for Cas9 (top panel) or unc-119 sgRNA (bottom panel). For all samples, control reactions were run in the absence of Reverse Transcriptase (-RT; lanes 1, 3, and 5).
We designed sgRNAs complementary to coding sequences in the unc-119 and dpy-13 genes. These genes were selected for targeting because loss-of-function alleles have been isolated at these loci that cause easily identifiable uncoordinated (Unc) or dumpy (Dpy) phenotypes, respectively17, 18. Studies have indicated that CRISPR-Cas guided double-strand breaks can be repaired through the process of non-homologous end joining (NHEJ), generating insertions and deletions (indels) in the vicinity of the cleavage site7–9, 11. We reasoned that indels disrupting the coding sequences of unc-119 and dpy-13 would mimic previously identified alleles causing Unc and Dpy phenotypes.
To verify expression of both Cas9-SV40 NLS and sgRNAs, we microinjected the gonads of wild type adults, generating transgenic progeny that carry each expression vector alone or both in stable extrachromosomal arrays19. Total RNA was isolated from these transgenic lines, and reverse transcription-polymerase chain reaction (RT-PCR) assays were performed to detect transcripts. These assays confirmed that Cas9-SV40 NLS and sgRNAs are transcribed in transgenic animals (Fig. 1c), indicating that the eft-3 and pol III promoters in our vectors are active.
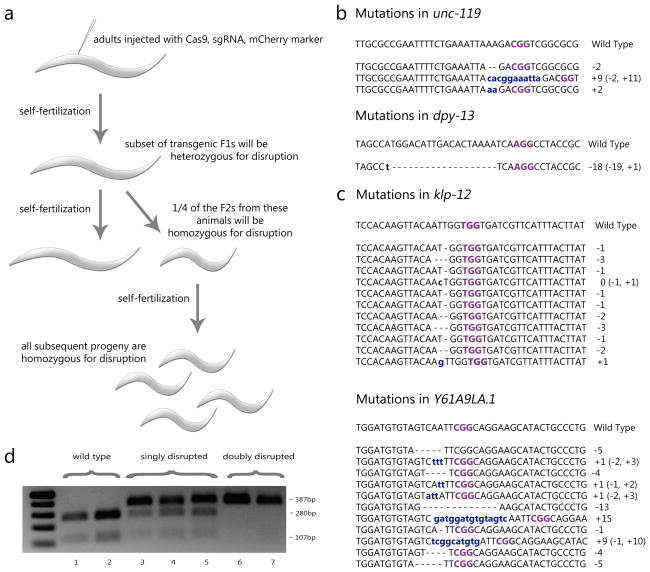
We next investigated whether our Cas9/sgRNA expression system could direct targeted cleavage and disruption of unc-119 and dpy-13 in the germline. We microinjected animals with vectors expressing Cas9, one of the two sgRNAs, and a vector driving expression of mCherry in body wall muscles to label transformed F1 progeny. No mCherry-positive F1 animals displayed Unc or Dpy phenotypes. We isolated these mCherry-positive animals and screened their F2 progeny for Unc or Dpy phenotypes (Fig. 2a). In two replicate experiments expressing Cas9 and the unc-119-specific sgRNA, we recovered Unc F2 progeny from 1/27 and 1/105 isolated F1 animals (Fig. 2b). In a third experiment targeting the unc-119 locus using higher concentrations of our expression vectors (see Supplementary Methods), Unc F2 progeny were recovered from 1/60 F1 animals. When targeting the dpy-13 locus, we recovered Dpy F2 progeny from 1/210 individual F1 animals (Fig. 2b). In all four experiments, when Unc and Dpy F2 progeny were identified, they were recovered at a frequency of 25% from singled F1 animals. All of the F3 progeny from Unc and Dpy F2 mutant animals displayed Unc and Dpy phenotypes, respectively (Fig. 2c and Supplementary Movies 1–3). These observed patterns of inheritance are consistent with recessive loss-of-function mutations originating in the germline of injected animals. We were unable to recover mutant animals from progeny of F1 animals not expressing our mCherry marker, or from animals injected with Cas9 or sgRNA alone (Supplementary Table 1), suggesting that both components are required for cleavage. To verify that disruptions targeted unc-119 and dpy-13, we isolated DNA from mutant animals and sequenced regions spanning the predicted sites of cleavage. The genomes of all Unc mutants and the Dpy mutant possessed a unique indel located within the expected target sequences, occurring three to four bases upstream of the PAM sequence (Fig. 2d). All of the identified indels are predicted to alter the coding sequence of each gene, and would lead to the production of truncated proteins. These molecular changes are consistent with the phenotypes we observe, resembling previously characterized loss-of-function mutants. These results indicate that our vector system enables the expression of Cas9 and sgRNAs in the germline to achieve targeted, heritable gene disruptions.
