Abstract
To understand local transmission of vector-borne diseases, it is important to identify potential vectors, characterize their host feeding patterns, and determine if vector-borne pathogens are circulating within the region. This study simultaneously investigated these aspects of disease transmission by collecting engorged mosquitoes within two rural study sites in the central Red River Valley of North Dakota. Mosquitoes were identified, midguts were excised, and the blood was expelled from the midguts. DNA was extracted from blood meals and subjected to PCR and direct sequencing to identify the vertebrate origin of the blood. Using different primer sets, PCR was used to screen for two types of vector-borne pathogens, filarioid nematodes and hemosporidian parasites. White-tailed deer were the primary source of blood meals for the eight aedine mosquito species collected. None of the 288 deer-derived blood meals contained filarioid or hemosporidian DNA. In contrast, 18 of 32 Culex tarsalis and three of three Cx. pipiens blood meals contained avian blood, representing eight different species of birds. Of 24 avian-derived blood meals examined, 12 contained Plasmodium DNA, three of which also contained Leucocytozoon DNA (i.e., dual infection). Potential confounding effects resulting from parasite acquisition and development from previous blood meals (e.g., oocysts) were eliminated because host blood had been removed from the midguts prior to DNA extraction. Thus, specific parasite lineages/species could be unequivocally linked to specific vertebrate species. By combining mosquito identification with molecular techniques for identifying blood meal source and pathogens, a relatively small sample of engorged mosquitoes yielded important new information about mosquito feeding patterns and hemosporidia infections in birds. Thorough analyses of wild-caught engorged mosquitoes and other arthropods represent a powerful tool in understanding the local transmission of vector-borne and zoonotic diseases.
Key Words: Mosquito, Blood meal, Xenomonitoring, Plasmodium, Leucocytozoon, PCR
Introduction
The Red River Valley (RRV) is plagued by mosquitoes every summer, yet knowledge of mosquito fauna and biology within the region is marginal. Within the city of Grand Forks, North Dakota, the mosquito fauna is dominated by three species—two floodwater species, Aedes vexans and Ae. dorsalis, which are present throughout the summer, and Culex tarsalis, which is abundant during mid to late summer and is the primary vector for West Nile virus (Deckert 1995, Bell et al. 2005, Bell et al. 2006). Preliminary studies suggest that species richness of mosquitoes is greater in the surrounding rural areas than within the city (Vaughan, unpublished data). Almost nothing is known about the blood-feeding patterns of mosquitoes within the rural RRV. Blood-feeding patterns can incriminate mosquito species involved in various mosquito-borne diseases. Similarly, the diversity of zoonotic pathogens in the RRV remains little studied. Surveying vertebrate populations for blood pathogens can be difficult and time consuming. Instead, we examined the blood meals of wild-caught mosquitoes. The DNA within each blood meal was analyzed by PCR and sequencing to identify its vertebrate host origin. PCR and sequencing have also been used to identify various types of hemoparasites within mosquito blood meals (Chanteau et al. 1994, Farid et al. 2007, Massey et al. 2007, Bartlett-Healy et al 2009, Chambers et al. 2009, Kim et al. 2009, Latrofa et al. 2012). This is known as “molecular xenomonitoring.” Using these techniques, we determined the blood-feeding patterns of local mosquito species as well as the prevalence and species identity of hemoparasites within birds fed on by mosquitoes.
Materials and Methods
Mosquito collection
Mosquitoes were collected from two sites within Steele County, North Dakota. The first site was a 40-acre hardwood forest with a closed canopy and thick underbrush located 8.4 km southwest of Hatton, ND. The site is typically flooded in early spring by the Goose River, leaving behind breeding pools within the forest throughout the summer. The second site was a farmstead located 1.6 km west of Hatton, ND. The farm was surrounded by open cropland and is more typical of the RRV landscape. Mosquito control was absent at both sites.
Host-seeking mosquitoes were collected using three battery operated, CO2-baited Mosquito Magnet X traps (MMX) (Woodstream Lititz, PA) spaced ≈200 meters apart. Traps were deployed two to three times per week and operated from 1800 to 0800 h, from late May through August. The MMX traps were operated at the forest site during 2009 and 2010, and at the farm site during 2011. In 2011, resting mosquitoes were collected at both sites from underbrush, tree holes, and around the bases of trees and buildings with a battery-operated vacuum aspirator. The aspirator was constructed of lightweight aluminum, with a fan and a 33-cm-diameter circular collection area, by personnel at the Metropolitan Mosquito Control District, St. Paul, MN. Insects were transported to the laboratory and placed in −20°C freezers for immobilization. Mosquitoes were identified to species (Darsie Jr. and Ward 2005) and stored at −80°C. Besides mosquitoes, the MMX traps also collected large numbers of host-seeking black flies (Simuliidae). Blackflies were identified to species (Adler et al. 2004) but were not processed or analyzed for vertebrate or hemoparasite DNA.
