SUMMARY
RAS genes are commonly mutated in cancer; however, RAS mutations are rare in breast cancer, despite the fact that Ras and ERK are frequently hyperactivated. Here we report that the RasGAP gene, RASAL2, functions as a tumor and metastasis suppressor. RASAL2 is mutated or suppressed in human breast cancer and RASAL2 ablation promotes tumor growth, progression, and metastasis in mouse models. In human breast cancer RASAL2-loss is associated with metastatic disease, low RASAL2 levels correlate with recurrence of luminal B tumors, and RASAL2 ablation promotes metastasis of luminal mouse tumors. Additional data reveal a broader role for RASAL2 inactivation in other tumor-types. These studies highlight the expanding role of RasGAPs and reveal an alternative mechanism of activating Ras in cancer.
INTRODUCTION
The Ras pathway is one of the most commonly deregulated pathways in human cancer (Downward, 2003). Mutations in RAS genes occur in a variety of tumor types (Karnoub and Weinberg, 2008; Pylayeva-Gupta et al., 2011); however the Ras pathway is also frequently activated as a consequence of alterations in upstream regulators and downstream effectors, underscoring the importance of this pathway in cancer (Downward, 2003).
Ras is negatively regulated by RasGAPs (Ras GTPase Activating Proteins), which catalyze the hydrolysis of Ras-GTP to Ras-GDP (Bernards, 2003). As such, RasGAPs are poised to function as potential tumor suppressors. Indeed, the NF1 tumor suppressor encodes a RasGAP and is mutated in the familial cancer syndrome neurofibromatosis type 1 (Cawthon et al., 1990). NF1 also is lost or suppressed in sporadic cancers, including glioblastoma (Cancer Genome Atlas Research Network, 2008; Parsons et al., 2008; McGillicuddy et al., 2009), non-small cell lung cancer (Ding et al., 2008), neuroblastoma (Hölzel et al., 2010), and melanoma (Krauthammer et al., 2012; Maertens et al., 2012). More recently the RasGAP gene, DAB2IP, has been shown to function as a potent tumor and metastasis suppressor in prostate cancer (Min et al., 2010). In total there are 14 RasGAP genes in the human genome (Bernards, 2003). All contain a RasGAP domain but exhibit little similarity elsewhere. It is currently unknown whether any of these other genes may also function as human tumor suppressors.
Breast cancer is the most common cancer in women worldwide (Kamangar et al., 2006). K-, H-, and N-RAS mutations are relatively rare in this tumor type and together have been detected in only ~3.2% of all breast lesions (Bamford et al., 2004). Nevertheless, the Ras/ERK pathway is hyperactivated in ≥50% of breast cancers and has been proposed to be involved in tumor progression and recurrence, suggesting that Ras may be more frequently activated by other mechanisms in these tumors (Sivaraman et al., 1997; Lintig et al., 2000; Mueller et al., 2000). In this study we demonstrate that the RasGAP gene, RASAL2, functions as a tumor suppressor in breast cancer. Through the analysis of human tumor samples, human xenografts, and genetically engineered mouse models we show that RASAL2 loss plays a causal role in breast cancer development and metastasis. Additional mouse modeling studies reveal a broader potential role for RASAL2 in other tumor types. Together these studies highlight the expanding role of RasGAP genes in cancer and reveal an important mechanism by which Ras becomes activated in breast tumors.
RESULTS
The RasGAP Gene, RASAL2, is a Candidate Tumor Suppressor
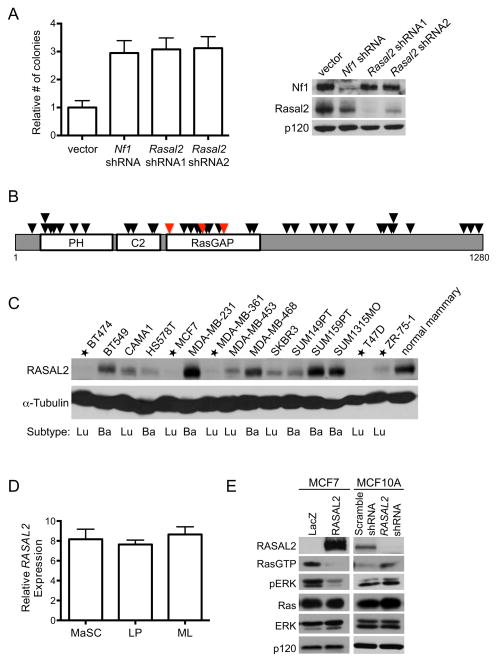
We previously developed a cell-based screen to identify additional RasGAPs that might function as tumor suppressors (Min et al., 2010). Distinct shRNAs that recognize individual RasGAP genes were introduced into immortalized mouse embryonic fibroblasts (MEFs) and cells were evaluated for the ability to grow in soft agar. Three genes scored in this screen: Nf1, a well-documented tumor suppressor gene, Dab2ip, which we have since shown is a tumor suppressor in prostate cancer, and a third RasGAP gene, Rasal2 (Min et al., 2010). Several Rasal2-specific shRNA sequences promoted colony growth in this assay and did so as well as Nf1- and Dab2IP-specific shRNAs (Figure 1A and (Min et al., 2010)). However, it should be noted that transformation is not generally promoted by the loss any RasGAP, suggesting that only a subset of RasGAPs may function as tumor suppressors (Min et al., 2010). Upon identifying RASAL2 as a candidate tumor suppressor, we searched publicly available databases and found mutations within the catalytic RasGAP domain in human breast cancers (Figure 1B and Table S1) (Sjöblom et al., 2006; Shah et al., 2012). Current genomic mutation databases indicate that RASAL2 is also mutated in several other cancers including colorectal, lung, and ovarian tumors (Figure 1B and Table S2). In total 42 non-synonymous mutations have been detected in RASAL2, 31% of which reside in the catalytic RasGAP domain, many of which are predicted to be deleterious (Table S2 and Table S3). Because the mechanism by which Ras becomes activated in breast cancer is largely unknown, and because mutations in breast tumors were among the first to be identified, we began by investigating a potential role for RASAL2 inactivation in breast cancer development.
Figure 1. RASAL2 is candidate tumor suppressor.
(A) Left: Immortalized MEFs were infected with lentiviral shRNAs targeting Rasal2, Nf1, or control, and plated in soft agar. Data are reported as relative number of colonies ± SEM. Inactivation of Nf1 or Rasal2 induced a statistically significant increase in anchorage-independent growth (p≤0.0001). Right: Western blot confirming knockdown.
(B) RASAL2 mutations in human tumor samples (Bamford et al., 2004). Each triangle represents a non-synonymous mutation. Red triangles indicate breast cancer mutations. See also Table S1 and S2.
(C) RASAL2 expression in a panel of human breast cancer cell lines in comparison to normal human mammary epithelial cells. Cell lines with very low or no RASAL2 are starred. Luminal (Lu) or basal (Ba) subtype categorization is indicated.
(D) Relative RASAL2 expression in subsets of sorted human mammary epithelial cells (Lim et al., 2009). Mammary stem cell-enriched: (CD49hi EpCAM−). LP: luminal progenitor (CD49f+ EpCAM+). ML: mature luminal (CD49f− EpCAM+). Data show relative expression ± SD. Similar results were obtained using two additional RASAL2 probes. There were no statistically significant differences in RASAL2 expression between subsets of cells.
(E) Left: Western blot of Ras-GTP and phospho-ERK (pERK) levels in MCF7 cells following expression of LacZ or RASAL2. Right: Western blot of Ras-GTP and phospho-ERK (pERK) levels in MCF10A cells following shRNA-mediated inactivation of RASAL2 or control (non-targeting “Scramble” shRNA).