Figure 2.
Heritable, targeted gene disruptions in the germline using CRISPR-Cas systems. (A) Wild type (Bristol N2) adults were injected with vectors expressing Cas9, sgRNA, and a body wall muscle-specific mCherry marker. mCherry-positive F1 animals were isolated, a small fraction of which were heterozygous for the disruption. Next, the F2 animals were screened for mutant phenotypes, reflecting homozygous disruption. All further progeny of these F2 mutants were homozygous for the disruption. (B) A table summarizing the results of the four experiments, in which 4 disruptions were found out of 402 mCherry-positive F1 animals. (C) Images of worms from our wild type background line, a disrupted unc-119 line, and a disrupted dpy-13 line. (D) Sequences of the indel mutations found in our mutant lines. Insertions are marked in blue, deletions are marked by dashes, and the PAM is marked in purple. *The third experiment targeting the unc-119 locus utilized five-fold higher concentrations of expression vectors (see Supplementary Methods for details).
To extend our initial results and test whether we could also recover animals carrying disruptions that do not lead to visible phenotypes, we selected two additional loci (klp-12 and Y61A9LA.1) with no known loss-of-function phenotypes and generated sgRNAs to target them. We microinjected these animals with Cas9, sgRNA, and mCherry expression vectors as above, and isolated mCherry-positive F1 progeny. We allowed these F1 animals to lay eggs and then genotyped these animals by sequencing regions of genomic DNA spanning expected cleavage sites. In two replicate experiments targeting klp-12 and one targeting Y61A9LA.1, we generated disruptions in 80.3%, 77. 1%, and 18.1% of the F1s screened, respectively (Fig. 3a, 3b). Interestingly, at the klp-12 locus, 27 out of 80 F1 animals carrying a disruption were homozygous for a single disruption while other animals carried two unique disruption alleles. We speculate that these doubly-targeted mutant F1s are generated through two sequential break and repair events. The first event may occur in the haploid oocyte, where NHEJ-mediated repair introduces an indel The second event likely occurs later in the sperm-contributed chromosome, where either NHEJ introduces a second, unique indel, or through homologous recombination uses the already-disrupted chromosome as a template and copies the error, yielding a homozygous mutant. We followed the inheritance of four klp-12 alleles identified in F1 animals by genotyping single mCherry-negative F2 animals and confirmed the heritability of all of these disruptions (Supplementary Fig. 3).
Figure 3.
Heritable, targeted gene disruptions in genes that lead to no obvious phenotypes. (A) A table summarizing the results of the three experiments, in which 93 disruptions were found out of 173 mCherry-positive F1 animals. (B) Sequences of the indel mutations found in several of our mutant lines. Insertions are marked in blue, deletions are marked by dashes, and the PAM is marked in purple. (C) Sequence at the klp-12 locus showing the target PAM site in purple and the MfeI restriction site in green. (D) An image of a 1% agarose gel showing a restriction digest of PCR amplicons spanning the klp-12 cleavage site from seven F1 animals. Wild type sequences in lanes 1 and 2 are cut into bands of 280bp and 107bp, while doubly disrupted sequences remain full length at 387bp in lanes 6 and 7. Lanes 3, 4, and 5 show all three bands, indicating worms that are singly disrupted.
To demonstrate an additional screening strategy capable of identifying disruptions that do not cause obvious phenotypes, we designed our klp-12 sgRNA targeting sequence to overlap with that of a restriction enzyme, MfeI. When CRISPR-Cas mediated cleavage occurs at this site, any indels spanning the restriction enzyme recognition sequence would lead to a restriction fragment length polymorphism in PCR amplicons generated from mutant genomic DNA (Figure 3C). Using this approach, we were able to distinguish between wild type animals, singly-disrupted animals, and doubly-disrupted animals (Figure 3C) that were confirmed by our sequencing analysis described above. These results indicate that when possible, this method can provide a convenient way to pre-screen a large number of candidate F1 progeny for sequence disruptions and reduce the number of animals requiring validation by sequencing.