DNA extraction
Mosquitoes were dissected in phosphate-buffered saline using jeweler's forceps. Midguts were excised, split open, and gently pressed against the inside of 1.5-mL microtubes to expel the midgut contents. Nucleic acids were extracted using a guanidine/ethanol protocol (Tkach and Pawlowski 1999).
Identification of vertebrate DNA
To identify the vertebrate origin of each blood meal, mitochondrial cytochrome b and cytochrome oxidase subunit I genes were amplified using previously described protocols (Townzen et al. 2008). Successful amplifications were detected by gel electrophoresis. PCR products were cleaned with ExoSAP-IT (Affymetrix, Santa Clara, CA) according to the manufacturer's protocol and sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc., ABI, Foster, CA) and a 3100 Genetic Analyzer (Applied Biosystems Inc., ABI, Foster, CA). Sequences were analyzed and trimmed to no fewer than 300 bp using the BioEdit program (Ibis Biosciences, Carlsbad, CA). Sequences were aligned to published sequences available via the National Center for Biotechnology Information (NCBI) using Basic Local Alignment Search Tool (BLAST). Host sequences were considered a match at or above 98% base pair matches.
Identification of parasite DNA
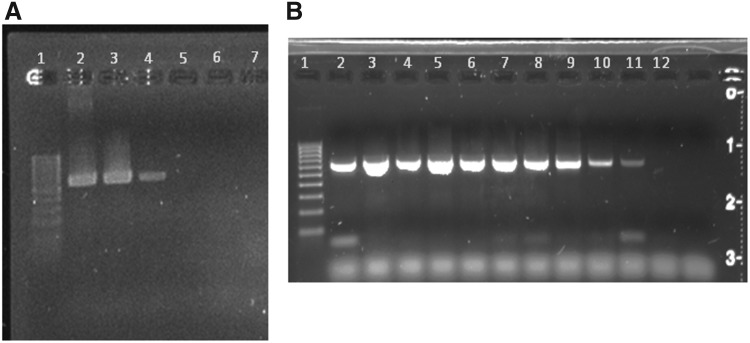
The DNA extracts were screened for filarioid nematode and hemosporidian DNA using different primer sets (Table 1). For filarioid nematodes, PCR primers were designed based on conserved sequences of the 18S ribosomal RNA genes of Loa loa, Brugia pahangi, Onchocerca cervicalis, and Chandlerella quiscali (GenBank accession numbers: DQ094173.1, EU496884.1, DQ094174.1). When tested, the primers successfully amplified 18S rRNA gene fragments of five different genera from four different subfamilies of filarioid nematodes, including Dirofilariinae (Dirofilaria immitis, Waltonella sp.), Onchocercinae (B.pahangi), Splendidofilariinae (C. quiscali), and Lemdaninae (Eufilaria sp.), indicating their utility for detecting a broad taxonomic range of filarioid species (Fig. 1A). The primers exhibited high sensitivity when tested against two-fold dilutions of DNA extracted from C. quiscali microfilariae recovered from the lungs of a Common Grackle (Quiscalus quiscula) (Fig. 1B). To screen for Babesia parasites, a set of seminested primers was developed using sequences of the 18S rRNA genes of Babesia spp. (GenBank accession numbers: AY046577.1, AY237638.1, AB190459.1, HQ184411.1) and tested using DNA from Babesia microti (a kind gift of S. Telford III). The thermocycling protocols for the filarioid and the Babesia PCR were identical; 95°C for 2 min for initial activation followed by 40 cycles of 95°C for 45 sec, 55°C for 30 sec, and 72°C for 45 sec with a one-time final extension at 72°C for 7 min. Detection of the cytochrome b gene from Plasmodium, Leucocytozoon, and Haemoproteus used the previously described protocol and primers of Hellgren et al. (2004).
Table 1.
