Abstract
Marine organisms in intertidal zones are subjected to periodical fluctuations and wave activities. To understand how microbes in intertidal biofilms adapt to the stresses, the microbial metagenomes of biofilms from intertidal and subtidal zones were compared. The genes responsible for resistance to metal ion and oxidative stresses were enriched in both 6-day and 12-day intertidal biofilms, including genes associated with secondary metabolism, inorganic ion transport and metabolism, signal transduction and extracellular polymeric substance metabolism. In addition, these genes were more enriched in 12-day than 6-day intertidal biofilms. We hypothesize that a complex signaling network is used for stress tolerance and propose a model illustrating the relationships between these functions and environmental metal ion concentrations and oxidative stresses. These findings show that bacteria use diverse mechanisms to adapt to intertidal zones and indicate that the community structures of intertidal biofilms are modulated by metal ion and oxidative stresses.
A biofilm is a complex aggregate of microorganisms, such as bacteria, diatoms, fungi, and protozoa, in extracellular polymeric substances (EPS) and can develop on almost all the surface1. The microbial biofilm is regarded as a preferred mode of life and is now viewed as a common adaptation, perhaps even a life stage, of most microbes in natural systems1. Microbial communities of natural biofilms in the marine environment change in response to various environmental factors, including salinity, temperature2, dissolved oxygen3, and habitat4.
In intertidal areas, which are considered as interfaces of the ocean, atmosphere, and terrestrial environments, the biofilm communities are subjected to fluctuations in metal ion concentrations, temperature, desiccation, UV irradiation, and wave activities5. The organisms in intertidal zones can spend half of their lives in extreme, arid conditions during emersion and half of their lives in stable, benign seawater. In contrast, subtidal systems are more stable because they are submerged most of the time and only exposed briefly during the extremely low tides of full and new moon events. In contrast to intertidal zones, this zone provides a habitat for a large diversity of plants and animals6,7,8.
Physiological adaptations are crucial for bacteria and higher organisms to adapt to changing environmental conditions and are considered to play a major role in determining which species will be the “winner” in response to global climate change. In highly variable environments such as intertidal zones, some marine organisms can utilize metabolic depression, anaerobic energy production, and stress-protective mechanisms to guard against environmental stresses4,9,10. Variations in biofilm components that develop at different vertical sites along the shore were detected11,12,13,4, which indicates that the structure of microbial communities in biofilm is shaped by environmental factors.
In our previous studies, we examined bacterial community succession (from 3 to 20 days) of subtidal biofilms, and biofilms of ≤9 days old demonstrated clearly different ribotypes from those of ≥12 days old14. However, the effects of tide levels on biofilms and the mechanisms employed by microbes in biofilms to cope with variations in the intertidal environment require further investigation. Metagenomic analysis is an ideal technique for addressing the issues above mentioned because it allows us to explore significant differences in metabolic potential in different environments. In the present study, we used a metagenomic analysis to identify differences in the microbial community composition and functions of 4 types of biofilms developed at 2 tidal levels (intertidal and subtidal) and for 2 different durations (6 and 12 days, defined as two different biofilm development stages by Chung et al., 201014). We discovered that the genes responsible for tolerance to oxidative and metal ion stresses were enriched in intertidal biofilms. In addition, the genes responsible for the biosynthesis of EPS, which is important in environmental stress responses, were also enriched in intertidal biofilms. Furthermore, we characterized the taxonomic and functional differences between intertidal and subtidal biofilms and provided a suitable model to elucidate how microbes in intertidal biofilms cope with stresses in such harsh environments.
Results
Comparison of clusters of orthologous group (COG) categories between intertidal and subtidal biofilm metagenomes
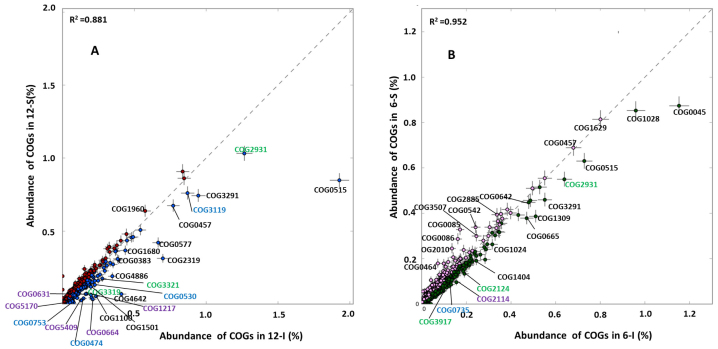
Pyrosequencing data for the microbial metagenomes of 4 biofilm samples are summarized in Table 1. The total number of sequences ranged from 451,100 for the 12-I biofilm (hereafter referred to as 6-I and 12-I for the 6- and 12-day intertidal biofilms, and 6-S and 12-S for the 6- and 12-day subtidal biofilms, respectively), and up to 534,336 for the 6-S biofilm, with an average length of ~500 bp. Functional analysis of the biofilm metagenomes by assignment of metagenomic sequences to the COG database was performed. To identify functions with significant differences among different biofilms, a quantitative comparison of 4,223 individual COGs among these 4 biofilm samples was generated by best classification. Parallel comparison of I-12 versus S-12 and I-6 versus S-6 by STAMP15 (Fisher's G-test, P < 0.01) revealed that nearly all of the significantly changed individual COGs belonging to the categories of inorganic ion transport and metabolism, secondary metabolite biosynthesis, transport and catabolism and signal transduction mechanisms were enriched in intertidal biofilms (Fig. 1). This result was consistent with the significantly changed individual COGs indicated by the binomial, hypergeometric and G-test (P < 0.01). Regarding functional diversity, all of the metagenomes displayed a similar Shannon index (6.26, 6.45, 6.47 and 6.42 for 12-I, 12-S, 6-I and 6-S, respectively).