To assess the possibility of CRISPR-Cas cleavage at off-target loci in our mutant strains, we searched for other sites in the genome that could potentially be targeted by our sgRNAs. Evidence suggests that the 12 nucleotides in the target sequence proximal to the PAM are the most critical determinants of cleavage specificity and may constitute a ‘seed’ region20. We scanned the genome for sequences of the form (N)12NGG, and selected candidate off-target sites for each sgRNA that contained the minimum number of mismatches within these sequences. We sequenced the genomic regions spanning these potential cleavage sites in several of our mutant strains and found no evidence of cleavage or indels at these loci (Supplementary Fig. 2). However, these results do not systematically assess the specificity of CRISPR-Cas guided cleavage in C. elegans and future work will be required to further investigate the potential for off-target cleavage.
The discovery that RNA-guided endonucleases can cleave target sequences in the nuclei of eukaryotic cells has enabled genome editing in cultured cells, yeast, vertebrates, and Drosophila. Here, through the use of a U6 snRNA pol III promoter to drive sgRNA expression, we demonstrate that CRISPR-Cas guided cleavage can introduce heritable mutations in C. elegans. In principle, the methodology described here could be applied to other model organisms in which efficient delivery of DNA to the germline is feasible. Our results suggest that CRISPR-Cas based systems possess great potential for heritable genome editing in a wide variety of multicellular eukaryotes.
Methods
Strains and maintenance
The Bristol N2 strain (kindly provided by the Caenorhabditis Genetics Center, University of Minnesota) was used in all experiments described. All animals were grown on NGM agar plates seeded with the E. coli bacterial strain OP50, and maintained using standard procedures21.
Identification of a conserved U6 snRNA pol III promoter
In order to develop a pol III promoter expression vector, we identified a conserved U6 snRNA locus by performing BLAT searches using the consensus U6 snRNA sequence22. One locus on Chromosome IV was selected for further analysis, and alignment and conservation tracks were extracted from the UCSC genome browser23. We identified approximately 80 base pairs of upstream sequence and 10 base pairs of downstream sequence conserved among several nematode species (see Supplementary Figure 1 for alignment). We therefore conservatively chose to include 500 bases of upstream sequence and 237 bases of downstream sequence flanking the snRNA sequence.
sgRNA targeting sequence identification and selection
Using the known sequence requirements of CRISPR-Cas guided cleavage, we searched for target sequences in the C. elegans genome on the basis of the following criteria:
Sequences had to be of the form G/A(N)19NGG, where the G/A(N)19 represents a 20 nucleotide sequence that will recognize a homologous stretch of double-stranded DNA in the genome, and the 3′ NGG sequence represents the essential protospacer-associated motif (PAM).
If a protein-coding gene knockout is desired, sequences contained within known open reading frames should be targeted. Although this is not a strict requirement, it likely ensures that a disruption will create an allele that shifts the canonical reading frame, often producing premature termination codons.
Where possible, it is also desirable to look for target sequences that possess a restriction enzyme recognition sequence a few bases upstream of the PAM. This will facilitate pre-screening F1 progeny by restriction digests.
To actually select these sgRNA target sequences, we copied the genomic sequence spanning all of the coding exons and intervening intronic sequences of a gene of interest from Wormbase into Microsoft Word and, using the asterisk character as a wildcard, searched for strings that met the above criteria.
Plasmid construction
To create the Cas9-SV40 NLS expression vector, a worm codon-optimized Cas9 open reading frame with an internal intron sequence and a 3′ end fused SV40 nuclear localization signal sequence (see Supplementary Table 2 for a full sequence) was synthetically produced (Genscript Inc.) and inserted into the vector pUC57. This intron containing open reading frame was PCR-amplified using the oligonucleotide primers cas9 start F/cas9 tbb-2 UTR R (see Supplementary Table 2 for a full list of primers used in this study). The promoter region from the eft-3 gene and 3′ UTR from the gene tbb-2 were PCR-amplified from plasmid pCFJ601 (obtained from Addgene through the kind gift of E. Jorgensen and C. Frokjaer-Jensen) using the primers pUC57 EcoRI Peft-3 F/Peft-3 cas9 start R and tbb-2 UTR F/tbb-2 UTR pUC57 R, respectively. These three PCR products (promoter, Cas9-SV40 NLS + intron, and 3′ UTR) were then inserted into an EcoRI/HindIII digested pUC57 plasmid using the Gibson assembly method as previously described24.