Work from our laboratory and others have shown that the RasGAP genes NF1 and DAB2IP are inactivated in cancer by genetic, epigenetic, and proteasomal mechanisms (Dote et al., 2004; McGillicuddy et al., 2009; Min et al., 2010). Moreover, in many instances the non-genetic mechanisms of inactivation of these tumor suppressors appear to be more prevalent than mutational events in sporadic tumors (McGillicuddy et al., 2009; Min et al., 2010; Maertens et al., 2012). Therefore we began by examining RASAL2 protein expression in a panel of breast cancer cell lines. In comparison to normal mammary epithelial cells, RASAL2 was absent or minimally expressed in at least 5 out of 15 breast cancer cell lines, suggesting that RASAL2 may be lost or suppressed in this tumor type (Figure 1C). RASAL2 levels were high in MDA-MB-231 and SUM159PT cells, which are known to harbor mutations in KRAS and HRAS, respectively (Hollestelle et al., 2007). We also noted that RASAL2 was frequently absent in cells derived from luminal cancers in this panel of lines. Cell sorting studies indicated that there were no inherent differences in RASAL2 expression in any specific cell population within the mammary cell hierarchy: luminal progenitor, mature luminal, or mammary stem cell-enriched, suggesting that the low RASAL2 levels associated with luminal cancer cell lines was not inherently associated with a pre-existing reduction in RASAL2 levels due to a specific cell of origin or fate, as has been suggested for other genes (Figure 1D) (Lim et al., 2009). When RASAL2 was reconstituted in MCF7 cells, which express little to no endogenous RASAL2, Ras-GTP and phospho-ERK levels were suppressed (Figure 1E). Conversely, acute inactivation of RASAL2 via shRNA sequences in immortalized mammary epithelial cells (MCF10A) increased Ras-GTP and phospho-ERK levels (Figure 1E). These data confirm that RASAL2 is a functional RasGAP and that its loss activates Ras and ERK in this tumor type.
RASAL2 Functions as a Tumor Suppressor in Breast Cancer
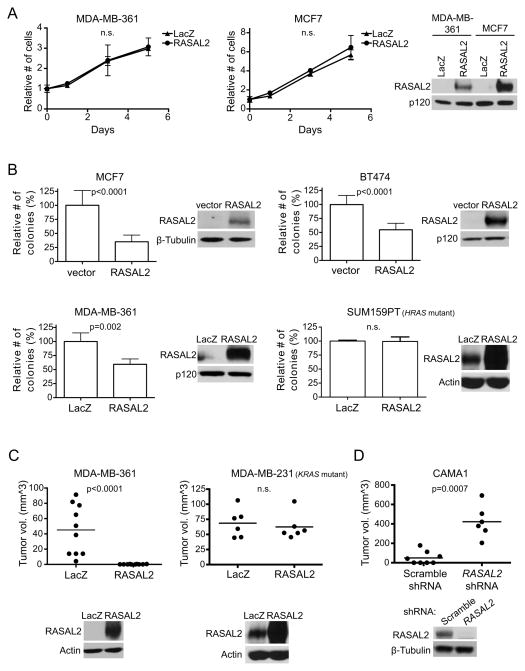
We next investigated the biological consequences of reconstituting or suppressing RASAL2 in breast cancer cell lines. When RASAL2 was introduced into human breast cancer cells that lack endogenous RASAL2, proliferation was largely unaffected (Figure 2A); however RASAL2 reconstitution significantly inhibited anchorage-independent colony growth (Figure 2B). In contrast, RASAL2 did not inhibit colony growth of SUM159PT cells, which retain RASAL2 expression but harbor an activating RAS mutation (Figure 2B). RASAL2 also potently suppressed the growth of RASAL2-deficient breast cancer xenografts in vivo, but again had no effect on RAS mutant tumors (Figure 2C). Conversely, shRNA-mediated suppression of endogenous RASAL2 in a breast cancer cell line that normally does not grow well as a xenograft promoted tumor growth in vivo (Figure 2D). Together, these gain- and loss-of function studies suggest that RASAL2 can function as a tumor suppressor in the mammary epithelium and that its inactivation or loss can contribute to mammary tumor development. Notably, like NF1 and DAB2IP, RASAL2 appears to restrict transformation and/or anchorage-independent growth, rather than generally suppressing cell proliferation in two- dimensional culture systems (Johannessen et al., 2005; Min et al., 2010).
Figure 2. RASAL2 functions as a tumor suppressor in breast cancer.
(A) Growth curves of MDA-MB-361 and MCF7 cells expressing RASAL2 or LacZ. Data points show triplicate averages ± SD. There were no statistically significant differences in proliferation. Western blot on right confirms ectopic RASAL2 expression.
(B) Soft agar colony formation of MCF7, BT474, MDA-MB-361, and SUM159PT cells expressing RASAL2 or LacZ. Data show relative number of colonies ± SD. There was a statistically significant decrease in anchorage-independent growth upon ectopic RASAL2 expression in RAS wild type cell lines (MCF7 and BT474 p<0.0001; MDA-MB-361 p=0.002) but not in the HRAS mutant cell line SUM159PT. Western blots confirm ectopic RASAL2 expression.
(C) Xenograft tumor formation of MDA-MB-361 and MDA-MB-231 cells expressing RASAL2 or LacZ. MDA-MB-361 cells were injected orthotopically into female NOD/SCID mice; MDA-MB-231 cells were injected subcutaneously into female nude mice. Horizontal bars indicate mean tumor volume. There was a statistically significant decrease in tumor growth upon ectopic RASAL2 expression (p<0.0001) in the RAS wild type cell line MDA-MB-361 but not in the KRAS mutant cell line MDA-MB-231. Western blots below confirm ectopic RASAL2 expression.
(D) Xenograft tumor formation of CAMA1 cells infected with shRNAs targeting RASAL2 or non-targeting control shRNA and injected subcutaneously into female NOD/SCID mice. Horizontal bars indicate mean tumor volume. There was a statistically significant increase in tumor growth upon RASAL2 inactivation (p=0.0007). Western blot confirms RASAL2 knockdown.
RASAL2 Functions Via Its Effects on Ras
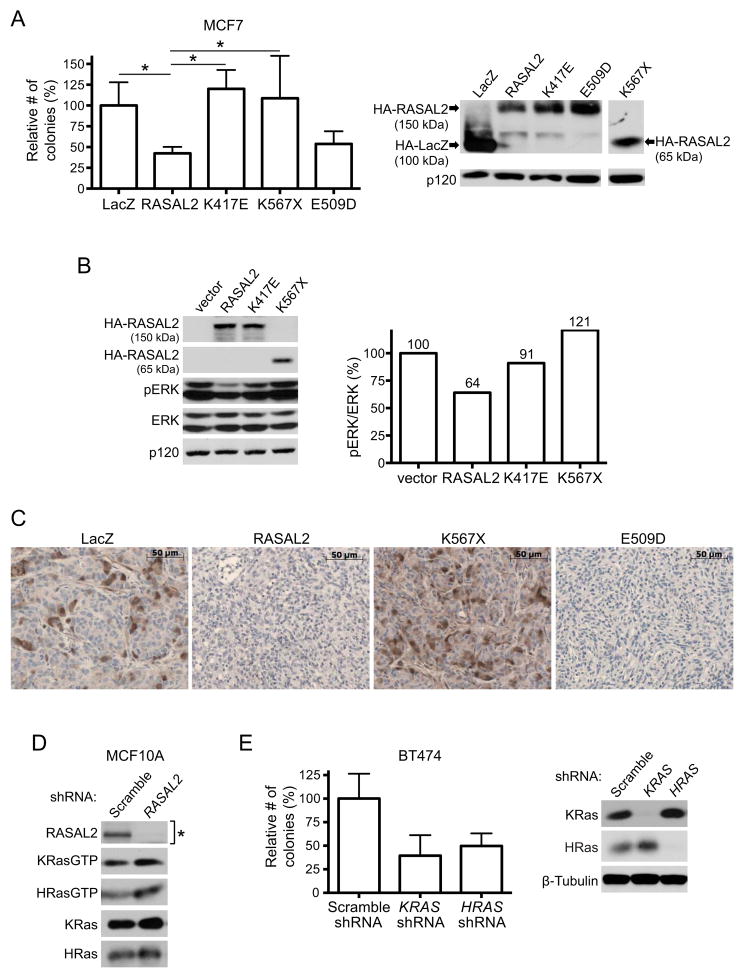
To determine whether the RasGAP domain of RASAL2 and effects on Ras were critical for tumor suppression, we first evaluated the effects of RASAL2 mutations identified in human breast cancer samples. Two of the three RasGAP domain mutants (K417E and K567X) failed to suppress anchorage-independent growth, demonstrating that these two mutations result in a clear loss-of-function (Figure 3A). The third mutation, which resulted in a more conservative amino acid change (E509D), still retained activity in this assay and therefore does not appear to be pathogenic; however a number of additional non-conservative mutations have been detected in the RasGAP domain in other tumor types (Table S3). Consistent with these biological observations, both the K417E and the K567X mutation were defective in their ability to suppress activation of the Ras/ERK pathway (Figure 3B). Phospho-ERK levels in xenograft tumors further illustrate the difference in activity between pathogenic and non-pathogenic mutations (Figure 3C). To complement these studies we investigated which Ras isoforms were activated in response to RASAL2 suppression and found that both K-Ras and H-Ras-GTP levels were elevated (Figure 3D). Accordingly ablation of either KRAS or HRAS suppressed colony growth by more than 50% (Figure 3E). Together these results demonstrate that the RasGAP domain is essential for RASAL2 tumor suppressor function and that both H- and K-Ras contribute to the pathogenesis caused by RASAL2 inactivation.