Table 1. A summary of the metagenomes of 6-day and 12-day biofilms from intertidal and subtidal locations in Hong Kong.
| Sample ID | 12-I | 12-S | 6-I | 6-S |
|---|---|---|---|---|
| No. of reads | 451,100 | 465,162 | 507,489 | 534,336 |
| Average length (bp) | 532 | 487 | 484 | 512 |
| Reads removed after quality control | 107,512 | 169,545 | 144,995 | 133,915 |
| Reads assigned to individual COGs | 173,676 | 164,814 | 239,876 | 221,765 |
| Reads assigned to KEGG EC numbers | 58,301 | 65,888 | 71,900 | 56,932 |
| Reads assigned to CAZy enzymes | 22,314 | 26,991 | 29,784 | 27,989 |
6-I and 12-I indicate 6- and 12-day intertidal biofilms, and 6-S and 12-S indicate 6- and 12-day subtidal biofilms, respectively.
Figure 1. Individual COGs for the intertidal and subtidal biofilm metagenomes.
Parallel comparison of I-12 vs. S-12 and I-6 vs. I-S by STAMP (Fisher's G-test, P < 0.01) revealed that COGs belonging to categories of inorganic ion transport and metabolism (indicated in blue),secondary metabolite biosynthesis, transport and catabolism (indicated in green) and signal transduction mechanisms (indicated in purple) were over-represented in intertidal biofilms. A complete list of significantly changed COGs and annotations for these three categories is provided in Figure S1, Figure S2 and Figure S3.
Metal ion transporter and secondary metabolism genes
As mentioned above, genes involved in inorganic ion transport and secondary metabolism were over-represented in the intertidal metagenomes (Fig. 1). Six genes involved in inorganic ion transport and metabolism were enriched in intertidal biofilm metagenomes compared with their subtidal counterparts, including cation transport ATPases, antiporters, pumps and catalase (Fig. S1). These genes are responsible for transporting a variety of ions: Ca2+, Zn2+, Fe2+, Na+, Co+ and Cd+. Cation transport ATPase is a large superfamily that can utilize the energy of ATP to transport ions against electrochemical gradients and to fuel many antiporters and cation pumps16,17. The enrichment for these transporters suggested that the microbes in intertidal biofilms adopted an energy-consuming mechanism to maintain ion homeostasis.
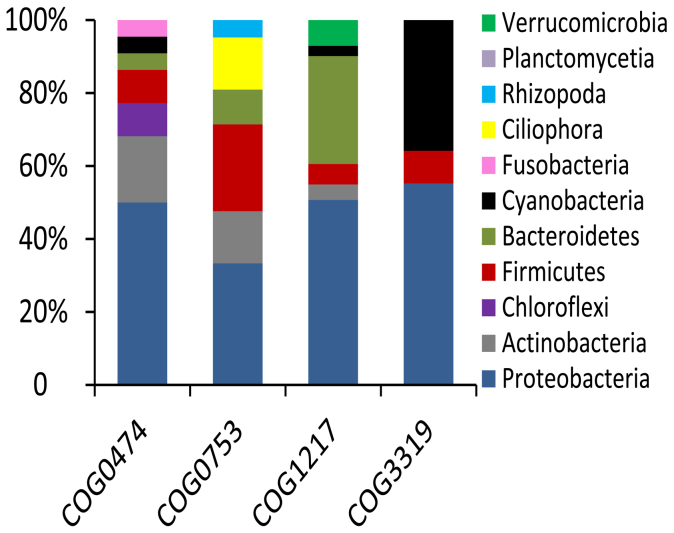
Catalase is an enzyme that uses hydrogen peroxide as a substrate to produce water and oxygen18. The abundance of catalase genes in 12-I biofilm was 8-fold higher than that in the 12-S biofilm (Fig. S1). Two polyketide synthase (PKS) COGs (COG3319 and COG3321) were enriched in the intertidal biofilm metagenomes. Non-ribosomal peptide synthetases (NRPSs) and PKSs are necessary to produce a variety of secondary metabolites, such as the siderophores involved in iron acquisition, to cope with intracellular oxidative stress19. The 6-I biofilm showed enrichment for anthracene-degrading 2-hydroxychromene-2-carboxylate isomerase20, which can induce oxidative stress in eukaryotes21,22. All of the above-mentioned genes were related to the oxidative stress response. Figure 2 shows the taxonomic assignment of catalase (COG0753), type I polyketide synthases (COG3319), cation transport ATPase (COG0474), and a predicted membrane GTPase involved in the stress response (COG1217) from the I-12 biofilm metagenome. Most of the reads for genes encoding catalase and cation transport ATPase belonged to the Proteobacteria, while type I polyketide synthases belonged to the Proteobacteria, Cyanobacteria and Firmicutes. These results suggested that Proteobacteria provided the largest contribution to the functional gene pool enriched in intertidal biofilms.
