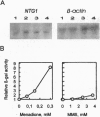
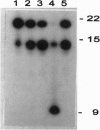
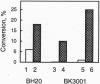
Abstract
One gene locus on chromosome I in Saccharomyces cerevisiae encodes a protein (YAB5_YEAST; accession no. P31378) with local sequence similarity to the DNA repair glycosylase endonuclease III from Escherichia coli. We have analyzed the function of this gene, now assigned NTG1 (endonuclease three-like glycosylase 1), by cloning, mutant analysis, and gene expression in E. coli. Targeted gene disruption of NTG1 produces a mutant that is sensitive to H2O2 and menadione, indicating that NTG1 is required for repair of oxidative DNA damage in vivo. Northern blot analysis and expression studies of a NTG1-lacZ gene fusion showed that NTG1 is induced by cell exposure to different DNA damaging agents, particularly menadione, and hence belongs to the DNA damage-inducible regulon in S. cerevisiae. When expressed in E. coli, the NTG1 gene product cleaves plasmid DNA damaged by osmium tetroxide, thus, indicating specificity for thymine glycols in DNA similarly as is the case for EndoIII. However, NTG1 also releases formamidopyrimidines from DNA with high efficiency and, hence, represents a glycosylase with a novel range of substrate recognition. Sequences similar to NTG1 from other eukaryotes, including Caenorhabditis elegans, Schizosaccharomyces pombe, and mammals, have recently been entered in the GenBank suggesting the universal presence of NTG1-like genes in higher organisms. S. cerevisiae NTG1 does not have the [4Fe-4S] cluster DNA binding domain characteristic of the other members of this family.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barton A. B., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the genes in the FUN38-MAK16-SPO7 region. J Bacteriol. 1994 Apr;176(7):1872–1880. doi: 10.1128/jb.176.7.1872-1880.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T., Tano K., Kasai H., Ohtsuka E., Nishimura S. Evidence for two DNA repair enzymes for 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in human cells. J Biol Chem. 1993 Sep 15;268(26):19416–19421. [PubMed] [Google Scholar]
- Boiteux S., Belleney J., Roques B. P., Laval J. Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res. 1984 Jul 11;12(13):5429–5439. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Gajewski E., Laval J., Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992 Jan 14;31(1):106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Huisman O. Isolation of a formamidopyrimidine-DNA glycosylase (fpg) mutant of Escherichia coli K12. Mol Gen Genet. 1989 Jan;215(2):300–305. doi: 10.1007/BF00339732. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem. 1992 Feb 14;201(1):134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- Cunningham R. P., Weiss B. Endonuclease III (nth) mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M., Laval J., Boiteux S. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry. 1993 Nov 16;32(45):12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- Doetsch P. W., Henner W. D., Cunningham R. P., Toney J. H., Helland D. E. A highly conserved endonuclease activity present in Escherichia coli, bovine, and human cells recognizes oxidative DNA damage at sites of pyrimidines. Mol Cell Biol. 1987 Jan;7(1):26–32. doi: 10.1128/mcb.7.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossett J., Lee K., Cunningham R. P., Doetsch P. W. Yeast redoxyendonuclease, a DNA repair enzyme similar to Escherichia coli endonuclease III. Biochemistry. 1988 Apr 5;27(7):2629–2634. doi: 10.1021/bi00407a054. [DOI] [PubMed] [Google Scholar]
- Hill J., Donald K. A., Griffiths D. E., Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991 Oct 25;19(20):5791–5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. F., McRee D. E., Fisher C. L., O'Handley S. F., Cunningham R. P., Tainer J. A. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992 Oct 16;258(5081):434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- Leadon S. A., Barbee S. L., Dunn A. B. The yeast RAD2, but not RAD1, gene is involved in the transcription-coupled repair of thymine glycols. Mutat Res. 1995 Nov;337(3):169–178. doi: 10.1016/0921-8777(95)00021-b. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993 Apr 22;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Melamede R. J., Hatahet Z., Kow Y. W., Ide H., Wallace S. S. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994 Feb 8;33(5):1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Cruz C., Grollman A. P., Miller J. H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Pham L., Nghiem Y., Cruz C., Miller J. H. MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res. 1990 Jul 11;18(13):3841–3845. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberg E., Eide L., Bjørås M. The base excision repair pathway. Trends Biochem Sci. 1995 Oct;20(10):391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- Seeberg E., Nissen-Meyer J., Strike P. Incision of ultraviolet-irradiated DNA by extracts of E. coli requires three different gene products. Nature. 1976 Oct 7;263(5577):524–526. doi: 10.1038/263524a0. [DOI] [PubMed] [Google Scholar]
- Tchou J., Bodepudi V., Shibutani S., Antoshechkin I., Miller J., Grollman A. P., Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J Biol Chem. 1994 May 27;269(21):15318–15324. [PubMed] [Google Scholar]
- Thayer M. M., Ahern H., Xing D., Cunningham R. P., Tainer J. A. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995 Aug 15;14(16):4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira R., van der Kemp P. A., Thomas D., Geiger A., Nehls P., Boiteux S. Formamidopyrimidine DNA glycosylase in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1994 Sep 11;22(18):3760–3764. doi: 10.1093/nar/22.18.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]