To create the pol III promoter expression vector, we ordered two overlapping gBlocks gene fragments (IDT) collectively containing the 500 upstream nucleotides flanking a conserved U6 snRNA locus, a target sequence with homology to a portion of the coding sequence of the unc-119 gene, remaining sequence corresponding to the sgRNA, and 237 nucleotides downstream of the U6 snRNA locus (see Supplementary Table 2 for full sequences). The two gBlocks were stitched together by PCR using the primers U6prom EcoRI F/U6prom HindIII R. This PCR product was then digested with EcoRI and HindIII and ligated into an EcoRI/HindIII digested pUC57 plasmid, creating vector pU6::unc-119 sgRNA.
To generate the dpy-13 sgRNA expression vector, we used the pU6::unc-119 sgRNA vector above as a template and amplified two overlapping PCR fragments using the primers U6prom EcoRI F/dpy-13 gRNA R and dpy-13 gRNA F/U6prom HindIII R. These PCR products were gel purified, and then mixed together in a second PCR reaction with primers U6prom EcoRI F/U6prom HindIII R. This final PCR product was digested with EcoRI and HindIII and ligated into an EcoRI/HindIII digested pUC57 plasmid, creating the vector pU6::dpy-13 sgRNA.
To generate the klp-12 sgRNA expression vector, we used the pU6::unc-119 sgRNA vector above as a template and amplified two overlapping PCR fragments using the primers U6prom EcoRI F/klp-12 gRNA R and klp-12 gRNA F/U6prom HindIII R. These PCR products were gel purified, and then mixed together in a second PCR reaction with primers U6prom EcoRI F/U6prom HindIII R. This final PCR product was digested with EcoRI and HindIII and ligated into an EcoRI/HindIII digested pUC57 plasmid, creating the vector pU6::klp-12 sgRNA.
To generate the Y61A9LA.1 sgRNA expression vector, we used the pU6::unc-119 sgRNA vector above as a template and amplified two overlapping PCR fragments using the primers U6prom EcoRI F/Y61A9LA.1 gRNA R and Y61A9LA.1 gRNA F/U6prom HindIII R. These PCR products were gel purified, and then mixed together in a second PCR reaction with primers U6prom EcoRI F/U6prom HindIII R. This final PCR product was digested with EcoRI and HindIII and ligated into an EcoRI/HindIII digested pUC57 plasmid, creating the vector pU6::Y61A9LA.1 sgRNA.
DNA microinjection
Plasmid DNA was microinjected into the germline of adult hermaphrodite animals using standard methods as described previously25. Injection solutions were prepared to contain a final concentration of 100ng/uL for two replicate unc-119 experiments and the dpy-13 experiment, and 500ng/uL for a third unc-119 experiment and all klp-12 and Y61A9LA.1 experiments. In all injections, we used the vector pCFJ104 (Pmyo-3::mCherry) as a co-injection marker. The vectors used in this study were present at the following final concentrations in injection mixes:
(Peft-3::Cas9-SV40 NLS::tbb-2 3′UTR) : 50 ng/uL
(pU6::unc-119 or dpy-13 sgRNA) : 45 ng/uL
pCFJ104: 5 ng/uL
and
(Peft-3::Cas9-SV40 NLS::tbb-2 3′UTR) : 250 ng/uL
(pU6::unc-119 or klp-12 or Y61A9LA.1 sgRNA) : 225 ng/uL
pCFJ104: 25 ng/uL
When vectors were injected separately for Figure 1C, the final concentration of DNA was adjusted to 100 ng/uL by adding DNA ladder.
We have noticed that injections with a total DNA concentration of 500 ng/uL can lead to sterility (up to 25%) of F1 adult progeny. At present it is difficult to conclude if the cause of this sterility is due to an overall increase in plasmid DNA delivered in injections, or due to an increased concentration of a particular plasmid in our injection mix. This increase in sterility did not significantly affect our ability to recover fertile animals carrying disruptions at the unc-119, klp-12 and Y61A9LA.1 loci. If sterility does become an issue, we suggest testing several concentrations of each plasmid when trying to generate targeted disruptions in genes of interest.