Figure 2. Taxonomy of functional genes in the 12-I biofilm metagenome.
Reads of catalase (COG0753), cation transport ATPase (COG0474), thioesterase domains of type I polyketide synthases or non-ribosomal peptide synthetase (COG3319) and GTPase involved in the stress response (COG1217) were extracted from the 12-I biofilm metagenome and BLASTed against the NCBI-nr database and assigned to different genera using MEGAN.
Signal transduction pathways
In addition to inorganic ion transport and secondary metabolism, another COG category enriched in intertidal biofilms was signal transduction (Fig. 1 and Fig. S3). Comparison of the 12-I biofilm with the 12-S biofilm metagenome revealed 18 signaling transduction genes with significantly differences in abundance; all of these genes were enriched in the 12-I biofilm and included several serine/threonine protein phosphatase (PSP), a GTPase involved in the stress response and cAMP-binding proteins23, all of which regulate the stress response in bacteria. Similarly, 5 genes had a significantly higher abundance in the 6-I biofilm compared with the 6-S biofilm, including the universal stress protein UspA24 and related nucleotide-binding proteins, adenylate cyclase and three histidine kinases. The enrichment for these signal transduction genes in intertidal biofilms suggested that they might play a protective role against environmental stresses. The microbes with GTPase genes (COG1217) involved in the stress response in the 12-I biofilm are illustrated in Figure 2, indicating most of these microbes with these genes were the Proteobacteria and Bacteroidetes.
EPS biosynthesis pathways
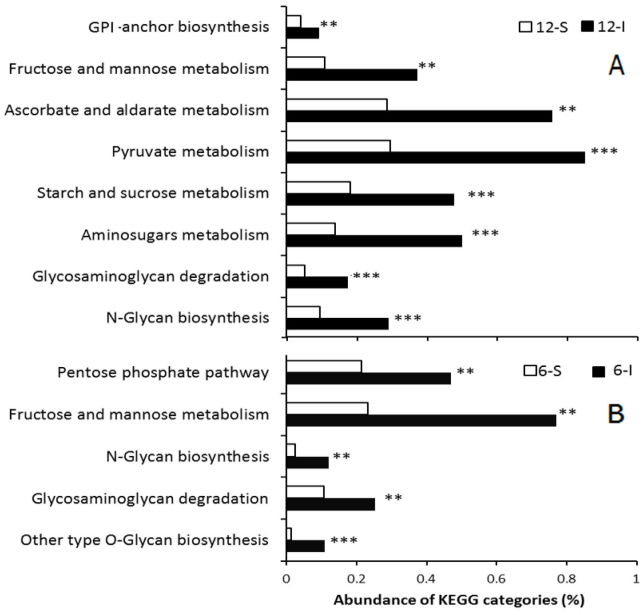
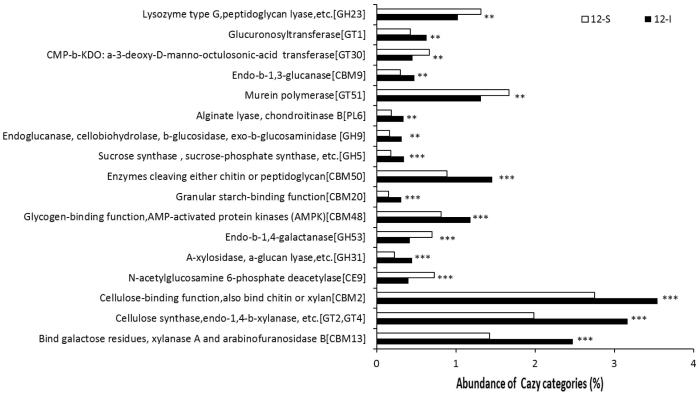
Biofilm's EPS, which contains several components including monosaccharides and polysaccharides, plays an important role in environmental stress responses, including the protective response against oxidative stress25,26 and ion chelation5,27. We hypothesized that enhancement of EPS synthesis might be a strategy employed by intertidal biofilms to cope with environmental stresses. Because the results obtained for the COG categories revealed a lack of congruency among intertidal and subtidal biofilms, metagenomic sequences were assigned to the KEGG database, which is particularly useful for comparison of metabolic pathways, with the goal of identifying EPS components associated with metabolic pathways. Figure 3 illustrates the pathways involved in carbohydrate metabolism and glycan biosynthesis and metabolism that were significantly changed between intertidal and subtidal biofilms. Compared with the 12-S biofilm, all of the sugar biosynthesis pathways were enriched in the 12-I biofilm e.g., metabolism of amino sugars, starch and sucrose, fructose and mannose and biosynthesis of N-glycan (Fig. 3A), which are common in bacterial EPS28. Similarly, compared to the 6-S biofilm, biosynthesis pathways associated with fructose and mannose metabolism as well as O-glycan biosynthesis were enriched in the 6-I biofilm (Fig. 3B). In addition, a BLAST search against the Carbohydrate-Active Enzymes (CAZy) specialist database confirmed the results and revealed detailed activities of carbon metabolism proteins. Comparison of the 12-I and 12-S biofilm metagenomes using STAMP (Fisher's G-test, P < 0.01) showed that a significant difference in all of the CAZy protein families (Fig. 4). In particular, the 12-I biofilm was enriched for polysaccharide biosynthesis gene families, including cellulose synthase, chitin synthase, xylanases and glucanase. These results suggested that enhancement of EPS might be a strategy utilized by microbes in intertidal biofilms to cope with environmental stresses.