Figure 3. RASAL2 functions as a tumor suppressor via its effects on Ras.
(A) Soft agar colony formation of MCF7 cells expressing HA-tagged LacZ, wild type, or mutant RASAL2 (see also Table S3). Data show relative number of colonies ± SD. * indicates p≤0.05. Western blot confirms expression of constructs.
(B) Western blot reflecting the relative activation of the Ras/ERK pathway in the presence of HA-tagged wild type or mutant RASAL2. The pERK/ERK ratio of each sample was calculated and normalized to the vector control.
(C) Phospho-ERK (pERK) expression in MDA-MB-361 xenograft tumors. LacZ, RASAL2, or mutant RASAL2 was expressed in MDA-MB-361 cells and cells were injected orthotopically into female NOD/SCID mice. pERK levels were assessed by immunohistochemistry.
(D) Western blot showing HRas-GTP and KRas-GTP levels in MCF10A cells following shRNA-mediated inactivation of RASAL2 or control shRNA. As indicated by the asterisk, the blot confirming RASAL2 knockdown is a duplicate from Figure 1E, as these immunoblots were generated from the same samples.
(E) Soft agar colony formation of BT474 cells infected with an shRNA targeting HRAS or KRAS or a non-targeting control. Data show relative number of colonies ± SD. Western blot confirms Ras isoform-specific knockdown.
RASAL2 Inactivation Promotes Migration, Invasion, and Tumor Progression
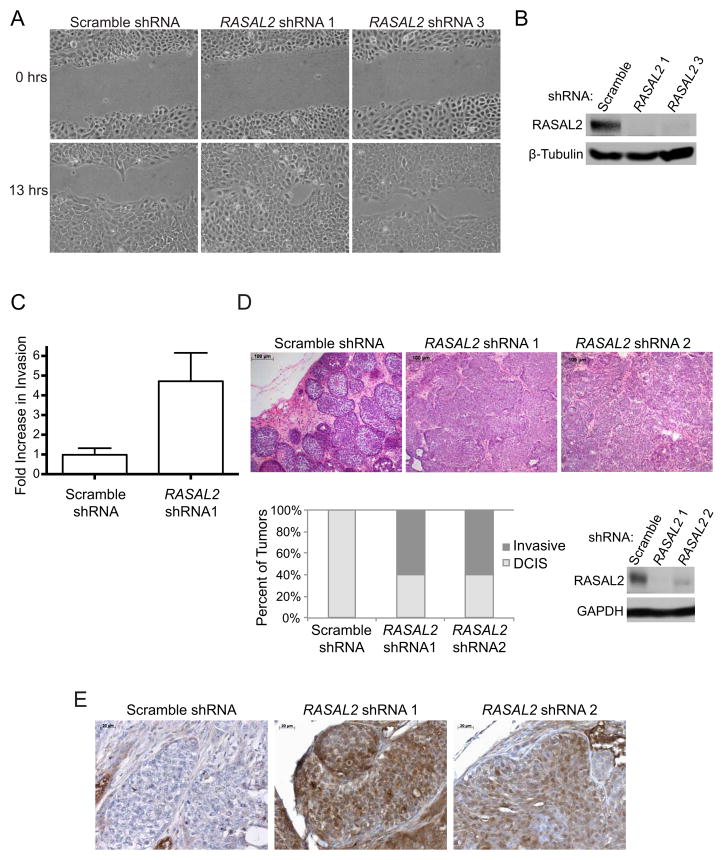
To fully characterize the oncogenic effects of RASAL2 loss we investigated whether RASAL2 suppression might also promote migration, invasion, and tumor progression. RASAL2 suppression promoted the migration of MCF10A cells in a wound-healing assay (Figure 4A,B), and significantly enhanced invasion through Matrigel (Figure 4C, p=0.002). Similar to what was observed in colony assays shown in Figure 3E, ablation of HRAS or KRAS reduced invasiveness (Figure S1). We also utilized a xenograft model of breast cancer progression that mimics the progression of ductal carcinoma in situ (DCIS) to invasive carcinoma (Miller et al., 2000; Hu et al., 2008). Specifically MCF10ADCIS cells, a derivative of MCF10A cells that are enriched for progenitor population of cells (Miller et al., 2000), develop into DCIS-like lesions when grown as xenografts in mice. However after a latency of approximately 8 weeks, they progress to invasive carcinoma, characterized by the loss of the myoepithelial cell layer and basement membrane (Hu et al., 2008). We acutely inactivated RASAL2 using two distinct shRNAs in these cells and found that RASAL2 inactivation accelerated tumor progression, resulting in a rapid disruption of the myoepithelium and basement membrane, and the development of invasive adenocarcinoma after just 3 weeks (Figure 4D). These results suggest that RASAL2 inactivation may also play a role in breast cancer progression. Notably, tumors in which RASAL2 had been depleted also exhibited a marked elevation of phospho-ERK as compared to control tumors (Figure 4E).
Figure 4. RASAL2 inactivation promotes migration, invasion, and tumor progression.
(A) Cell migration of MCF10A cells infected with shRNAs targeting RASAL2 or a non-targeting control.
(B) Western blot confirming RASAL2 knockdown in MCF10A cells used in (A) and (C).
(C) Transwell invasion of MCF10A cells infected with an shRNA targeting RASAL2 or a non-targeting control. Invasion was measured after 24 hours and reported as average ± SD (p=0.002).
(D) Xenograft tumor progression of MCF10ADCIS cells infected with shRNAs targeting RASAL2 or a non-targeting control Top: H&E images of xenograft tumors. Bottom left: Quantification of xenograft tumor progression. Bottom right: Western blot confirming RASAL2 knockdown.
(E) Phospho-ERK (pERK) expression in MCF10ADCIS xenograft tumors from (D) as assessed via immunohistochemistry.