RNA isolation and RT-PCR assays
Total RNA was isolated from lines stably carrying plasmids as extrachomosomal arrays using Tri reagent (Sigma) as recommended by the manufacturer. RT-PCR assays were performed using the OneStep RT-PCR kit (Qiagen) according to the protocol described by the manufacturer. Thirty nanograms of total RNA was used as input for each reaction. The sequences of primers used are provided in Supplementary Table 2.
Screening for disruptions in animals with no obvious phenotypes and genotyping
To screen for disruptions in the klp-12 gene, we placed F1 animals in 5 uL of single worm lysis buffer (10 mM Tris pH 8.0, 50 mM KCl, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween-20, 100 ug/mL proteinase K) and lysed the animals for one hour at 60°C, followed by incubation at 95°C to inactivate the proteinase K. We then amplified a region of genomic DNA spanning the predicted disruption site by PCR using Phusion High fidelity polymerase (Thermo Scientific) as recommended by the manufacturer, using all 5 uL of worm lysate as a template (see Supplementary Table 2 for a list of all primers used for PCR amplification and genotyping). PCR amplicons were then cleaned using the Genejet PCR purification kit (Thermo Scientific) as recommended by the manufacturer. 5 uL of PCR product were then digested with the restriction enzyme MfeI (NEB) per manufacturer recommendations, and digestion products were resolved on a 1% agarose gel, stained with 100 ug/mL ethidium bromide, and detected using a UV transilluminator.
To genotype all other animals and loci of interest, single animals were lysed, relevant regions were amplified by PCR, and PCR products were cleaned as described above. Cleaned PCR products were then sequenced by Sanger Sequencing methods (Genewiz).
To monitor inheritance of targeted disruptions at the klp-12 locus, we followed the F2 progeny of three F1 animals carrying four alleles with disrupted sequences (two animals carrying a homozygous mutation and one animal carrying two independent disruptions). We sequenced single F2 progeny from these animals (Five F2s from each of the homozygous mutants and 18 F2s from the animal carrying two independent disruptions). We demonstrated that for all four alleles, the allele found in the F1 generation was passed on faithfully to the F2 generation. In the case of the F1 carrying two independent disruptions, we were able to isolate homozygous F2 mutant animals carrying each independent mutant allele at the expected Mendelian frequencies of 25%.
Supplementary Material
Supplementary Fig. 1: A putative U6 snRNA locus found on C. elegans Chromosome IV and aligned with similar sequences from three other Caenorhabditis species. In green is the conserved snRNA sequence; our promoter conservatively includes 500 bases of upstream sequence and 237 bases of downstream sequence that flank it.
Supplementary Fig. 2: Genotyping results at off-target loci. Mismatches between targeting sgRNAs and off-target loci are highlighted in red. In all examined cases, no indels were found at the off-target loci.
Supplementary Fig. 3: Sequencing results of F1 and F2 progeny from animals injected with cas9 and sgRNAs specific for the klp-12 locus. (A) The sequence and traces of a wild-type F1 and five of its F2 progeny. (B) The sequence and traces of a disrupted F1, carrying two copies of the same allele, and five of its F2 progeny, all of which were also homozygous for the same disruption. (C) The sequences and traces of a heterozygous, doubly disrupted F1 (each deciphered mutant sequence is listed above the sequencing trace) and eight of its F2 progeny. Four of these eight F2 progeny are similarly heterozygous, while two are homozygous for one disrupted allele and the last two are homozygous for the other disrupted allele. In all cases, the final three nucleotides (TGG) represent the PAM sequence.
Supplementary Table 1: Summary of results from control microinjection experiments. No mutants were recovered.
Supplementary Table 2: List of all oligonucleotides and relevant sequences used in this study.
A movie of a wild-type (N2 strain) worm
A movie of an unc-119 worm created by Cas9-mediated gene disruption.
A movie of a dpy-13 worm created by Cas9-mediated gene disruption.
