Abstract
HTm4 (MS4A3) is a member of a family of four-transmembrane proteins designated MS4A. MS4A proteins fulfil diverse functions, acting as cell surface signalling molecules and intracellular adapter proteins. Early reports demonstrated that HTm4 is largely restricted to the haematopoietic lineage, and is involved in cell cycle control, via a regulatory interaction with the kinase-associated phosphatase, cyclin A and cyclin-dependent kinase 2 (CDK2). Here we describe the expression pattern of HTm4 in peripheral blood cells using gene expression microarray technology, and in normal foetal and adult human tissues, as well as adult human cancers, using tissue microarray technology. Using oligonucleotide microarrays to evaluate HTm4 mRNA, all peripheral blood cell types demonstrated very low levels of HTm4 expression; however, HTm4 expression was greatest in basophils compared to eosinophils, which showed lower levels of HTm4 expression. Very weak HTm4 expression is found in monocytes, granulocytes and B cells, but not in T cells, by lineage specific haematopoietic cell flow cytometry analysis. Interestingly, phytohaemagglutinin stimulation increases HTm4 protein expression in peripheral blood CD4-T-lymphocytes over nearly undetectable baseline levels. Western blotting and immunohistochemical studies show strong HTm4 expression in the developing haematopoietic cells of human foetal liver. Immunohistochemical studies on normal tissue microarrays confirmed HTm4 expression in a subset of leucocytes in nodal, splenic tissues and thymic tissue, and weak staining in small numbers of cell types in non-haematopoietic tissues. Human foetal brain specimens from 19 to 31 gestational weeks showed that the strongest-staining cells are ventricular zone cells and the earliest-born, earliest-differentiating ‘pioneer’ neurons in the cortical plate, Cajal-Retzius and, to a lesser extent, subplate-like neurons. Malignant tissue microarray analysis showed HTm4 expression in a wide variety of adenocarcinomas, including breast, prostate and ovarian. These findings warrant the further study of the role of HTm4 in the cell cycle of both haematopoietic and tumour cells.
Keywords: HTm4, cell cycle regulator, cancer, KAP, CDK2
Introduction
HTm4 is also called membrane-spanning 4-domains, subfamily A, member 3 (MS4A3) [1–5], which belongs to a newly defined, extensive family of human and murine proteins that have similar structure, highly homologous amino acid sequence and close chromosomal location. They all contain a distinctive four-transmembrane structure with N- and C-terminal cytoplasmic regions. CD20 (MS4A1) [1, 6, 7] and FcɛRI [8] antigen receptor β chain (MS4A2) are the most extensively studied genes within the MS4A family. The other MS4A genes might have their roles as cell surface signalling receptors and intracellular adapter proteins [6, 9–11].
HTm4, in human beings, is a 214 amino acid protein whose expression was reportedly restricted to cells of the haematopoietic lineage in adult human beings [1]. We previously showed that HTm4 forms a functionally relevant complex with cyclin-dependent kinase-associated phosphatase (KAP) and CDK2 [4, 5]. Exogenous expression of HTm4 stimulates KAP activity leading to the dissociation of cyclin A, the dephosphorylation of CDK2 and subsequent cell cycle arrest [3]. These studies were the first to identify HTm4 as a novel modulator of the cell cycle. Interestingly, despite these in vitro studies that provided evidence that overexpression of HTm4 resulted in cell cycle arrest, we also confirmed the in vivo coexpression of HTm4, KAP and CDK2 in the proliferating cell populations of germinal centres within secondary follicles of human tonsils and showed that these proteins are all highly expressed in actively cycling cells [4]. This observation could be explained by the expression of an inhibitory cell cycle regulator such as HTm4 in response to proliferation or an effect that is modified by the concentrations of other cell cycle regulators, both known and unknown.
Furthermore, we recently found that HTm4 expression is tightly regulated during the differentiation of haematopoietic stem cells [4] and murine central nervous system [5]. These findings highlight the importance of further elucidating the function of the HTm4-KAP-CDK2 interaction in normal and dysregulated haematopoiesis, as well as during embryogenesis.
Regulation of haematopoietic cell cycle progression is critical in controlling the constant self-renewal, differentiation and homeostasis of the haematopoietic system [12, 13]. Dysregulation of this process has profound consequences, as demonstrated by the development of autoimmune disorders, blood dyscrasias and haematological malignancies with all their associated morbidity and mortality [14, 15].