Figure 3. KEGG pathways associated with carbohydrate metabolism and glycan biosynthesis and metabolism that were significantly different between intertidal and subtidal biofilm metagenomes.
Comparison of the pathways was based on the abundance of corresponding EC numbers. **P < 0.01 and ***P < 0.001. A.12-I biofilm vs. 12-S biofilm. B. 6-I biofilm vs. 6-S biofilm.
Figure 4. CAZy families with significant differences between the 12-I and 12-S biofilms (Fisher's G-test, P < 0.01).
**P < 0.01 and ***P < 0.001.
Comparison between the 12-I and 6-I biofilm metagenomes
Because genes that cope with metal ion and oxidative stress were enriched in intertidal biofilms, we tested whether the enrichment intensified during biofilm development. The 12-I and 6-I biofilm metagenomes were compared. As indicated in Figure S4, most of the COGs mentioned above displayed a significantly increased abundance in the 12-I biofilm in comparison with the 6-I biofilm, as indicated by the comparison of catalase, polyketide synthases, cation transporters and stress regulators. Consistently, a Cu/Zn superoxide dismutase (Cu/Zn-SOD, COG2032) was present only in the 12-I biofilm metagenome. This enzyme is important for protection against oxidative stress29. Moreover, anti-sigma factor (COG3806), which is a well-known regulator of the stress response30, was also significantly enriched in the 12-I biofilm. These results indicate a community shift driven by increasing stress during the development of intertidal biofilm development.
Discussion
In this study, a metagenomic analysis was conducted to examine functional differences between intertidal and subtidal biofilms that developed for 6 and 12 days, respectively. The functional diversity of the intertidal and subtidal biofilms was similar, whereas the spectra of metabolic functions differed between them. The COG categories of secondary metabolism, inorganic ion transport and metabolism and signaling transduction were enriched in intertidal biofilms. The higher frequency of cation transport and pump-related genes, which regulate metal ion homeostasis, might reflect the elevated levels of metal ion stress faced by intertidal biofilms, which are more persistent than that of subtidal zones and subjected to salt accumulation over time. The enrichment of catalase in the 12-I biofilm metagenome indicated that intertidal biofilms might be exposed to oxidative stress. In addition, we found that the enzyme commission (EC) number of genes encoding superoxide dismutase (SOD, EC1.15.1.1) was more abundant in the 12-I biofilm metagenome compared with the 12-S metagenome (0.08% vs. 0.05%, P < 0.05). Reactive oxygen species are commonly produced in the ocean, and oxidative stress is an important component of the stress response in marine organisms exposed to a variety of insults such as thermal stress, UV radiation, and pollution31. Thus, apart from catalase, other genes that contribute to resistance to oxidative stress, such as special secondary metabolism genes, were also enriched in both 12-day and 6-day intertidal biofilms. The production of secondary metabolites is often associated with environmental stresses, including metal ion and oxidative stresses32,33,34. For example, the polyketides are a large class of secondary metabolites that are present in bacteria, fungi and plants and contribute to virulence and responses to oxidative stress19. Following Cyanobacterium anabaena exposure to oxidative stress, the gene cluster encoding polyketide synthase is activated and siderophore release is increased to acquire Fe3+ and enhance electron transport19. The observed enrichment for these genes supported the hypothesis that oxidative stress plays an important role in shaping the functional structure and adaptability of intertidal biofilms. In fact, metal ion homeostasis and oxidative stress are considered to be coupled. For instance, iron, in its reduced form, potentiates oxygen toxicity by converting, via the Fenton reaction, the less reactive hydrogen peroxide to the more reactive oxygen species, hydroxyl radical and ferryl iron (Fe2+ + H2O2 + H+ → Fe3+ + O2- + H2O)35. Furthermore, toxic metals such as lead, cadmium and mercury, among others, deplete the cells' major antioxidants and increase the production of reactive oxygen species (ROS) such as hydroxyl radical (H[Odot]), superoxide radical (O2-) and hydrogen peroxide (H2O2)36. The simultaneous enrichment of ion transport genes and oxidative response genes also suggests a correlation between responses to these two types of stresses.