Loss of Rasal2 Promotes Metastasis and Ras Activation in a Genetically Engineered Mouse Model of Luminal Breast Cancer
As a rigorous and complementary means of investigating the biological consequences of Rasal2 inactivation in vivo we generated genetically engineered mice that lack Rasal2. Mouse embryonic stem cells that contain a gene-trap cassette within the third intron of Rasal2 were used to generate Rasal2-deficient mice (Figure 5A). Appropriate integration and loss of Rasal2 expression were confirmed in heterozygous and homozygous mutant animals (Figure 5B,C). Rasal2 −/− mice were viable and fertile and born at Mendelian ratios. We found that mutant animals did exhibit shorter overall survival as compared to control animals (77.8 compared to 95.6 weeks; p=0.007, Figure S2A). However there was no obvious difference in phenotype between wild type and Rasal2 −/− mice. A subset of animals from both cohorts developed tumors associated with old age. While Rasal2 mutant mice developed these tumors earlier, the tumor spectrum was similar to wild type animals and they did not develop mammary lesions (Figure S2B). These results indicate that Rasal2 loss is not sufficient to drive breast cancer in mice, but may play a more general role in enhancing the development of other spontaneous tumors.
Figure 5. Loss of Rasal2 promotes metastasis and Ras activation in a genetically engineered mouse model of breast cancer.
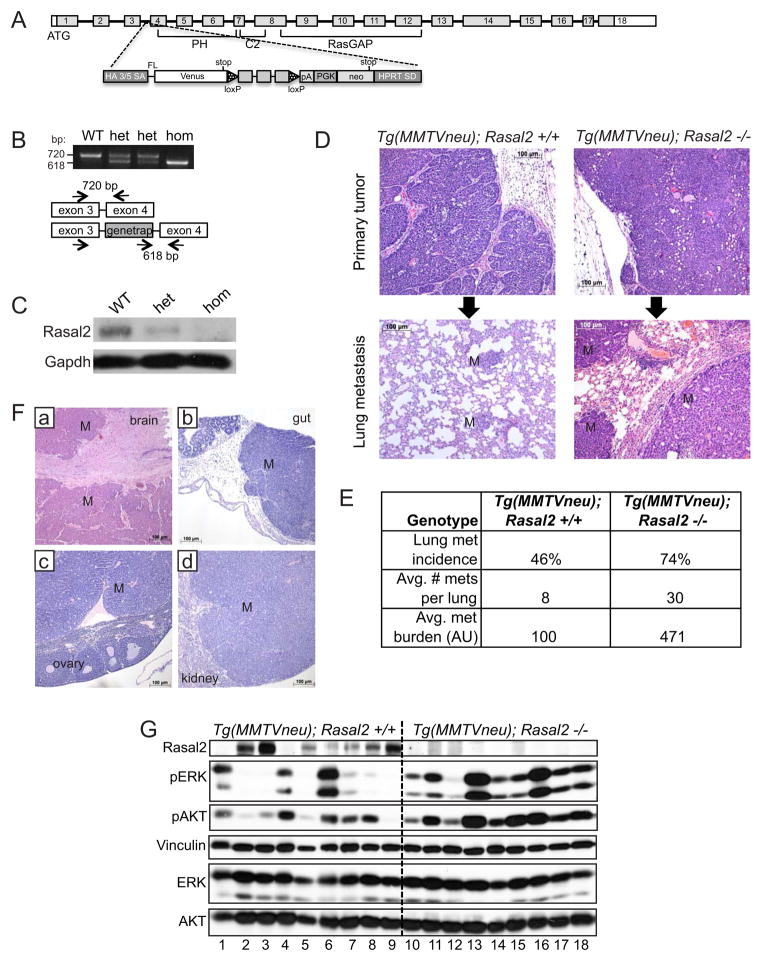
(A) Schematic of Rasal2 genomic locus and pNMDi4 genetrap cassette. Un-shaded regions in exons 1 and 18 mark 5′ and 3′ UTRs, respectively. Known domains of Rasal2 are noted (PH, C2, and RasGAP). See Experimental Procedures for detailed description of pNMDi4. The genetrap cassette targets the third intron of Rasal2.
(B) Genotyping of Rasal2 mice to distinguish wild type (WT), heterozygous mutant (het), and homozygous mutant (hom).
(C) Western blot confirming loss of Rasal2 protein in genetrap animals (mammary gland tissue). WT: wild type, het: heterozygous, hom: homozygous mutant.
(D) Top: H&E images of primary mammary adenocarcinomas from MMTVneu; Rasal2 +/+ and MMTVneu; Rasal2 −/− animals. Bottom: H&E images of lung metastases from MMTVneu; Rasal2 +/+ and MMTVneu; Rasal2 −/− animals. M indicates metastases.
(E) Lung metastasis burden in MMTVneu; Rasal2 +/+ and MMTVneu; Rasal2 −/− animals. Lung metastasis incidence: percent of tumor-bearing females with lung metastases at sacrifice (p=0.05; n=24 MMTVneu; Rasal2 +/+, n=23 MMTVneu; Rasal2 −/−). Average number of lung metastases per animal: counted per representative section of lungs for each tumor-bearing female (p=0.04). Average metastasis burden per animal: average total area of metastasis in a representative section of lung for each tumor-bearing female (arbitrary units; p=0.04). See also Figure S2.
(F) H&E images of metastases to brain (a), gut (b), ovary (c), and kidney (d) in compound tumor-bearing females. M indicates regions of metastasis.
(G) Western blot analysis of phospho-ERK (pERK) and phospho-AKT (pAKT) levels in primary mammary tumors from MMTVneu; Rasal2 +/+ animals (numbers 1–9) and MMTVneu; Rasal2 −/− animals (numbers 10–18).
To examine the effects of RASAL2 loss on mammary tumorigenesis, we crossed Rasal2 −/− mice to animals that constitutively overexpress a wild type Her2 (Erbb2) transgene in the mammary epithelium (MMTVneu mice) (Guy et al., 1992): a mouse model of luminal tumors (Herschkowitz et al., 2007). These tumors exhibit some differences from human luminal cancers, as they do not express estrogen receptor; however, unsupervised hierarchical clustering analysis and tumor pathology demonstrate that lesions from these animals recapitulate many of the key features of human luminal tumors (Guy et al., 1992;Herschkowitz et al., 2007). As such these animals are currently the best available genetically engineered mouse model for luminal cancer (Guy et al., 1992;Herschkowitz et al., 2007). Female MMTVneu mice develop focal luminal mammary tumors and a fraction of tumor-bearing females develop lung metastases (Guy et al., 1992). As expected, MMTVneu and MMTVneu; Rasal2 −/− compound mice developed mammary adenocarcinomas, and did so at a similar high frequency (Figure 5D, top panels, and Figure S2E). Strikingly however, we found that MMTVneu; Rasal2 −/− mice developed substantially more metastases than MMTVneu animals. First, a higher fraction of compound mutant mice developed lung metastases (Figure 5E, 74% versus 46%, p=0.05). Second, compound mutant mice developed more metastases per lung than MMTVneu animals (Figure 5E, 30 per mouse versus 8 per mouse, p=0.04). Finally, the metastases were significantly larger in MMTVneu; Rasal2 −/− mice as compared to MMTVneu mice (Figure 5D, bottom panels and Figure 5E, p=0.04). Interestingly, a subset of compound mutant mice developed tumors that metastasized to other organs, including brain, kidney, ovary, and gastrointestinal tract, which has not been observed in MMTVneu animals historically or in our cohort (Figure 5F). Moreover, in most autochthonous mouse models of mammary adenocarcinoma, metastasis is typically limited to the lung and occasionally lymph nodes (Kim and Baek, 2010). However, human breast cancers do frequently metastasize to the brain and these other distal sites, underscoring the significance of these observations and the potential utility of this mouse model (Weigelt et al., 2005). Despite the dramatic increase in metastasis, no differences in primary tumor incidence, growth rate, or tumor size were observed in MMTVneu versus MMTVneu; Rasal2 −/− mice, indicating that the differences in metastatic burden were not due to underlying differences in primary tumor onset or growth (Figure S2C,D,F). Importantly, when we compared primary tumors from MMTVneu; Rasal2 −/− mice and MMTVneu animals, higher levels of phospho-ERK and phospho-AKT were more consistently observed in MMTVneu; Rasal2 −/− lesions (Figure 5G). In addition, we found that Rasal2 was spontaneously lost or suppressed in a subset of MMTVneu tumors and that this was accompanied by a substantial increase in phospho-ERK and phospho-AKT levels (Figure 5G). Finally, the primary tumor that spontaneously lost/suppressed Rasal2 and exhibited the most robust activation of the Ras pathway was a metastatic outlier within the MMTVneu cohort (Tumor 6, Figure 5G, and Figure S2G). Taken together these findings indicate that Rasal2 loss enhances Ras activity in mammary tumors and that its loss promotes tumor progression, invasion, and metastasis in both autochthonous mouse models of breast cancer and human xenografts.