In this study, we confirmed that HTm4 is expressed in selective subsets of human haematopoietic cells and foetal central nervous system. We also discovered that HTm4 is expressed in a wide range of malignancies, which highlights the importance of HTm4 as a cell cycle regulator in various malignancies.
Materials and methods
Immunohistochemical studies for HTm4
All staining was performed by standard immunoperoxidase methods as previously described [4].
Purification of leucocytes and human lung mast cells
All persons in this study provided written, informed consent, and the Ethical Review Boards at the relevant hospitals (National Center for Child Health and Development, and Jikei University School of Medicine) approved the study [16]. The persons used in this study were all healthy volunteers, especially having no allergic diseases. Human granulocytes, mononuclear cells (CD14+ cells), basophils, eosinophils, T cells (CD4+ and CD8+), peripheral B cells and mast cells were purified as reported earlier [16].
GeneChip expression analysis
Human genome-wide gene expression was examined using the Human Genome U133A probe array (GeneChip, Affymetrix, Santa Clara, CA, USA), which contains the oligonucleotide probe set for 22,000 full-length genes. Experiments were performed in accordance with the manufacturer’s protocol (Expression Analysis Technical Manual) and results were normalized as previously reported [16].
Phytohaemagglutinin (PHA) stimulation assay
Human peripheral blood cells were collected and incubated with or without 1 μg/ml PHA (Sigma, St Louis, MO, USA) for 72 hrs. After 72 hrs, cells were collected and analysed for HTm4 expression by flow cytometry within the CD4+ T-cell population.
Flow cytometry analysis
Human red blood cells in peripheral blood were lysed with hypotonic shock using ACK Lysis Buffer (BioWhittaker, Walkersville, MD, USA). White blood cells were fixed with 4% (w/v) paraformaldehyde and then permeablized by staining buffer with 0.1% (w/v) saponin. Cells were labelled with 2 μg/ml of polyclonal anti-HTm4 for 20 min., followed by the incubation with anti-rabbit IgG Fab fluorescein isothiocyanate (FITC) (Caltag, Burlingame, CA, USA), along with CD4-PE, CD8-PE, CD14-PE, CD15-PE and CD19-PE (BD Pharmingen, San Diego, CA, USA), respectively. The flow cytometry analyses were performed on a FACScan flow cytometer (Becton Dickinson Immunocytometry Systems, San Diego, CA, USA). In the control group, bone marrow cells were labelled with 2 μg/ml of rabbit IgG1 (Sigma, St. Louis, MO) for 20 min., followed by the incubation with anti-rabbit IgG Fab FITC (Caltag).
Electrophoresis and Western blotting
After 10% SDS-PAGE, the 25 μg of protein lysates (Medley, BD Clontech, Palo Alto, CA, USA) were transferred to a polyvinylidene fluoride membrane by electroblotting and the membrane was blocked for 1 hr using 5% bovine serum albumin in PBS pH 7.0. The membrane was then probed for either 2 hrs at RT using 2.0 mg/ml human HTm4 polyclonal antibody. After extensive washing (4 × 5 min. in PBS/0.5% Tween-20), a developing antibody was applied (1 hr at RT, 1:10,000 dilution of donkey anti-rabbit (Amersham, Piscataway, NJ, USA). After further washing bands were visualized using ECL according to the manufacturer’s instructions (Amersham). As a positive control, an HTm4 transfected U937 cell line was also used.
Results
Expression pattern of HTm4 in normal human tissues
Expression pattern of HTm4 transcripts in normal human peripheral blood by oligonucleotide microarray
We examined the mRNA expression levels of the most extensively studied MS4A members, CD20 (MS4A1), FcɛRIb (MS4A2) and HTm4 (MS4A3) in different haematopoietic cell subtypes. As expected, we found CD20 had highest expression in CD191 B cells. FcɛRIb was highly expressed in basophils and mast cells. HTm4 was predominantly expressed in basophils, with only very low levels of mRNA expression in other granulocytes, and B- and T-cell populations (Fig. 1). The preferential expression of HTm4 in basophils is in keeping with previously reported microarray, RT-PCR and immunofluorescence studies [16].
Fig 1.