A BLAST analysis against the KEGG database revealed that genes associated with sugar metabolism and biosynthesis (typically found in bacterial EPS) were enriched in intertidal biofilm metagenomes. This finding was further supported by results of the CAZy protein activity comparison, demonstrating enrichment of polysaccharide metabolism activity in the 12-I biofilm. For example, in enzymology, cellulose synthase [GT2] is an enzyme that catalyzes the chemical reaction UDP-glucose + (1,4-beta-D-glucosyl)n → UDP + (1,4-beta-D-glucosyl)n+137. The presence of both polysaccharide biosynthesis and degradation enzymes in the same biofilm might be attributed to the physiological heterogeneity in biofilms. The enrichment for these genes indicated that enhancement of EPS might be another strategy adopted by microbes to resist the harsh intertidal environment. Similar results were obtained in a previous study in which exposure of Pseudomonas fluorescens biofilm to copper-ion-induced oxidative stress increased biofilm-specific exopolysaccharide-related metabolism38. In addition, taxonomic assignment of the genes enriched in intertidal biofilm metagenomes (Fig. 2) indicated that stress resistance in intertidal biofilms could be partly attributed to Bacteroidetes, which specialize in degrading complex organic matter such as polysaccharides39 and their presence also supports the hypothesis that intertidal biofilms contain abundant EPS.
Intertidal biofilms had enriched signal transduction genes. PSP is a form of phosphoprotein phosphatase that acts upon serine/threonine residues and serves as an “on-off” signal transduction switch that is triggered by environmental stresses40. Members of the GTPase superfamily are extremely important in regulating membrane signaling pathways in all cells41 and have been reported to regulate the formation of bacterial biofilms42. Alternatively, cAMP-binding proteins can also mediate biofilm formation through environmental stress responses43,44. It has been reported that in Escherichia coli, an increase in cellular cAMP levels results in overexpression of catE genes43, which encode catalase. The UspA domain has been detected in more than 1,000 different genes and is often fused with other domains, as exemplified in kdpD, the sensor kinase that regulates high-affinity K+ transport45. The UspA gene family, adenylate cyclase and histidine kinase can be induced by oxidative stress. These enzymes activate downstream signaling and regulate the stress response. Therefore, in the present study, one of the underlying mechanisms of stress resistance might be the transcriptional control of genes involved in oxidative and metal ion stresses in intertidal biofilms by these signal transduction regulators.
Laboratory studies have documented extensively that the process of biofilm development is coupled with improved stress-coping ability. The results from the present study demonstrated that enrichment of genes responsible for resistance to metal ion and oxidative stresses in the 12-I biofilm in comparison with the 6-I biofilm, indicating that in response to these stresses, species sorting and a shift in community structure occurred during intertidal biofilm development. In a previous study, micro-scale differentiation was observed between the base and streamers of stream biofilms where the Proteobacteria and Bacteroidetes were strong competitors in terms of their effective responses to the environmental dynamics46. In the present study, although we did not perform architectural microscopy to observe differentiation during biofilm development, our results indicated the presence of a functional shift during biofilm development in response to environmental stresses and that Proteobacteria and Bacteroidetes also contribute to the stress resistance in intertidal biofilms.
In summary, findings from the present study elucidated the stresses that shape the community structures of intertidal biofilms compared with subtidal biofilms. We hypothesize that intertidal biofilm signaling regulators are activated by metal ion and oxidative stresses present in this highly variable environment. Subsequently, the expression of EPS, secondary metabolites and ion transporters may occur (Fig. 5). Thus, the community is shaped by the two stresses, and environmental selection causes differences between intertidal and subtidal biofilm structures, as supported by the enrichment of genes responsible for coping with the two stresses in the 12-I biofilm compared with the 6-I biofilm. The pursuit of nutrients may be considered to play a more important role in shaping community structure than stresses imposed by the changing environment, as some organisms in intertidal zones suffer from nutrient limitation47. However, genes encoding sugar metabolism enzymes were more strongly represented by reads in the intertidal biofilms, and energy-dependent ion transporters and pumps were highly abundant in intertidal biofilms, which indicated that energy-saving mechanisms were not used and nutrient deficiencies did not occur in intertidal biofilms. Thus, the enrichment for these sugar metabolism genes should be considered as an adaptive mechanism to environmental stress. In conclusion, adaptation of the intertidal biofilm community is produced by metal ion and oxidative stresses. There is evidence that secondary metabolites and bacterial cell-to-cell signaling mediate interactions between bacteria and eukaryotes. Therefore, in addition to stress tolerance, the enrichment for secondary metabolism and signal transduction genes might also explain why intertidal microbial biofilms are more attractive to eukaryotes for settlement. However, direct linkage between these phenomena is still lacking. Finally, because our conclusions are based on analysis of metagenomes from only one intertidal site, additional data from a broader range of sites and during different seasons are required to evaluate whether the present results occur consistently between intertidal and subtidal biofilms.