RASAL2 in Primary Human Breast Cancers
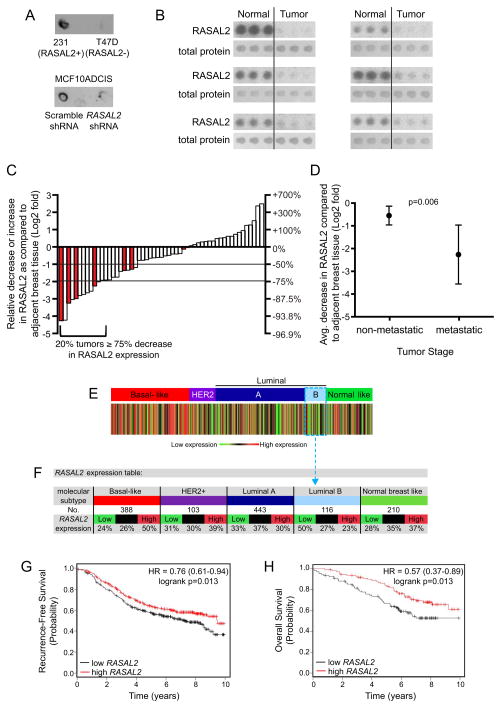
Genomic analyses demonstrate that RASAL2 mutations do occur in human breast cancer but are relatively rare (Bamford et al., 2004; Sjöblom et al., 2006; Shah et al., 2012, Table S1). However, the two other known RasGAP tumor suppressors appear to be more frequently inactivated in cancer via non-genetic mechanisms. To more accurately determine how frequently RASAL2 is lost or suppressed in human breast cancers we directly examined RASAL2 protein levels in primary human tumors. Existing RASAL2 antibodies cannot be used for immunohistochemistry; therefore we obtained breast cancer arrays comprised of 55 sets of protein lysates (in triplicate) from matched primary breast tumors and adjacent normal mammary tissue taken from naïve patients (Mueller et al., 2010). These tumor samples were histologically verified to contain at least 80% cancer cells and the normal tissue is cancer cell-free. We first validated our purified RASAL2 antibody in this assay and found that dot blots from RASAL2-expressing and non-expressing human breast cancer cell lines exhibited the expected pattern of expression (Figure 6A, top). RASAL2-specific shRNA sequences also effectively ablated expression in this assay (Figure 6A, bottom). Using the tumor arrays we found that RASAL2 expression was decreased by 75%–100% in 20% of human breast tumors as compared to adjacent normal mammary tissue (Figure 6B,C). These results confirm our findings in breast cancer cell lines, suggesting that RASAL2 expression is lost or suppressed in a significant fraction of human breast cancers at a frequency that is much greater than indicated by mutation analysis alone. More importantly however, low RASAL2 protein levels were significantly associated with metastasis (Figure 6C,D, p=0.006).
Figure 6. RASAL2 expression is lost/low in primary human breast cancers and low levels are associated with metastasis and recurrence.
(A) RASAL2 dot blot of whole cell RIPA extracts from human breast cancer cell lines with high or low RASAL2 expression (MDA-MB-231 “231” and T47D, respectively) (top) or MCF10ADCIS cells infected with control or RASAL2-targeting shRNAs (bottom).
(B) Dot blot images from human breast tumor lysate array. Six sets of RASAL2 and total protein stains are shown. Each set contains triplicate spots of tumor lysate (right) and triplicate spots of paired normal tissue lysate (left).
(C) Quantification of RASAL2 expression in tumor lysate arrays. Each bar depicts the change in RASAL2 expression in one sample as compared to its matched normal control as described in Experimental Procedures. Shaded bars indicate metastatic samples.
(D) RASAL2 protein expression in tumor versus normal in non-metastatic (Stages I, II, III) versus metastatic (Stage IV) tumors. Graph shows the Log2 fold change in RASAL2 protein expression in tumor versus normal. Data are reported as average ± 95% CI. p=0.006.
(E) Heatmap of RASAL2 gene expression as a function of robust molecular subtype predictor classification, which is based on patients classified in the same tumor subtype. Percentages of tumors with high, intermediate, and low RASAL2 expression per molecular subtype are given in the gene expression table.
(F) RASAL2 expression table. For each breast cancer subtype, the number of samples and percentage of samples with low, intermediate, or high RASAL2 mRNA expression are indicated.
(G) Kaplan-Meier curve showing recurrence-free survival of luminal B tumors with high or low RASAL2 expression (logrank p = 0.013).
(H) Kaplan-Meier curve showing overall survival of luminal B tumors with high or low RASAL2 expression (logrank p = 0.013).
Because the cell line analysis indicated that RASAL2 expression was low or undetectable in a subset of luminal breast cancer cell lines (Figure 1), and mouse modeling studies further demonstrated that RASAL2 loss promoted the metastasis of luminal tumors, we evaluated RASAL2 expression in different breast cancer subtypes. Human breast cancers can be molecularly classified into five distinct subtypes: basal-like, HER2-positive, luminal A, luminal B, and normal breast-like (Perou et al., 2000; Sørlie et al., 2001; Hu et al, 2006; Parker et al, 2006). Notably, molecular subtype association analysis of transcriptional profiles from primary breast cancers revealed that RASAL2 expression was low in luminal B breast cancers; 50% of luminal B tumors expressed the lowest levels of RASAL2, consistent with a potential role for RASAL2 loss in this subtype (Figure 6E,F). Moreover, low RASAL2 expression was also associated with both increased tumor recurrence (Figure 6G, logrank p=0.0133) and decreased overall survival (Figure 6H, logrank p=0.0131) in patients with luminal B cancers. Finally, using publicly available TCGA Methylation 450 data, we found that two CpG sites in the RASAL2 promoter region are differentially methylated in primary breast tumors. Specifically, RASAL2 promoter methylation is enriched in luminal B tumors (p<0.05, Mann-Whitney U test). These luminal B tumors also showed the lowest expression of RASAL2. Notably, both sites exhibit a significant increase in methylation when comparing luminal B samples with the lowest expression of RASAL2 (bottom 33%) to luminal B samples with highest expression (top 33%) (p<0.05, t-test). Taken together, cellular, xenograft, mouse modeling, and human tumor studies suggest that RASAL2 loss promotes breast cancer development and metastasis, and may play a particularly important role in the progression of luminal B tumors. In this context it is notable that KRAS amplifications are generally not present in luminal tumors but frequently occur in basal-like breast cancers (Cancer Genome Atlas Network, 2012), suggesting that RASAL2 suppression may provide an alternative mechanism of Ras activation in the luminal subtype. Nevertheless, while the models used in this study provide functional evidence to support a role for RASAL2 inactivation in luminal B tumors, these data do not preclude its involvement in a fraction of other subtypes.