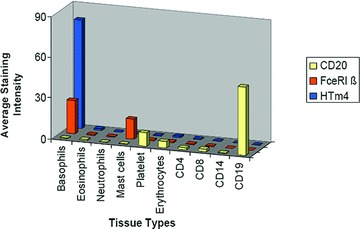
HTm4 expression in human normal peripheral blood cells by mRNA expression microarray profiling. X-axis indicates the cell types. Y-axis is the average intensity.
Expression pattern of HTm4 in normal human peripheral blood by FACS
HTm4 expression in major human peripheral blood cell population (CD4, CD8, CD14, CD15 and CD19) was examined by flow cytometry (Fig. 2). HTm4 surface expression was not found in unstimulated CD4+ T helper cells or CD8+ cytotoxic T cells, and only very weak expression was found in CD14+ monocytes, CD15+ granulocytes and CD19+ B cells.
Fig 2.

HTm4 expression in human normal peripheral blood cells by flow cytometry. Cells were labelled with 2 μg/ml of polyclonal anti-HTm4 for 20 min., followed by the incubation with anti-rabbit IgG Fab FITC (Caltag), along with CD4-PE, CD8-PE, CD14-PE, CD15-PE and CD19-PE (BD Pharmingen), respectively. The flow cytometry analyses were performed on a FACScan flow cytometer (Becton Dickinson Immunocytometry Systems). In the control group, bone marrow cells were labelled with 2 μg/ml of Rabbit IgG1 (Sigma) for 20 min., followed by the incubation with anti-rabbit IgG Fab FITC (Caltag). Tracing represent the staining intensity for HTm4 (green tracing) or control Ig (blue tracing) for the respective gated haematopoietic cell populations.
HTm4 is up-regulated in normal human peripheral blood CD4+ T cells after PHA stimulation
PHA is powerful T lymphocyte mitogen. It binds to oligosaccharide receptor localized on T-cell surface and stimulates the T cells to proliferate. After 72 hrs of PHA stimulation on normal human peripheral blood cells, we found HTm4 expression was weakly up-regulated in CD4+ T cells (Fig. 3) compared to the CD4– population of cells. The same phenomenon could not be found in CD4+ T cells without PHA stimulation (Fig. 3).
Fig 3.
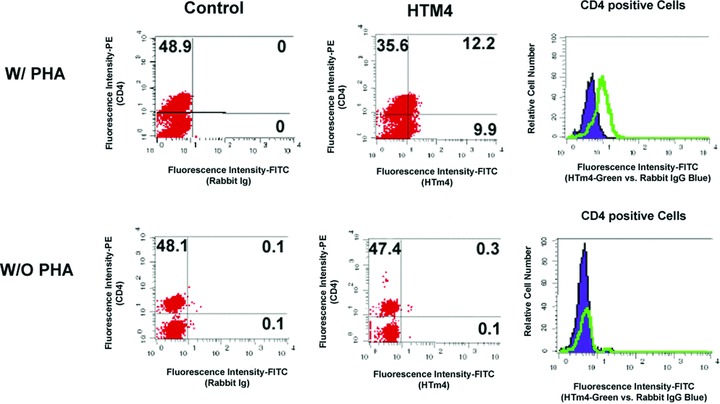
HTm4 up-regulation in human CD4+ T cells after PHA stimulation. Human peripheral blood cells were collected and incubated with or without PHA (1 μg/ml) for 72 hrs. The right-hand histograms show an increase in HTm4 staining (green tracing) in the CD4+ cells after PHA stimulation (upper panels), compared to CD4 cells without PHA stimulation (lower panels). Control Ig staining is depicted in blue.
Expression pattern of HTm4 in normal human tissues by Western blot
Normal human adult and foetal tissue lysates from Clontech was analysed by anti-human HTm4 polyclonal antibody. We found a significant positive staining of HTm4 in human foetal liver (Fig. 4). Weak staining was found in some haematopoietic tissues, such as adult lymph nodes and thymus, as well as some none haematopoietic tissues, such as ovary. Trace staining could also be found in liver, foetal brain and prostate. Interestingly, spleen and brain failed to show HTm4 expression that likely is related to the focal nature of the HTm4 expressing cells in those tissues (see immunohistochemical studies below and Table 1).
Fig 4.