Primers Used in Molecular Analyses of Mosquito Bloodmeals
| Target organism | Target gene | Primer name | 5′-3′ Primer sequence | Amplicon size (bp) |
|---|---|---|---|---|
| Vertebratea | mtDNA cytochrome b | Forward- Cyt b | GAGGMCAAATATCATTCTGAGG | 457 |
| Vertebratea | mtDNA cytochrome b | Reverse- Cyt b | TAGGGCVAGGACTCCTCCTAGT | 457 |
| Vertebratea | mtDNA cytochrome oxidase 1 | Forward- COI_long | AACCACAAAGACATTGGCAC | 663 |
| Vertebratea | mtDNA cytochrome oxidase 1 | Reverse- COI_long | AAGAATCAGAATARGTGTT | 663 |
| Vertebratea | mtDNA cytochrome oxidase 1 | Forward- COI_short | GCAGGAACAGGWTGAACCG | 324 |
| Vertebratea | mtDNA cytochrome oxidase 1 | Reverse- COI_short | AATCAGAAYAGGTGTTGGTATAG | 324 |
| Filaria | 18S rRNA | Forward- Chand FO | GAGACCGTTCTCTTTGAGGCC | 580 |
| Filaria | 18S rRNA | Reverse- Chand RO | GTCAAGGCGTANNTTTACCGCCGA | 580 |
| Babesia | 18S rRNA | Forward- BabF 1102–1122 | GACTAGGGATTGGAGGTCGTC | 739 and 240 |
| Babesia | 18S rRNA | Reverse- BabR 1841–1818 | GACCACCACCCAAAGAATCAA | 739 |
| Babesia | 18S rRNA | Reverse- BabR 1342–1318 | GGTCCGAATAATTCACCGGATCAC | 240 |
| Haemosporidiab | mtDNA cytochrome b | Forward- HaemNFl | CATATATTAAGAGAAITATGGAG | 570 |
| Haemosporidiab | mtDNA cytochrome b | Reverse- HaemNR3 | ATAGAAAGATAAGAAATACCATTC | 570 |
| Leucocytozoonb | mtDNA cytochrome b | Forward- HaemFL | ATGGTGTTTTAGATACTTACATT | 478 |
| Leucocytozoon*b | mtDNA cytochrome b | Reverse- HaemRL2 | CATTATCTGGATGAGATAATGGIGC | 478 |
| Plasmodium/Haemoproteusb | mtDNA cytochrome b | Forward- HaemF | ATGGTGCTTTCGATATATGCATG | 480 |
| Plasmodium/Haemoproteub | mtDNA cytochrome b | Reverse- HaemR2 | GCATTATCTGGATGTGATAATGGT | 480 |
FIG. 1.
Xenomonitoring PCR using an 18S rRNA gene. (A) Lane 1, ladder; lane 2, avian filarid (Eufilaria sp.); lane 3, frog filarid (Waltonella sp.); lane 4, avian filarid (Chandlerella sp.); lane 5, engorged Cx. pipiens (host=mouse); lane 6, mouse blood; lane 7, unengorged Ae. vexans. (B) Sensitivity of PCR to Ch. quiscali microfilaria in sample. Lane 1, ladder; lanes 2–12 representing 128, 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, and 0.125 microfilaria, respectively. PCR is sensitive to detect less than 1 microfilaria in each 5 μL sample.
Results
MMX traps collected 8855 and 9687 mosquitoes from the forest and farm sites, respectively, but few were blood-fed (1.3% and 0.1%, respectively). Vacuum aspiration collected less mosquitoes (4898 and 810 from the forest and farm sites, respectively), but the proportion blood-fed was greater (30% and 31%, respectively). Of 1841 engorged mosquitoes collected, DNA was extracted and PCRs were run on 770 individual mosquitoes to determine blood meal origin. Of these, usable DNA was recovered from 523 (68%) of the extracts. The low recovery rate may be due to the digestion of blood as well as low levels of DNA because partial blood meals were also categorized as engorged. Of 523 extracts with usable DNA, 416 were selected for sequencing. Of the 416 DNA extracts that were sequenced, 391 (94%) provided sufficiently high-quality sequences to be aligned with vertebrate host sequences in the NCBI database.
Mosquito host-feeding patterns
The majority of engorged mosquitoes (75%) collected at the forest site were Ae. excrucians, most of which (94%) fed on white-tailed deer (WTD) (Table 2). Similarly, the host feeding patterns observed for Ae. canadensis, Ae. flavescens, Ae. triseriatus, Culiseta inornata, and Coquillettidia perturbans suggest that the main blood source for mosquitoes at the forest site was WTD.
Table 2.
Mosquito Feeding Patterns at The Forest Site in 2009 and 2011, Steele County, North Dakota
| Host | Ae. excrucians | Ae. triseriatus | Ae. vexans | Ae. canadensis | Ae. flavescens | Cs. inornata | Co. perturbans | Total |
|---|---|---|---|---|---|---|---|---|
| White-tailed deer | 104 | 17 | 3 | 2 | 1 | 6 | 3 | 136 |
| Cow | 2 | 2 | 1 | 5 | ||||
| Human | 1 | 1 | 2 | |||||
| Raccoon | 2 | 2 | ||||||
| American Mink | 1 | 1 | ||||||
| Common Yellow-throat | 1 | 1 | ||||||
| Total | 110 | 17 | 3 | 2 | 1 | 8 | 6 | 147 |
Vertebrate species determined by PCR and sequencing. See Table 1 for primers and target genes.
Most (74%) of the engorged mosquitoes collected at the farm site were Ae. vexans, the majority of which (70%) fed on WTD, although other blood sources were also used, including cow, dog, rabbit, and human (Table 3). With the notable exception of the two Culex species, most of the mosquitoes collected at the farm site were decidedly mammophilic. Cx. tarsalis was the second most numerous mosquito species collected at the farm site and used a wide range of host animals (Table 3). Only 16% of Cx. tarsalis blood meals were from WTD. The majority of Cx. tarsalis blood meals (56%) were avian derived. Robins, house sparrows, and grackles constituted >60% of all avian-derived blood meals taken by Cx. tarsalis. Even so, the bird species composition (n=9) within Cx. tarsalis blood meals was diverse. Culex pipiens also seem to feed preferentially on birds although the numbers collected were low (Table 3).