Rasal2 Mutations Promote Tumor Development and Widespread Metastasis in p53 Mutant Mice
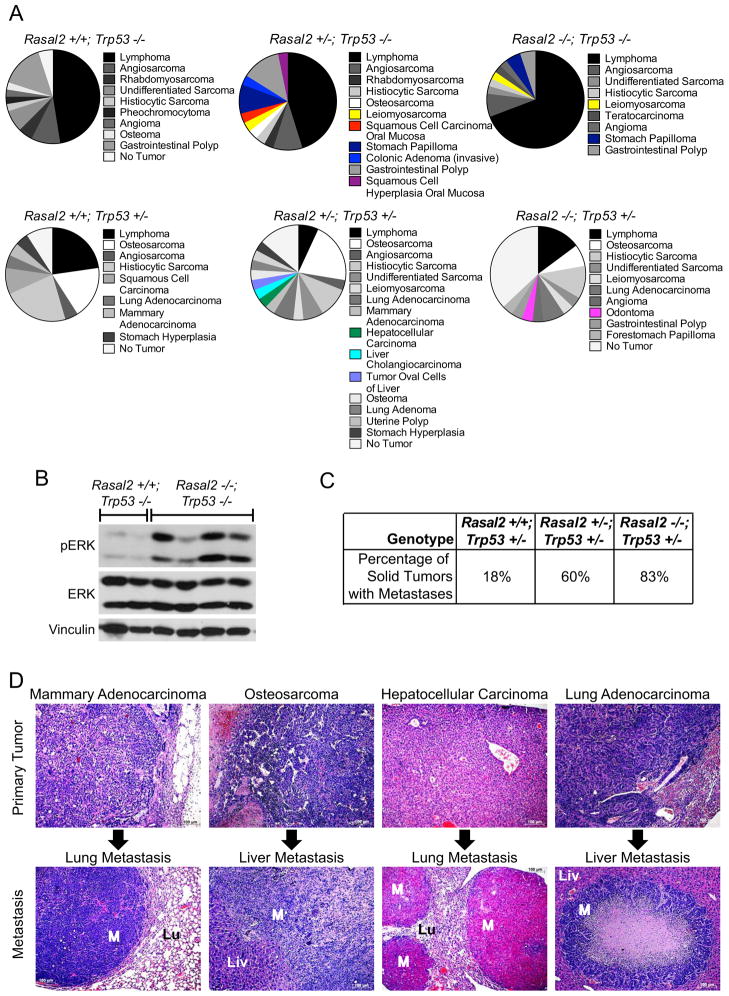
To determine whether RASAL2 inactivation might also contribute to the development of other sporadic tumors, Rasal2 −/− mice were crossed to mice mutant for p53, one of the most commonly inactivated tumor suppressors in human cancer (Vousden and Lane, 2007). Trp53 mutant mice develop a spectrum of lymphomas and sarcomas and some carcinomas arise in heterozygotes (Donehower et al., 1992; Jacks et al., 1994). In addition to the classical tumors observed in Trp53 mutant mice, Rasal2/Trp53 compound mutant mice developed several other lesions that were not found in Trp53 mutant controls, historically or in our cohort (Figure 7A). Specifically, Rasal2/Trp53 mutant mice developed hepatocellular carcinomas and other liver tumors, colonic adenomas, and oral and stomach tumors (Figure 7A). These findings are of particular interest because RASAL2 mutations have been found in related human cancers, namely hepatocellular carcinoma, colorectal carcinoma, head and neck squamous cell carcinoma, and stomach cancer (Table S2). Notably, the Rasal2 −/−; p53 −/− tumors exhibited higher levels of pERK as compared to Rasal2 +/+; p53 −/− tumors (Figure 7B).
Figure 7. Rasal2, Trp53 compound mutant mice develop highly metastatic tumors.
(A) Phenotypes in Rasal2/Trp53 compound mutant mice. Pie charts display the array of phenotypes in each genotype. New phenotypes in Rasal2 mutant compound mice are shown in color. n=21 Rasal2 +/+; Trp53 −/−, 18 Rasal2 +/−; Trp53 −/−, 31 Rasal2 −/−; Trp53 −/−, 16 Rasal2 +/+; Trp53 +/−, 21 Rasal2 +/−; Trp53 +/−, 21 Rasal2 −/−; Trp53 +/−.
(B) Western blot analysis of phospho-ERK (pERK) levels in primary tumors from Rasal2 +/+; Trp53 −/− and Rasal2 −/−; Trp53 −/− compound mice.
(C) Percentage of metastatic solid tumors in Rasal2 +/+, +/−, and −/−; Trp53 +/− compound mice. Increased metastasis in compound animals is statistically significant (p=0.003). The paucity of metastatic solid tumors in Trp53 +/− mice is supported by historical data.
(D) Images of metastatic lesions (bottom) and their primary tumors (top). From left to right: mammary adenocarcinoma and lung metastasis, osteosarcoma and liver metastasis, hepatocellular carcinoma and lung metastasis, and lung adenocarcinoma and liver metastasis. M: metastasis, Lu: lung, Liv: liver.
However the most striking phenotype in Rasal2/Trp53 mutant mice was that Rasal2 loss potently promoted metastasis. The Trp53 −/− mice, either with or without functional Rasal2, typically died from the primary tumor, which was frequently lymphoma. Nevertheless 60% of the solid tumors that developed in Rasal2 +/−; Trp53 +/− mice and 83% of the solid tumors from Rasal2 −/−; Trp53 +/− mice were metastatic, as compared to 18% of tumors in Trp53 +/− mice (Figure 7C, p=0.003). Specifically, Rasal2/Trp53 mutant animals developed highly metastatic mammary adenocarcinomas, hepatocellular carcinomas, lung adenocarcinomas, and various sarcomas, again tumor types in which RASAL2 mutations have been detected in humans (Figure 7D). Thus these findings further underscore the role of RASAL2 loss as a driver of metastasis and suggest that its inactivation may play a role in the progression of breast and other human cancers.
DISCUSSION
The Ras pathway plays a well-established role in cancer (Downward, 2003). However the primary mechanism(s) by which Ras becomes activated in breast cancers has remained elusive. Here we report that RASAL2, which encodes a RasGAP, functions as a tumor and metastasis suppressor in breast and other cancers. Specifically, we have shown that loss-of-function mutations in RASAL2 are found in human breast cancers and other tumor types; however, like other RasGAP genes RASAL2 appears to be more frequently inactivated by non-genetic mechanisms and it is substantially repressed in at least 20% of primary human breast cancers. We also showed that RASAL2 ablation promotes tumor growth and progression in two different human xenograft models, while RASAL2 reconstitution suppresses mammary tumor growth. Notably, RASAL2 mutations activate Ras and dramatically enhance metastasis in a genetically engineered mouse model of luminal mammary cancer. RASAL2 mutations also cooperate with p53 mutations to promote the development and metastasis of several tumor types, including mammary tumors, in a second mouse model. Finally, we show that low RASAL2 levels are associated with metastasis in human breast cancer.
Notably, the lowest RASAL2 mRNA expression levels are most frequently observed in luminal B human breast cancers and are associated with recurrence and reduced survival of patients with this tumor subtype. Collectively, these data suggest that RASAL2 loss plays a causal role in breast cancer pathogenesis. It should be noted that the breast cancer xenograft studies and the overall increase in tumor incidence in Rasal2/p53 mice suggest that RASAL2 may play a role in primary tumor development, whereas the dramatic metastatic phenotype in Rasal2/MMTVneu and Rasal2/p53 mutant animals suggests a role for RASAL2 loss in metastasis. Similarly, the Ras pathway has been shown to play a role in both primary tumor development as well as metastasis, depending on context. As such we hypothesize that RASAL2 inactivation may play a role in one or both processes depending on the presence of other mutations in a given tumor. Nevertheless, the human breast cancer data presented in this study suggest that RASAL2 loss may play a more prominent role in progression and metastasis in this tumor type.