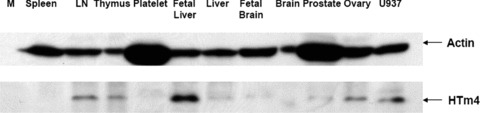
HTm4 expression in human normal tissues by Western blotting. Medley human tissue lysate (Clontech) was loaded in each lane. The membrane was detected by 2 ug/ml anti-HTm4 antibody as primary antibody at RT for 1 hr. After four 15 min. wash, membrane was incubated with 1:5000 diluted anti-rabbit-HRP secondary antibody for 1 hr (Actin was used for normalization). M = marrow; LN = lymph node; U937 are HTm4 transfected control U937 cell line.
Table 1.
HTm4 expression in normal tissues by immunohistochemistry
| Tissue types | HTm4 expression | Cell types (intensity) |
|---|---|---|
| Spleen | + | Red pulp macrophages (3.5) |
| Lymph node | + | Germinal centres (2) |
| Lung | + | Alveolar macrophages (3) |
| Stomach | + | Fundic glands (3) |
| Liver | + | Bile ducts (2.5) |
| Pancreas | + | Pancreatic ductal cells (2.0) |
| Testis | + | Epididymis (1.0); seminiferous (1.5); rete (2.5) |
| Prostate | + | Glandular epithelium (0.5) |
| Breast | + | Ducts (2.0) |
| Placenta | + | Cytotrophoblast (1.0) |
| Brain | + | Glia and neurons (2.0) |
| Thyroid | 0 | – |
| Oesophagus | 0 | – |
| Small bowel | 0 | – |
| Colon | 0 | – |
| Gallbladder | 0 | – |
| Kidney | 0 | – |
| Heart | 0 | – |
| Skin | 0 | – |
| Aorta | 0 | – |
HTm4 staining Intensity Criteria:
0: absent; 1: positive; 0.5: trace; 1: weak; 1.5: weak positive;
2: moderate; 2.5: moderate positive; 3: strong; 3.5: strong positive.
HTm4 expression in foetal human brain ventricular-zone and ‘pioneer’ cells
Limited sections from two human foetal brain specimens from 19 gestational weeks (mid- to late second trimester) to 31 gestational weeks (mid-third trimester) were immunohistochemically stained with anti-human HTm4 (Fig. 5). Overall, compared to the murine brain, the same degree of intense, diffuse staining of cortical plate, diencephalic and brainstem structures was not seen [5]. Interestingly, the strongest-staining cells are in the ventricular zone, and in the earliest-born, earliest-differentiating ‘pioneer’ neurons in the cortical plate, the Cajal-Retzius and, to a lesser extent, the subplate-like neurons. Specifically, in the earlier gestation foetus, Cajal-Retzius neurons of cerebral cortical layer I are distinctly stained, although subplate neurons are negative. In the remainder of the cortical plate, a very faint neuropil background staining is seen, although distinct cytoplasmic or nuclear positivity in cortical plate cells is not detectable. The caudate, putamen and white matter show possible faint staining of glia-like cells. Cells of the ventricular and subventricular zones are strongly stained. At 31 weeks, Cajal-Retzius cells and occasional subplate neurons are positive. Scattered positive cells are now seen in all layers of the cortical plate, and in the caudate nucleus, putamen, globus pallidus, thalamus and hypothalamus. Scattered positive cells are seen in the ependyma, ventricular and subventricular zones. Expression is seen in glia-like cells in the cerebral white matter, in the hippocampus, and in the pons and medulla. Hippocampal pyramidal and dentate neurons, Purkinje and granule cells of the cerebellar cortex and cerebellar dentate neurons are negative.
Fig 5.
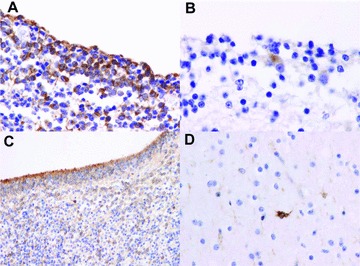
HTm4 expression in human foetal brain. Representative sections of formalin fixed paraffin embedded human foetal brain stained for HTm4. (A) Ventricular zone at 19 post-conceptual weeks (400×), (B) Cajal-Retzius cell at 19 post-conceptual weeks (600×), (C) ventricular zone at 31 post-conceptual weeks (200×), (D) Subplate-like cells at 31 post-conceptual weeks (400×).