Table 3.
Mosquito Feeding Patterns at The Agriculture Site In 2010 and 2011, Steele County, North Dakota
| Host | Cx. tarsalis | Cx. pipiens | Ae. canadensis | Ae. dorsalis | Ae. excrucians | Ae. flavescens | Ae. triseriatus | Ae. vexans | Cs. inornata | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Deer | 5 | 11 | 1 | 3 | 125 | 4 | 149 | |||
| Cow | 1 | 1 | 21 | 23 | ||||||
| Dog | 5 | 2 | 1 | 1 | 14 | 23 | ||||
| Cat | 2 | 1 | 2 | 1 | 1 | 7 | ||||
| Rabbit | 2 | 11 | 13 | |||||||
| Human | 1 | 1 | 4 | 6 | ||||||
| AmericanRobin | 5 | 1 | 6 | |||||||
| House Sparrow | 5 | 5 | ||||||||
| Cedar Waxwing | 3 | 3 | ||||||||
| Mourning Dove | 1 | 1 | ||||||||
| AmericanGoldfinch | 1 | 1 | ||||||||
| Chipping | ||||||||||
| Sparrow | 1 | 1 | ||||||||
| Cliff | ||||||||||
| Swallow | 1 | 1 | ||||||||
| CommonGrackle | 1 | 3 | 4 | |||||||
| Green | ||||||||||
| Heron | 1 | 1 | ||||||||
| Total | 32 | 3 | 14 | 2 | 1 | 2 | 8 | 177 | 5 | 244 |
Vertebrate species determined by PCR and sequencing. See Table 1 for primers and target genes.
Parasite detection
From the 255 blood meals derived from WTD, we suspected we might find DNA evidence for two parasite species; namely, Setaria yehi, a mosquito-borne filarioid nematode of North American cervids, and Babesia odocoilei, a tick-borne hemosporidian of WTD. We found no DNA evidence of either parasite in mosquito blood meals.
From the 24 blood meals derived from birds, we suspected we might find DNA evidence for two parasite groups; namely, filarioid nematodes (i.e., blood microfilariae) and hemosporidians (i.e., Plasmodium, Leucocytozoon, or Haemoproteus). None contained filarioid or Haemoproteus DNA. However, three (12%) of the 24 bird-derived blood meals contained Leucocytozoon DNA and 12 (50%) contained Plasmodium DNA (Table 4). Leucocytozoon DNA was present in two of the four blood meals taken from Common Grackles and one of the five blood meals taken from American Robins (Turdus migratorius). Plasmodium DNA was present in three of the six blood meals taken from American Robins, all four blood meals taken from Common Grackles, one of the three blood meals from Cedar Waxwings (Bombycilla cedrorum), three of the five blood meals from House Sparrows, and the single blood meal taken from Green Heron (Butorides virescens). When the 12 Plasmodium DNA sequences were compared with DNA sequences available from the NCBI database, two separate Plasmodium lineages emerged, both without formal species status. One lineage was present in a Cx. tarsalis blood meal taken from an American Robin. This lineage was 99% identical to the sequence of a Plasmodium isolate recovered from a White-throated Thrush (Turdus assimilis) in Costa Rica (accession no. JN819347). The other 11 Plasmodium sequences were identical to each other and to the sequence of a Plasmodium isolate recovered from a Barred Owl (Strix varia) in Wisconsin (accession no. EU627827). The fact that the second Plasmodium lineage was recovered from blood meals derived from four different families of passerine birds, a heron, and an owl indicates that this particular Plasmodium lineage has a broad host range. Three of the 12 blood meals (25%) contained both Plasmodium and Leucocytozoon DNA, including one blood meal from an American Robin and two blood meals from Common Grackles. This suggests that hemosporidian polyparasitism among some passerine species within the RRV may be common.
Table 4.