It should be noted that while RAS mutations are rare in breast cancer, they do occur. Moreover amplifications of wild type RAS are frequently observed in basal breast cancers, the most aggressive subtype of human breast cancer, underscoring the connection between Ras activation and breast cancer progression (Cancer Genome Atlas Network, 2012). Our data indicate that overall RASAL2 is suppressed or lost in at least 20% of human breast cancers. However expression and DNA methylation analysis of different breast cancer subtypes suggests that RASAL2 loss may play a particularly important role in the progression of luminal B tumors. The observation that RASAL2 ablation promotes metastasis in a mouse model of luminal tumors provides important functional data to support this conclusion. In this respect it is notable that luminal B tumors have poorer outcomes than luminal A tumors, however the mechanism(s) that drive the progression of these tumors are largely unknown. Our data suggest that RASAL2 loss/suppression may play a causal role in the progression of this subtype, although these observations do not preclude its potential involvement in other subtypes.
Finally, while RASAL2 is mutated in breast cancer and in other human tumors, it appears to be more commonly inactivated via non-mutational mechanisms. Notably, the other RasGAP tumor suppressors, NF1 and DAB2IP, are also inactivated by both genetic and several non-genetic mechanisms (McGillicuddy et al., 2009; Min et al., 2010). Similarly, PTEN and INPP4B, two other tumor suppressors that negatively regulate an overlapping set of signals, are also suppressed by multiple mechanisms in cancer, some of which have not yet been elucidated (Gewinner et al., 2009; Song et al., 2012). As such, loss of PTEN protein expression, rather than mutational status or copy number, is often evaluated in clinical samples during clinical trials and for pathological staging (Thomas et al., 2004). The observation that RASAL2 loss plays a causal role in breast cancer progression and metastasis in animal models and that its expression is lowest in primary human tumors that ultimately progress or recur, suggests that RASAL2 could be useful as a prognostic biomarker, in at least a subset of breast cancers, such as luminal B tumors. Regardless, these studies have identified an important tumor suppressor involved in breast cancer progression and have revealed an alternative mechanism by which Ras becomes activated in this disease.
EXPERIMENTAL PROCEDURES
Cell Culture and DNA Constructs
MEFs were immortalized as described (Johannessen et al., 2005). MCF7, MCF10A and MDA-MB-361 cells were purchased from ATCC. BT549, HS578T, MDA-MB-231, MDA-MB-453, MDA-MB-468, SKBR3, T47D, and ZR-75-1 cells were obtained from Dr. William Hahn (Dana-Farber Cancer Institute). SUM149PT, SUM159PT, SUM1315MO, and BT474 cells were obtained from Dr. Frank McCormick (University of California San Francisco). CAMA1 cells were obtained from Dr. Marcia Haigis (Harvard Medical School). MCF1ADCIS were provided by Dr. Fred Miller (Karmanos Cancer Institute) and the Polyak laboratory.
shRNAs from The RNAi Consortium (Broad Institute, MIT) with the following sequences were utilized: NF1 shRNA (5′-TTATAAATAGCCTGGAAAAGG-3′), RASAL2 shRNA1 (5′-CCCTCGTGTTCTTGCTGATAT-3′), RASAL2 shRNA2 (5′-GCCTTCCACCTCTTCATAGTA-3′), KRAS shRNA (5′-CAGTTGAGACCTTCTAATTGG-3′) and HRAS shRNA (5′-GACGTGCCTGTTGGACATCCT-3′). A scrambled shRNA was purchased from Addgene (5′-CCTAAGGTTAAGTCGCCCTCG-3′). A RASAL2-targeting shRNA was cloned into the pLKO vector (shRNA3) (5′-ATGGAGTGCAATAGGACATTG-3′). The Mammalian Gene Collection fully sequenced human RASAL2 cDNA was purchased from Open Biosystems (cat. # MHS4426-99623118) and was cloned into the pHAGE-N-Flag-HA lentiviral expression vector (Dr. J. Wade Harper, Harvard Medical School) for expression in cell lines. Infections and transfections were performed as described in Supplemental Experimental Procedures.
Proliferation, Soft Agar, Migration, and in vitro Invasion Assays are described in Supplemental Experimental Procedures.
Xenograft Assays
Female nude and NOD/SCID mice were purchased from Charles River Laboratories (cat. #s 088 and 394, respectively) for subcutaneous xenograft experiments. Cells were injected with Matrigel (BD Biosciences cat. # 354234) as follows: MCF10ADCIS (1×105 cells, 50% matrigel, nude mice), MDA-MB-231 (1×106 cells, 50% matrigel, nude mice), CAMA1 (2×106 cells, 50% matrigel, NOD/SCID mice). For mammary fat pad orthotopic xenograft experiments, 1×106 MDA-MB-361 cells were injected in 50% matrigel bilaterally into the 4th mammary glands of female NOD/SCID mice (Jackson Laboratories). Tumor size was measured by caliper and tumor volume was calculated using the formula volume = (length × width2) × π/6.
MCF10ADCIS Invasion assay
RASAL2 expression was ablated in MCF10ADCIS cells using two distinct shRNAs and loss of RASAL2 expression was confirmed by immunoblot. 100,000 shRASAL2 or shScramble control cells were injected subcutaneously into the flanks of female nude (nu/nu) mice. Five or six tumors of each genotype (Scramble shRNA, RASAL2 shRNA1, or RASAL2 shRNA2) were harvested after 3 weeks, fixed, and stained with H&E to assess tumor morphology. A tumor was scored as an invasive carcinoma by a pathologist, which was accompanied by an obvious loss in its well-circumscribed appearance and a disruption of the myoepithelium and basement membrane.
Rasal2 Mutant Mice
All animal procedures were approved by the Center for Animal and Comparative Medicine at Harvard Medical School in accordance with the NIH Guildelines for the Care and Use of Laboratory Animals and the Animal Welfare Act.
A mouse embryonic stem cell line in which the pNMDi4 genetrap cassette targets Rasal2 was purchased from the Toronto Centre for Phenogenomics/Canadian Mouse Mutant Repository (clone CMHD 463C12). The pNMDi4 genetrap cassette contains the following elements as depicted in Figure 4A: HA 3/5 SA (splice acceptor), FL (flexible linker), Venus (enhanced yellow fluorescent gene) with stop codon, loxP sites, pA (polyadenylation signal), PGK (promoter), neo (neomycin resistance gene) with stop codon, HPRT SD (splice donor). The neomycin resistance gene stop codon prevents translation of 3′ exons. The presence of the genetrap cassette within the third intron of Rasal2 was confirmed using cDNA PCR. Chimeric mice were generated and crossed to C57BL/6-E animals (Charles River Laboratories) and pups were tested for presence of the genetrap. Two additional copies of the genetrap cassette elsewhere in the genome were discovered in the mouse ES cell line and genetrap mice. Genetrap-positive mice were crossed to wild type animals and Southern blotting was used to identify pups that had the genetrap cassette only within the Rasal2 locus (data not shown). These animals were used as founders for all cohorts and subsequent crosses.
Rasal2 Genotyping
Primers for PCR of the genetrap cassette were NMD.F (5′-CATGGTCCTGCTGGAGTTC-3′) and NMD.R (5′-TGCCTTTAGACCTTTTTGTGG-3′). Total RNA was extracted from homogenized tails using QiaShredder and RNeasy kits (Qiagen), and cDNA was synthesized using qScript cDNA Synthesis Kit (Quanta). PCR was performed on cDNA with primers NeoL (5′-GCTATCAGGACATAGCGTTGGCTAC-3′), GT463C12_F3 (5′-TCGGATCCTTCTGGAGTCAG-3′), and GT463C12_R1 (5′-CTCTCTCGGAGGCAGAGCTA-3′) to detect wild type (F3/R1) and mutant (NeoL/R1) transcripts.
Compound Mutant Mice
Rasal2 genetrap mice were crossed to FVB/N-Tg(MMTVneu)202Mul/J mice (Jackson Laboratories cat. # 002376) (Guy et al., 1992) or to B6.129S2-Trp53Tm1Tyj/J mice (Jackson Laboratories cat. # 002101) (Jacks et al., 1994). Cohorts of Rasal2 mutant mice and controls were on a 129-enriched background (75% 129SvImJ, 25% C57BL6). Cohorts of Trp53; Rasal2 compound mice and controls were on a mixed 129/B6 background (62.5% C57BL6, 37.5% 129SvImJ). Cohorts of MMTVneu; Rasal2 compound mice and controls were on a background of 56% 129SvImJ, 25% FVB, and 19% C57BL6.