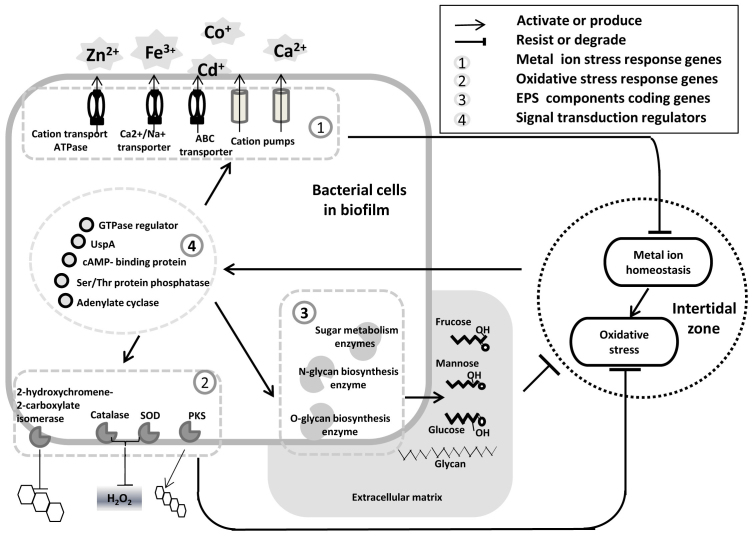
Figure 5. Schematic model of microbial reactions triggered by oxidative and metal ion stresses in intertidal biofilms.
Stress signals activate signal transduction pathways within bacterial cells, resulting in transporter activation, secondary metabolism and EPS production.
Methods
Development and collection of biofilms
Multispecies biofilms were developed on polystyrene Petri dishes in Port Shelter Bay, Hong Kong (22°19′43″N, 114°16′24″E) at 2 different tidal levels–intertidal and subtidal–in July 2012. Petri dishes (60-mm diameter, Falcon #3002, Becton Dickinson & Co. USA) attached to large plastic panels were covered with a 125-μm mesh to prevent settlement of marine invertebrates. As mentioned in the results part, 6 and 12 days were defined as two different biofilm development stages. Thus, the panels were submerged for 6 or 12 days, resulting in the collection of 4 treatments at 2 tidal levels. After submersion for 6 or 12 days, the Petri dishes were retrieved and transported back to the laboratory in a cooler with in situ seawater. The Petri dishes were washed twice using autoclaved 0.22 μm-filtered seawater (AFSW) to remove loosely attached particles and bacteria and then used for DNA extraction.
DNA extraction and pyrosequencing
For each tidal level and duration, biofilms developed in 40 Petri dishes were harvested using sterile cell scrapers and suspended in Tris-HCl buffer. Before nucleic acid extraction, microbial cells were pelleted by centrifugation at 4000 rpm for 10 min and then lysed with lysozyme, proteinase K and 10% SDS. Total nucleic acid was extracted and purified using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The quality and quantity of DNA were checked by agarose gel electrophoresis and with a Nanodrop device (ND-1000 spectrophotometer, DiaMed China Limited, Hong Kong) at 260 nm.
To characterize the microbial community in the biofilm samples and to determine major pathways in the different biofilms, ~3 μg of DNA was extracted from each of the 4 biofilm samples and pyrosequenced individually using the ROCHE 454 FLX Titanium platform.
Metagenomic analysis
Metagenomic analysis was performed as previously described48,49. Quality control for the pyrosequencing reads was conducted using the next generation sequencing (NGS) QC toolkit50. Reads <100 bp were removed, and then AmbiguityFiltering was performed to remove sequences containing 5 or more repetitive Ns before applying the CDHIT-454 to identify and discard duplicates and nearly identical duplicates in the pyrosequencing reads. The longest representatives among the duplicates were retained for further analysis.
The 16S rRNA gene fragments were extracted based on the prediction using Meta-RNA. Using the program “Filtering by length”, 16S rRNA fragments of <150 bp were removed. Taxonomic classification was conducted using the online RDP Classifier version 2.5 (http://rdp.cme.msu.edu) with a confidence threshold of 50% for classification at the KEGG (version 67.0)51, CAZy52 and COG (version 9.05)53 prokaryote databases on local servers, using an E-value of <10−5. The results of the BLAST analysis are summarized. The COG numbers, KEGG frequencies and EC numbers were annotated using an in-house program. Statistical analyses were conducted separately using STAMP and ShotgunfunctionalizeR54 in the R project. In the STAMP analysis, the Fisher's G-test and P < 0.01 were used to identify significant differences in gene abundance between 12-I vs. 12-S and 6-I vs. 6-S, respectively. In the ShotgunfunctionalizeR analysis, binomial, hypergeometric and G-test and P < 0.01 were used. Shannon diversities were calculated using ShotgunfunctionalizeR.
For taxonomic assignment of the COG genes, the reads for COG0753, COG0474, COG1217 and COG3319 were extracted from the 12-I metagenome data using an in-house program. Similar to the BLAST analysis against the KEGG and COG databases, BlASTx was performed against the NCBI nr database using an E-value of <10−5 to reveal the top 10 hits. Subsequently, the taxonomies were summarized using MEGAN55 and genera with an identification score >80 were retained.