Detection of Hemosporidian Dna Within Mosquito Bloodmeals of Avian Origin. Steele County, North Dakota
| Month | Day | Year | Mosquito Species | Avian Host Species | Plasmodium | Leucocytozoon |
|---|---|---|---|---|---|---|
| June | 9 | 2010 | Ae. vexans | Mourning Dove | ||
| 11 | 2011 | Cx. pipiens | Common Grackle | + | + | |
| 11 | 2011 | Cx. pipiens | Common Grackle | + | ||
| 17 | 2011 | Cx. pipiens | Common Grackle | + | ||
| 17 | 2011 | Ae. canadensis | American Robin | + | + | |
| July | 1 | 2010 | Cx. tarsalis | Common Grackle | + | + |
| 1 | 2010 | Cx. tarsalis | Green Heron | + | ||
| 7 | 2010 | Cx. tarsalis | Cliff Swallow | |||
| 7 | 2010 | Cx. tarsalis | American Robin | + | ||
| 8 | 2010 | Cx. tarsalis | American Robin | |||
| 8 | 2010 | Cx. tarsalis | House Sparrow | |||
| 15 | 2010 | Cx. tarsalis | American Robin | + | ||
| 15 | 2010 | Cx. tarsalis | American Robin | |||
| 26 | 2010 | Cx. tarsalis | American Robin | |||
| August | 2 | 2010 | Cx. tarsalis | Chipping Sparrow | ||
| 6 | 2010 | Cx. tarsalis | American Goldfinch | |||
| 6 | 2009 | Ae. excrucians | Common Yellowthroat | |||
| 6 | 2010 | Cx. tarsalis | Cedar Waxwing | + | ||
| 6 | 2010 | Cx. tarsalis | Cedar Waxwing | |||
| 6 | 2010 | Cx. tarsalis | Cedar Waxwing | |||
| 6 | 2010 | Cx. tarsalis | House Sparrow | + | ||
| 6 | 2010 | Cx. tarsalis | House Sparrow | + | ||
| 6 | 2010 | Cx. tarsalis | House Sparrow | |||
| 8 | 2010 | Cx. tarsalis | House Sparrow | + |
Vertebrate and parasite species determined by PCR and sequencing. See Table 1 for primers and target genes.
Discussion
Molecular analyses of blood meals from wild-caught mosquitoes yielded new information on local zoonotic disease ecology. First, PCR and sequencing of vertebrate mitochondrial genes yielded information on the blood-feeding patterns of local mosquito species. This is important in defining the transmission ecology of mosquito-borne diseases within a region and helps predict the likelihood of success for exotic mosquito-borne pathogens that may move into a region. For example, we found that deer were the main blood source for many aedine mosquito species in the RRV. This presents a favorable ecological setting for certain arboviruses, such as Jamestown Canyon virus (see Andreadis et al. 2008) or perhaps even Rift Valley fever virus (see Iranpour et al. 2011), should these arboviruses become introduced in the RRV.
Second, once the host origin of a blood meal was identified, it was then possible to target the types of hemoparasites that might be present within a particular blood meal. By using different primer sets on the same DNA extracts, we were able to link the presence of certain hemoparasites to specific vertebrate species (i.e., molecular xenomonitoring). Although mosquitoes are the raw material for analyses, it is important to remember that molecular xenomonitoring does not provide information on a mosquito's ability to transmit the hemoparasites it has ingested (i.e., molecular xenomonitoring is not vector competence).
Molecular xenomonitoring merely substitutes for the direct capture and bleeding of vertebrates and thus it should be interpreted as an indirect way of estimating the prevalence and diversity of hemoparasites circulating within specific vertebrate populations. Because it is an indirect measurement, the accuracy and reliability of molecular xenomonitoring can be influenced by several factors, including obvious things such as the age of the blood meal and the sample size, as well as less-obvious factors such as sample processing and the unique biology of the hemoparasite and host under investigation.
For example, on the basis of the assays of 288 deer-derived blood meals, we found no evidence of Babesia or filaroid nematode infections in the local WTD population. This may indeed be the case for B. odocoilei because the vector tick Ixodes scapularis has only recently become established in the RRV (Russart and Vaughan, unpublished data). However, the reliability of our xenomonitoring to assess microfilarial infections in WTD is less certain. Reported prevalences of the cervid filarioid S. yehi in WTD are 16% (n=84) and 27% (n=1045) from Illinois and the southeastern United States, respectively (Prestwood and Pursglove 1977, Cook et al. 1979). The prevalence of S. yehi in California black-tailed deer is 40% (n=488) (Weinmann et al. 1973). Thus, one would expect that xenomonitoring of 288 deer-fed mosquitoes would result in detecting at least some S. yehi parasites. However, there are two confounding factors related to the biology of the parasite and the size of the host. First, S. yehi microfilariae quickly penetrate the midgut after ingestion by Aedes spp. mosquitoes (Lee 1971). Engorged mosquitoes in our studies, many of which likely fed at night, were collected in the morning and transported to the laboratory prior to dissections. The delay between mosquito ingestion of deer blood and mosquito dissection may have allowed microfilariae sufficient time to exit the midgut and escape detection. Second, deer are large animals, and a single animal can serve as the blood source for many mosquitoes. Thus, the prevalence of pathogens within the blood meals of mosquitoes feeding on large herding animals (particularly if mosquitoes were collected at the same time and location) may not directly correlate with the prevalence of pathogens within the vertebrate population in general. This is probably of lesser concern when considering pathogens within blood meals derived from small or solitary host species.