Table 1.
| Experiment | Gene | Injected Worms | Fls | Disruptions | Frequency |
|---|---|---|---|---|---|
| 1 | unc-119 | - | 27 | 1 | 1/27 (3.7%) |
| 2 | unc-119 | - | 105 | 1 | 1/105 (0.9%) |
| 3 | unc-119* | - | 60 | 1 | 1/60 (1.7%) |
| 4 | dpy-13 | - | 210 | 1 | 1/210 (0.5%) |
| 5 | klp-l2* | 12 | 66 | 53 | 53/66 (80.3%) |
| 6 | klp-l2* | 14 | 35 | 27 | 27/35 (77.1%) |
| 7 | Y61A9LA.l* | 11 | 72 | 13 | 13/72 (18.1%) |
Acknowledgments
We thank Bodo Stern, Andrew Murray, Arneet Saltzman, Joe Calarco, and members of the Calarco lab for comments on the manuscript. This work was supported by an National Institutes of Health (NIH) Early Independence Award (1DP5OD009153) and additional support from Harvard University to J.A.C., by NIH grant R01GM072551 to M.P.C, and an NHGRI CEGS grant to G.M.C.. A.E.F. is supported by a Ralph Ellison/American Federation for Aging Research postdoctoral fellowship.
Footnotes
Author Contributions
A.E.F., K.M.E., J.A.C. conceived of and designed experiments, with help from Y.B.T.; A.E.F. and J.A.C. assembled vectors; A.E.F. and J.A.C. performed microinjections and screened mutants; A.E.F., J.A.C. and Y.B.T. performed off-target genotyping analysis; A.E.F., K.M.E., J.A.C wrote the manuscript, with input from Y.B.T., M.P.C, and G.M.C.
References
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Current opinion in microbiology. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dicarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic acids research. 2013 doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 8.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013 doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nature methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nature biotechnology. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 15.Fruscoloni P, Zamboni M, Panetta G, De Paolis A, Tocchini-Valentini GP. Mutational analysis of the transcription start site of the yeast tRNA(Leu3) gene. Nucleic acids research. 1995;23:2914–2918. doi: 10.1093/nar/23.15.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zecherle GN, Whelen S, Hall BD. Purines are required at the 5′ ends of newly initiated RNAs for optimal RNA polymerase III gene expression. Molecular and cellular biology. 1996;16:5801–5810. doi: 10.1128/mcb.16.10.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Mende N, Bird DM, Albert PS, Riddle DL. dpy-13: a nematode collagen gene that affects body shape. Cell. 1988;55:567–576. doi: 10.1016/0092-8674(88)90215-2. [DOI] [PubMed] [Google Scholar]
- 18.Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. The EMBO journal. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas J, Lea K, Zucker-Aprison E, Blumenthal T. The spliceosomal snRNAs of Caenorhabditis elegans. Nucleic acids research. 1990;18:2633–2642. doi: 10.1093/nar/18.9.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic acids research. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 25.Kadandale P, Chatterjee I, Singson A. Germline transformation of Caenorhabditis elegans by injection. Methods Mol Biol. 2009;518:123–133. doi: 10.1007/978-1-59745-202-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: A putative U6 snRNA locus found on C. elegans Chromosome IV and aligned with similar sequences from three other Caenorhabditis species. In green is the conserved snRNA sequence; our promoter conservatively includes 500 bases of upstream sequence and 237 bases of downstream sequence that flank it.
Supplementary Fig. 2: Genotyping results at off-target loci. Mismatches between targeting sgRNAs and off-target loci are highlighted in red. In all examined cases, no indels were found at the off-target loci.
Supplementary Fig. 3: Sequencing results of F1 and F2 progeny from animals injected with cas9 and sgRNAs specific for the klp-12 locus. (A) The sequence and traces of a wild-type F1 and five of its F2 progeny. (B) The sequence and traces of a disrupted F1, carrying two copies of the same allele, and five of its F2 progeny, all of which were also homozygous for the same disruption. (C) The sequences and traces of a heterozygous, doubly disrupted F1 (each deciphered mutant sequence is listed above the sequencing trace) and eight of its F2 progeny. Four of these eight F2 progeny are similarly heterozygous, while two are homozygous for one disrupted allele and the last two are homozygous for the other disrupted allele. In all cases, the final three nucleotides (TGG) represent the PAM sequence.
Supplementary Table 1: Summary of results from control microinjection experiments. No mutants were recovered.
Supplementary Table 2: List of all oligonucleotides and relevant sequences used in this study.
A movie of a wild-type (N2 strain) worm
A movie of an unc-119 worm created by Cas9-mediated gene disruption.
A movie of a dpy-13 worm created by Cas9-mediated gene disruption.