Author Contributions
W.P.Z., Y.W. and P.Y.Q. wrote the main manuscript. O.O.L., Y.X., R.M.T., Z.M.G., Y.X.L. and P.Y.Q. designed and performed the experiments. W.P.Z., Y.W. and H.L.C. analyzed the data. W.P.Z. and L.Y. prepared samples. All authors discussed and reviewed the manuscript.
Supplementary Material
SUPPLEMENTARY INFO
Acknowledgments
This study was supported by the Nationa Basic Research Program of China (973 program, 2012CB417304), the Development Program of China (863 program, 2012AA092103), The Sanya Institute of Deep-Sea Science and Engineering (SIDSSE201206), General Research Fund (661611) of HKSAR Government and Global Collaborative Research Award from King Abdullah University of Science and Technology (SA-C0040/UK C0016).
References
- Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R. & Lappin-Scott H. M. Microbial biofilms. Annu Rev Microbiol 49, 711 (1995). [DOI] [PubMed] [Google Scholar]
- Lau S. C., Thiyagarajan V., Cheung S. C. & Qian P. Y. Roles of bacterial community in biofilms as mediator for larval settlement of marine invertebrates. Aquat Microb Ecol 38, 41–51 (2005). [Google Scholar]
- Nocker A., Lepo J. E., Martin L. L. & Snyder R. A. Response of estuarine biofilm microbial community development to changes in dissolved oxygen and nutrient concentrations. Microb Ecol 54, 532–542 (2007). [DOI] [PubMed] [Google Scholar]
- Qian P. Y., Thiyagarajan V., Lau S. C. K. & Cheung S. C. K. Relationship between bacterial community profile in biofilm and attachment of the acorn barnacle Balanus amphitrite. Aquat Microb Ecol 33, 225–237 (2003). [Google Scholar]
- Decho A. W. Microbial biofilms in intertidal systems: an overview. Contin Shelf Res 20, 1257–1273 (2000). [Google Scholar]
- Ayling A. M. The Role of Biological Disturbance in Temperate Subtidal Encrusting Communities. Ecology 62, 830–847 (1981). [Google Scholar]
- Candela J., Beardsley R. C. & Limeburner R. Separation of tidal and subtidal currents in ship-mounted acoustic Doppler current profiler observations. J Geophys Res 97, 769 (1992). [Google Scholar]
- Tokeshi M. in Aquatic Ecology: Scale, Pattern and Processes (eds Giller P. S., Hildrew A. G., & Raffaelli D. G.) 63–92 (Blackwell, Oxford, 1994). [Google Scholar]
- Somero G. N. The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J Exp Biol 213, 912–920 (2010). [DOI] [PubMed] [Google Scholar]
- Marshall D. J. & McQuaid C. D. Warming reduces metabolic rate in marine snails: adaptation to fluctuating high temperatures challenges the metabolic theory of ecology. P Roy Soc B-Biol Sci 278, 281–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keough M. & Raimondi P. Responses of settling invertebrate larvae to bioorganic films: effects of large-scale variation in films. J Exp Mar Biol Ecol 207, 59–78 (1996). [Google Scholar]
- Thomason J. C., Hills J. M., Clare A. S., Neville A. & Richardson M. Hydrodynamic consequences of barnacle colonization. Hydrobiologia 375, 191–201 (1998). [Google Scholar]
- Miron G., Boudreau B. & Bourget E. Intertidal barnacle distribution: a case study using multiple working hypotheses. Mar Ecol Prog Ser 189, 205–219 (1999). [Google Scholar]
- Chung H. C. et al. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J 4, 817–828 (2010). [DOI] [PubMed] [Google Scholar]
- Parks D. H. & Beiko R. G. Identifying biologically relevant differences between metagenomic communities. Bioinform 26, 715–721 (2010). [DOI] [PubMed] [Google Scholar]
- Silver S. Bacterial resistances to toxic metal ions-a review. Gene 179, 9–19 (1996). [DOI] [PubMed] [Google Scholar]
- Green N. M. ATP-driven cation pumps: alignment of sequences. Biochem Soc Trans 17, 970–972 (1989). [DOI] [PubMed] [Google Scholar]
- Fita I. & Rossmann M. G. The active center of catalase. J Mol Biol 185, 21–37 (1985). [DOI] [PubMed] [Google Scholar]
- Jeanjean R. et al. A large gene cluster encoding peptide synthetases and polyketide synthases is involved in production of siderophores and oxidative stress response in the cyanobacterium Anabaena sp. strain PCC 7120. Environ Microbiol 10, 2574–2585 (2008). [DOI] [PubMed] [Google Scholar]
- Kim E. et al. Evidence for the role of 2-hydroxychromene-2-carboxylate isomerase in the degradation of anthracene by Sphingomonas yanoikuyae B1. FEMS Microbio Let 153, 479–484 (1997). [DOI] [PubMed] [Google Scholar]
- Debiane D. et al. In vitro evaluation of the oxidative stress and genotoxic potentials of anthracene on mycorrhizal chicory roots. Environ Exp Bot 64, 120–127 (2008). [Google Scholar]
- Sander C. S., Chang H., Hamm F., Elsner P. & Thiele J. J. Role of oxidative stress and the antioxidant network in cutaneous carcinogenesis. Int J Dermatol 43, 326–335 (2004). [DOI] [PubMed] [Google Scholar]