We also failed to detect filarioid nematode DNA in avian-derived blood meals. In this case, concerns over microfilariae exiting the midgut and escaping detection are unwarranted because only a small fraction of ingested passerine microfilariae successfully exit the midguts of local Culex mosquitoes (Vaughan et al. 2012). Failure to detect filarioid DNA in avian-derived blood meals was most likely due to the low number of samples represented per bird species. The greatest numbers of conspecific blood meals were from American Robins (n=6), House Sparrows (n=5), and Common Grackles (n=4) (Table 4). But the estimated prevalence of filarioid infections for these bird species within the RRV is less than 20% (Vaughan et al. 2012). Such small samples sizes fall below the theoretical limit of detection. Obviously, the use of molecular xenomonitoring to detect low prevalence infections among a diverse community of host species requires larger sample sizes than used here.
In contrast, molecular xenomonitoring detected hemosporidian infections in 12 of the 24 avian-derived blood meals examined (Table 4), indicating a high prevalence of hemosporidian infections in local bird populations. The dominant hemosporidian consisted of a single lineage of Plasmodium capable of infecting a broad taxonomic range of birds. Active transmission of this Plasmodium lineage almost certainly occurred at this site as indicated by the high abundance of an ornithophilic and competent vector (e.g., Cx. tarsalis; see Work et al. 1990) and the fact that three of the five blood meals containing House Sparrow blood also contained Plasmodium DNA. Unlike other bird species identified in this study, House Sparrows are nonmigratory residents and thus are unlikely to have acquired their infections elsewhere.
In addition, a substantial proportion (25%) of hemosporidian infections within mosquito blood meals consisted of Plasmodium and Leucocytozoon, indicating that polyparasitism of local birds was common. The MMX traps used to collect mosquitoes also collected enormous numbers of black flies. Black flies are the vectors of Leucocytozoon parasites (Valkiunas 2005). A large sample (>800) of black flies was examined. All belonged to a single ornithophilic species, Simulium johannseni. Their appearance in late May and early June corresponded temporally with the detection of Leucocytozoon in avian-derived blood meals (Table 4). Thus in addition to Plasmodium, active transmission of Leucocytozoon may also have occurred at the site.
One of our objectives was to use molecular xenomonitoring to link specific parasites to specific host species. In the case of hemosporidia in avian-derived blood meals, the link was definitive (see Table 4) because of the way in which mosquitoes were processed—i.e., blood meal contents were removed from mosquito midgut tissue prior to nucleic acid extraction. Alternative ways in which engorged mosquitoes can be processed for DNA extraction include using whole mosquitoes or whole abdomens separated from the head/thorax region. The optimal method depends on the object of the study. If the object is merely to identify the vertebrate host origin of the blood, then the quickest method (i.e., whole mosquito) may be preferred. However if the object is to link blood meal pathogens to the vertebrate species from which the blood originated, then the method of sample preparation should be dictated by the type of pathogen being monitored.
There are five types of pathogens that can be taken up in a mosquito blood meal—arboviruses, bacteria, trypanosomes, hemosporidians, and microfilariae. With bacteria and trypanosomes, it does not matter if the mosquito is dissected. That is because blood-borne bacteria (e.g., rickettsiae, spirochetes, etc.) and most trypanosomes either never leave the mosquito digestive tract or, if they do (e.g, Salivaria trypanosome Trypanosoma brucei), they do not exit the gut until the blood meal has been digested beyond the point of accurately determining its vertebrate origin.
With hemosporidia and arboviruses, it is crucial to rid the sample of mosquito midgut tissue prior to extraction. As part of their normal development, hemosporidian oocysts stay attached to the outside of the gut for many days. Similarly, arborviruses infect midgut epithelium long after the viremic blood meal has been digested and the mosquito has oviposited. If an oocyst- or arbovirus-infected mosquito takes a second blood meal from a different host species, then confusion may arise as to the host origin of the pathogen. For example, Ejira et al. (2011a, b) identified avian Plasmodium DNA in the severed abdomens of engorged mosquitoes containing blood identified molecularly as being from cow and sitka deer. Avian Plasmodium DNA could not have originated from the blood of a ruminant. Instead it probably originated from oocysts attached to the outside of the midguts (i.e., previous blood meal from a bird), not from the blood inside the midguts. Similarily, Santiago-Alarcon et al. (2012) analyzed severed abdomens of 105 engorged Culicoides gnats. Four were concurrently positive for the DNA of humans and the DNA of avian Plasmodium and Haemoproteus parasites. Because avian hemosporidians do not infect humans, the only logical sequence of events that could account for such a result would be if the gnats had first fed on gametocytemic birds, the parasites underwent sporogonic development to oocysts, and then the infected gnats fed on a human. Without first separating the ingested blood from midgut tissue, it is impossible to unequivocally link a hemosporidian species (or an arbovirus) identified in an engorged abdomen to the vertebrate host species represented within the blood meal.
The filarioid nematodes are the most problematic because the nematodes' location within the mosquito can vary. In some filarioid/mosquito combinations, most, if not all, ingested microfilariae exit the blood meal into the hemocoel within hours after being ingested (Wharton 1957, Laurence and Pester 1961). In other combinations, few if any succeed in leaving the midgut (Ewert 1966, Zielke 1992). To account for such variability, severing abdomens from thorax might be the preferred method.