Protein Lysates and Western Blot Analyses
Protein extracts were isolated from cells or homogenized tissue in 1% SDS boiling lysis buffer. Ras-GTP levels were determined using a Ras Activation Assay kit (EMD Millipore). The following antibodies were used for immunoblots: Actin (Sigma cat. # A2066), phospho-AKT (Ser473, Cell Signaling cat. # 4060), AKT (Cell Signaling cat. # 9272), ER (Thermo cat. # R9101-SO), phospho-ERK (Thr202/Thr204, Cell Signaling cat. # 4370), ERK (Cell Signaling cat. # 9102), GAPDH (Cell Signaling cat. # 2118), HA (Covance cat. # MMS-101P), HER2 (Cell Signaling cat. # 2242), NF1 (UP69 C-terminal polyclonal antibody) (McGillicuddy et al., 2009), p120RasGAP (BD Transduction Laboratories cat. # 610040), HRas (Santa Cruz cat. # SC-520), KRas (Santa Cruz cat. # SC-30), panRas (Upstate cat. # 05-516), α-Tubulin (Sigma cat. # T5168), β-Tubulin (Sigma cat. # T4026), and Vinculin (Cell Signaling cat. # 4650). A peptide antigen (NP_773793 amino acids 1111–1130) was used to generate and affinity purify an anti-RASAL2 rabbit polyclonal antibody (Covance ImmunoTechnologies). For RASAL2 reconstitution studies cells were typically plated in 5% serum overnight 36 hours post transfection. pERK and ERK levels were assessed by Western Blot and quantified using ImageJ software.
Immunohistochemistry was performed as described in Supplemental Experimental Procedures
Mouse Tumor and Tissue Analysis
Tumors and tissues were fixed in buffered formalin, stored in 70% ethanol, paraffin embedded, and sectioned. Sections were stained with hematoxylin and eosin.
Human Tumor Lysate Array Analysis
Qualitative breast cancer tumor lysate arrays were purchased from Protein Biotechnologies (cat. # PMA2-001-L). Samples were de-identified and are not considered human subject research. Arrays were probed with the affinity purified RASAL2 antibody. The RASAL2 antibody was validated for this assay using tumor lysate arrays by probing nitrocellulose membranes spotted with 1μg/μl RIPA lysates from human breast cancer cell lines with or without RASAL2. Developed film was scanned and quantified using ImageJ software. Arrays were stained with Colloidal Gold, scanned, and total protein was quantified using ImageJ. RASAL2 levels in each spot were normalized to the Colloidal Gold level in the same spot. Triplicate spots were averaged, and the ratio between the tumor normalized triplicate and normal normalized triplicate was calculated and reported as a Log2 fold change value.
Molecular Subtype Association and Survival Analysis
Gene expression correlations targeted analysis was applied on published genomic data on patients classified in the same molecular subtype with the six molecular subtype predictors (Sorlie et al, 2003, Hu et al, 2006, and Parker et al, 2006) using GenExMiner as previously described (Jezequel et al. 2012). Samples were de-identified and are not considered human subject research. A gene expression map was determined by molecular subtype predictors (single sample predictors (SSPs) and/or subtype clustering models (SCMs). A gene expression table was also provided for robust classifications, indicating for each subtype the proportion of patient with low, intermediate and high gene expression; gene expression values being split in order to form 3 equal groups.
Gene expression data and relapse free and overall survival information were analyzed as previously described (Gyorffy et al., 2010). Data were downloaded from GEO (Affymetrix HGU133A and HGU133+2 microarrays), EGA, and TCGA. The background database integrates gene expression and clinical data simultaneously. To analyze the prognostic value of RASAL2, the patient samples are split into two groups according to median expression of RASAL2.
Supplementary Material
Highlights.
RASAL2 is inactivated in breast cancer and its loss is associated with progression
RASAL2 ablation promotes tumor growth, progression, and metastasis in cancer models
RASAL2 may play a particularly important role in luminal B breast cancer
RASAL2 loss may promote the progression of other cancers
Significance.
The RasGAPs are direct negative regulators of Ras and are therefore poised to function as potential tumor suppressors. Here we identify a RasGAP gene, RASAL2, as a tumor suppressor within this gene family. Our data suggest that RASAL2 loss plays a causal role in the development, progression, and metastasis of breast cancer and may play a broader role in the metastasis of other solid tumors as well. Collectively, these data reveal an alternative mechanism by which Ras becomes activated in cancer and identify a role for RASAL2 and Ras in breast cancer progression and metastasis.
Acknowledgments
This work was supported by grants from the Ludwig Center at DF/HCC (K.C.), the DOD (W81XWH-11-1-0140) (S.K.M.), and NIH National Heart, Lung, and Blood Institute (T32-HL07623) (S.K.M.).
Footnotes
The Sanger Institute Catalogue Of Somatic Mutations In Cancer http://www.sanger.ac.uk/cosmic (Wellcome Trust Sanger Institute).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, Robertson M, Dunn D, Gesteland R, O’Connell P. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dote H, Toyooka S, Tsukuda K, Yano M, Ouchida M, Doihara H, Suzuki M, Chen H, Hsieh JT, Gazdar AF, et al. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in breast cancer. Clin Cancer Res. 2004;10:2082–2089. doi: 10.1158/1078-0432.ccr-03-0236. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollestelle A, Elstrodt F, Nagel JHA, Kallemeijn WW, Schutte M. Phosphatidylinositol–3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol Cancer Res. 2007;5:195–201. doi: 10.1158/1541-7786.MCR-06-0263. [DOI] [PubMed] [Google Scholar]
- Hölzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, Nijkamp W, Xie J, Callens T, Asgharzadeh S, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, Richardson A, Violette S, Nikolskaya T, Nikolsky Y, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–75. doi: 10.1007/s10549-011-1457-7. http://bcgenex.centregauducheau.fr. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci USA. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Baek SH. Mouse models for breast cancer metastasis. Biochem Biophys Res Commun. 2010;394:443–447. doi: 10.1016/j.bbrc.2010.03.070. [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- von Lintig FC, Dreilinger AD, Varki NM, Wallace AM, Casteel DE, Boss GR. Ras activation in human breast cancer. Breast Cancer Res Treat. 2000;62:51–62. doi: 10.1023/a:1006491619920. [DOI] [PubMed] [Google Scholar]
- Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, Roberts B, Zhang T, Chenard M, Haines B, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med Genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messaien L, Bronson RT, McMahon M, Granter S, Flaherty KT, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SMG, Nghiemphu P, Liau LM, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92:1185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, Beroukhim R, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C, Liotta LA, Espina V. Reverse phase protein microarrays advance to use in clinical trials. Molecular Oncology. 2010;4:461–481. doi: 10.1016/j.molonc.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller H, Flury N, Eppenberger-Castori S, Kueng W, David F, Eppenberger U. Potential prognostic value of mitogen-activated protein kinase activity for disease-free survival of primary breast cancer patients. Int J Cancer. 2000;89:384–388. doi: 10.1002/1097-0215(20000720)89:4<384::aid-ijc11>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaraman VS, Wang H, Nuovo GJ, Malbon CC. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J Clin Invest. 1997;99:1478–1483. doi: 10.1172/JCI119309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GV, Horvath S, Smith BL, Crosby K, Lebel LA, Schrage M, Said J, De Kernion J, Reiter RE, Sawyers CL. Antibody-based profiling of the phosphoinositide 3-kinase pathway in clinical prostate cancer. Clin Cancer Res. 2004;10:8351–8356. doi: 10.1158/1078-0432.CCR-04-0130. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Peterse JL, van t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.