- Barth E. et al. Interplay of cellular cAMP levels, σS activity and oxidative stress resistance in Escherichia coli. Microbiology 155, 1680–1689 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele D. A. Universal stress proteins in Escherichia coli. J Bacteriol 187, 6253–6254 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lee S. M. & Mao Y. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157: H7 to osmotic and oxidative stress. Int J Food Microbiol 93, 281–286 (2004). [DOI] [PubMed] [Google Scholar]
- Keith L. M. W. & Bender C. L. AlgT (σ22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J Bacteriol 181, 7176–7184 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech I. B. & Cheung C. S. Interactions of exopolymers produced by sulphate-reducing bacteria with metal ions. Int Biodeterior Biodegr 35, 59–72 (1995). [Google Scholar]
- Kenne L. & Lindberg B. Bacterial polysaccharides. The polysaccharides 2, 287–363 (1983). [Google Scholar]
- Bordo D., Djinovic K. & Bolognesi M. Conserved patterns in the Cu, Zn superoxide dismutase family. J Mol Biol 238, 366–386 (1994). [DOI] [PubMed] [Google Scholar]
- Hughes K. T. & Mathee K. The anti-sigma factors. Annu Rev Microbiol 52, 231–286 (1998). [DOI] [PubMed] [Google Scholar]
- Lesser M. P. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol 68, 253–278 (2006). [DOI] [PubMed] [Google Scholar]
- Zobayed S. M. A., Afreen F. & Kozai T. Temperature stress can alter the photosynthetic efficiency and secondary metabolite concentrations in St. John's wort. Plant Physiol Biochem 43, 977–984 (2005). [DOI] [PubMed] [Google Scholar]
- Heeb S., Valverde C., Gigot-Bonnefoy C. & Haas D. Role of the stress sigma factor RpoS in GacA/RsmA-controlled secondary metabolism and resistance to oxidative stress in Pseudomonas fluorescens. FEMS Microbiol Let 243, 251–258 (2005). [DOI] [PubMed] [Google Scholar]
- Hanlon A. Stress and release: chemical modulation of secondary metabolite production in Aspergillus sp. Massachusetts Institute of Technology. (2006). [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch biochem biophys 373, 1–6 (2000). [DOI] [PubMed] [Google Scholar]
- Nuran E., Gurer-Orhan H. & Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem 6, 529–539 (2001). [DOI] [PubMed] [Google Scholar]
- Colvin J. R. Synthesis of cellulose in ethanol extracts of Acetobacter xylinum. Nature 183, 1135–1136 (1959). [DOI] [PubMed] [Google Scholar]
- Booth S. C. et al. Differences in metabolism between the biofilm and planktonic response to metal stress. J Proteome Res 10, 3190–3199 (2011). [DOI] [PubMed] [Google Scholar]
- Thomas F., Hehemann J. H., Rebuffet E., Czjzek M. & Michel G. Environmental and gut Bacteroidetes: the food connection. Front Microbiol 2, 93 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Kang C. M., Brody M. S. & Price C. W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Gene Dev 10, 2265–2275 (1996). [DOI] [PubMed] [Google Scholar]
- March P. E. Membran-associated GTPases in bacteria. Mol Microbiol 6, 1253–1257 (1992). [DOI] [PubMed] [Google Scholar]
- Chun M. J., Park K. J. & Ohk S. H. Putative down-stream signaling molecule of GTPase in Porphyromonas gingivalis. Appl Biochem Microbiol 48, 350–354 (2012). [PubMed] [Google Scholar]
- Zhang X. S., García-Contreras R. & Wood T. K. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol 189, 3051–3062 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C. M. et al. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor. PLoS pathog 5, e1000303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walderhaug M. O. et al. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J Bacteriol 174, 2152–2159 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer K., Hödl I., Singer G. & Battin T. J. Architectural differentiation reflects bacterial community structure in stream biofilms. ISME J 3, 1318–1324 (2009). [DOI] [PubMed] [Google Scholar]
- Davison I. R. & Pearson G. A. Stress tolerance in intertidal seaweeds. J Phycol 32, 196–211 (1996). [Google Scholar]
- Wang Y. et al. Autotrophic microbe metagenomes and metabolic pathways differentiate adjacent Red Sea brine pools. Sci Rep. 3, 01748; 10.1038/srep01748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lee O. O., Yang J. K., Li T. G. & Qian P. Y. Artifactual pyrosequencing reads in multiple-displacement-amplified sediment metagenomes from the Red Sea. Peer J 1, 69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. K. & Jain M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 7, e30619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37, 233–238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L. et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29, 22–28 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson E., Hugenholtz P. & Dalevi D. ShotgunFunctionalizeR: An R-package for functional comparisons of metagenomes. Bioinform 25, 2737–2738 (2009). [DOI] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J. & Schuster S. C. MEGAN analysis of metagenomic data. Genome Res 17, 377–386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY INFO