In summary, we successfully used DNA recovered from the midguts of wild-caught mosquitoes to simultaneously determine the host feeding patterns of local mosquitoes and conduct a survey of blood-borne parasites within local wildlife. Careful examination of relatively few specimens yielded important new information about the local transmission of avian hemosporidian parasites. Application of this technique can contribute to an increasing understanding of the transmission ecology of zoonotic diseases. However, certain considerations should be taken into account regarding how samples from the field are prepared prior to the extraction of nucleic acids.
Acknowledgments
We thank Dr. Sam Telford III, Tufts University, for the generous gift of Babesia microti samples used a as positive control in the development of our PCR. Sarina Bauer, Stephen Greiman, Alex Droske, Bo Chung, Jeffrey Bell, and Max Tkach assisted in setting traps and sorting and identifying mosquitoes. We thank Dr. Vasyl Tkach for assisting in primer development. We thank the Metropolitan Mosquito Control District, St. Paul, MN for the loan of their mosquito aspirator. Financial support was provided by UND Graduate School Fellowship (J.O.M.), ND EPSCoR Graduate Student Research Assistantship (J.O.M.), and National Institutes of Health (NIH) grant R03 AI092306 (J.A.V.).
Author Disclosure Statement
No competing financial interests exist.
References
- Adler P. Currie DC. Wood M. Zettler L. Janzen DH. The Black Flies (Simuliidae) of North America. Ithaca: Cornell University Press; 2004. [Google Scholar]
- Andreadis TG. Anderson JF. Armstrong PM. Main AJ. Isolations of Jamestown Canyon virus (Bunyaviridae: Orthobunyavirus) from field-collected mosquitoes (Diptera: Culicidae) in Connecticut, USA: A ten-year analysis, 1997–2006. Vector Borne Zoonotic Dis. 2008;8:175–188. doi: 10.1089/vbz.2007.0169. [DOI] [PubMed] [Google Scholar]
- Bartlett-Healy K. Crans W. Gaugler R. Vertebrate hosts and phylogenetic relationships of amphibian trypanosomes from a potential invertebrate vector, Culex territans Walker (Diptera: Culicidae) J Parasitol. 2009;95:381–387. doi: 10.1645/GE-1793.1. [DOI] [PubMed] [Google Scholar]
- Bell JA. Mickelson NJ. Vaughan JA. West Nile virus in host-seeking mosquitoes within a residential neighborhood in Grand Forks, North Dakota. Vector Borne Zoonotic Dis. 2005;5:373–382. doi: 10.1089/vbz.2005.5.373. [DOI] [PubMed] [Google Scholar]
- Bell JA. Brewer CM. Mickelson NJ. Garmen GW. Vaughan JA. West Nile virus epizootiology, Central Red River Valley, North Dakota and Minnesota, 2002–2005. Emerg Infect Dis. 2006;8:1245–1247. doi: 10.3201/eid1208.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers EW. McClintock SK. Avery MF. King JD, et al. Xenomonitoring of Wuchereria bancrofti and Dirofilaria immitis infections in mosquitoes from American Samoa: Trapping considerations and comparison of polymerase chain reaction assays with dissection. Am J Trop Hyg. 2009;5:774–781. [PubMed] [Google Scholar]
- Chanteau S. Luguiaud P. Failloux AB. Williams SA. Detection of Wuchereria bancrofti larvae in pools of mosquitoes by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1994;6:665–666. doi: 10.1016/0035-9203(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Cook TW. Ridgeway BT. Andrews R. Hodge J. Gastro-intestinal helminths in white-tailed deer (Odocoileus virginianus) of Illinois. J Wildl Dis. 1979;15:405–408. doi: 10.7589/0090-3558-15.3.405. [DOI] [PubMed] [Google Scholar]
- Darsie RF., Jr Ward RA. Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico. Gainesville, FL: University Press of Florida; 2005. [Google Scholar]
- Deckert WA. Dissertation. Grand Forks: University of North Dakota; 1995. The occurrence of mosquitoes in the central Red River Valley area in 1992–1993. [Google Scholar]
- Ejiri H. Sato Y. Kim KS. Hara T. Tsuda Y. Imura T. Murata K. Yukawa M. Entomological study of transmission of avian malarial parasites in a zoological garden in Japan: bloodmeal identification and detection of avian malarial parasite DNA from blood-fed mosquitoes. J Med Entomol. 2011a;48:600–607. doi: 10.1603/me10197. [DOI] [PubMed] [Google Scholar]
- Ejiri H. Sato Y. Kim KS. Tsuda Y. Murata K. Saito K. Watanabe Y. Shimura Y. Yukawa M. Blood meal identification and prevalence of avian malaria parasites in mosquitoes collected at Kushiro wetland, a subarctic zone of Japan. J Med Entomol. 2011b;48:904–908. doi: 10.1603/me11053. [DOI] [PubMed] [Google Scholar]
- Ewert A. Comparative migration of microfilariae and development of Brugia pahangi in various mosquitoes. Am J Trop Med Hyg. 1966;14:254–259. doi: 10.4269/ajtmh.1965.14.254. [DOI] [PubMed] [Google Scholar]
- Farid HA. Morsy ZS. Helmy H. Ramzy RM, et al. A critical appraisal of molecular xenomonitoring as a tool for assessing progress toward elimination of lymphatic filariasis. Am J Trop Med Hyg. 2007;4:593–600. [PMC free article] [PubMed] [Google Scholar]
- Hellgren O. Waldenstrom J. Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- Iranpour M. Turell MJ. Lindsay LR. Potential for Canadian mosquitoes to transmit Rift Valley fever virus. J Am Mosq Control Assoc. 2011;27:363–369. doi: 10.2987/11-6169.1. [DOI] [PubMed] [Google Scholar]
- Kim KS. Tsuda Y. Sasaki T. Kobayashi M. Hirota Y. Mosquito blood-meal analysis for avian malaria study in wild bird communities: Laboratory verification and application to Culex sasai (Diptera:Culicidae) collected in Tokyo, Japan. Parasitol Res. 2009;5:1351–1357. doi: 10.1007/s00436-009-1568-9. [DOI] [PubMed] [Google Scholar]
- Latrofa MS. Ciocchetta S. Annoscia G. Dantas-Torres F, et al. Molecular xenomonitoring of Dirofilaria immitis and Dirofilaria repens in mosquitoes from north-eastern Italy by real-time PCR coupled with melting curve analysis. Parasit Vectors. 2012;5:76. doi: 10.1186/1756-3305-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence BR. Pester FRN. The behavior and development of Brugia patei (Buckley, Nelson and Heisch, 1958) in a mosquito host, Mansonia uniformis (Theobold) J Helmintol. 1961;35:285–300. doi: 10.1017/s0022149x00004661. [DOI] [PubMed] [Google Scholar]
- Lee D. Setaria yehi. Berkeley: University of California; 1971. The role of the mosquito, Aedes sierrensis, in the epizootiology of the deer body worm. Thesis. [Google Scholar]
- Massey B. Gleeson DM. Slaney D. Tompkins DM. PCR detection of Plasmodium and blood meal identification in a native New Zealand mosquito. J Vector Ecol. 2007;1:154–156. doi: 10.3376/1081-1710(2007)32[154:pdopab]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Prestwood AK. Pursglove SR. Prevalence and distribution of Setaria yehi in southeastern white-tailed deer. J Am Vet Med Assoc. 1977;171:933–935. [PubMed] [Google Scholar]
- Santiago-Alarcon D. Havelka P. Schaefer HM. Segelbacher G. Bloodmeal analysis reveals avian Plasmodium infections and broad host preferences of Culicoides (Diptera: Ceratopogonidae) vectors. PLoS One. 2012;7:e31098. doi: 10.1371/journal.pone.0031098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach V. Pawlowski J. A new method of DNA extraction from the ethanol-fixed parasitic worms. Acta Parasitol. 1999;44:147–148. [Google Scholar]
- Townzen JS. Brower AV. Judd DD. Identification of mosquito bloodmeals using mitochondrial cytochrome oxidase subunit I and cytochrome b gene sequences. Med Vet Entomol. 2008:386–393. doi: 10.1111/j.1365-2915.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- Valkiunas G. Avian Malaria Parasites and Other Haemosporidia. Washington DC: CRC Press; 2005. [Google Scholar]
- Vaughan JA. Mehus JO. Brewer CM. Kvasager DK, et al. Theoretical potential of passerine filariasis to enhance the enzoonotic transmission of West Nile virus. J Med Entomol. 2012;6:1430–1441. doi: 10.1603/me12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann CJ. Anderson JR. Longhurst WM. Connolly G. Filarial worms of Columbian black-tailed deer in California. 1. Observations in the vertebrate host. J Wildl Dis. 1973;9:213–220. doi: 10.7589/0090-3558-9.3.213. [DOI] [PubMed] [Google Scholar]
- Wharton RH. Studies on filariasis in Malaya: The efficiency of Mansonia longipalpis as an experimental vector of Wuchereria malayi. Ann Trop Med Parasitol. 1957;51:422–439. doi: 10.1080/00034983.1957.11685832. [DOI] [PubMed] [Google Scholar]
- Work TM. Washino RK. Van Riper C. Comparative susceptibility of Culex tarsalis, Anopheles franciscanus and Culiseta inornata (Diptera: Culicidae) to Plasmodium relictum (Haemosporidia: Plasmodiiae) J Med Entomol. 1990;27:68–71. doi: 10.1093/jmedent/27.1.68. [DOI] [PubMed] [Google Scholar]
- Zielke E. Observations on the midgut penetration by Wuchereria bancrofti microfilariae in mosquitoes of varying susceptibilities to infection. Mosq Borne Dis Bull. 1992;9:60–63. [Google Scholar]