Expression pattern of HTm4 in normal adult human tissues by TMA
Immunohistochemical studies on normal human tissue microarrays were performed to evaluate for HTm4 expression. Both the immunoreactive cell type and the intensity of staining were noted and are detailed in Table 1. In general, HTm4 expression was restricted to a limited number of cell types including subset of leucocytes (primarily macrophages) in nodal and splenic tissues, and, in addition, small numbers of cell types in some non-haematopoietic tissues including ductal epithelium of the breast, testis (seminiferous tubules and rete), pancreas, prostate, stomach, thymus and the haematopoietic cells within foetal liver (Fig. 6A–I). The strong staining of immature haematolymphoid cells in the foetal liver (Fig. 6I) and cortical lymphoblasts of the thymus (Fig. 6J) are in keeping with a role for HTm4 in haematopoietic cell development. Absent staining was observed in thyroid, oesophagus, small bowel, colon (Fig. 6J), gallbladder, kidney, heart, skin or aorta. Control immunoglobulin staining failed to show any reactivity with these tissues (data not shown). As we have previously reported, germinal centre B cells within secondary follicles also showed HTm4 expression [4]. Interestingly, adult brain also showed moderate levels of HTm4 expression in focal glial elements and neurons (Table 1), but to a lesser extent than in the foetal brain, in keeping with the weak level of expression noted in the Western blot in foetal brain and inability to detect the protein in adult brain (Fig. 4).
Fig 6.
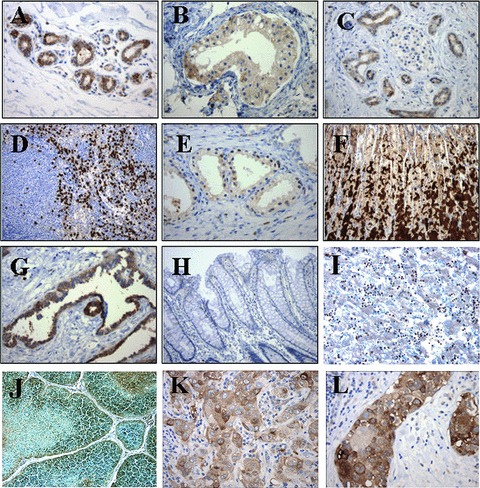
HTm4 expression in human normal and cancer tissues. Representative sections of formalin-fixed paraffin embedded adult human normal (A–H) or cancer tissues (I and J) were stained with anti-HTm4 antibody. (A–H: normal tissues; I–J: cancer tissues) (A) Breast 2003; (B) Seminiferous tubules, testis 2003; (C) Pancreas 200×; (D) Spleen 100×; (E) Prostate 200×; (F) Stomach 100×; (G) Rete testis 200×; (H) Colon 200×; (I) foetal Liver 200×; (J) Thymus 200×; (K) Breast Carcinoma 200×; (L) Breast DCIS 200×.
Expression pattern of HTm4 in human malignancies
The pattern of HTm4 expression was characterized in a wide variety of human malignancies by standard immunohistochemical methods on tissue microarrays containing multiple samples of each tumour type (Table 2). The intensity of staining was recorded on a scale from 0 to 3.5 and the average and standard deviation of staining were calculated. The highest level of expression was noted in seminomas of the testis with the mean intensity of staining being in the moderate range (average 1.93). Moderate to weak staining for HTm4 was observed in adenocarcinomas of the endometrium, ovary, breast (Fig. 6K–L) and prostate, with only weak staining observed in the basal cell carcinomas of the skin, transitional cell carcinomas of the bladder and glioblastomas of the brain (Table 2). No readily discernible staining was observed in the remaining tumours tested including adenocarcinomas of the colon, lung, pancreas and kidney, and squamous cell carcinomas of the gallbladder, cervix, head and neck and oesophagus (Table 2).
Table 2.
HTm4 expression in human cancers by immunohistochemistry
| Tissue type | Histology | Case | Average HTm4 intensity |
|---|---|---|---|
| (N) | Mean ±S.E. | ||
| Testis | Seminoma | 7 | 1.93 ±0.37 |
| Endometrium | Adenocarcinoma | 11 | 1.50 ±0.32 |
| Ovary | Adenocarcinoma | 10 | 1.40 ±0.31 |
| Breast | Ductal Carcinoma | 28 | 1.38 ±0.18 |
| Prostate | Adenocarcinoma | 18 | 1.36 ±0.28 |
| Skin | Basal cell carcinoma | 4 | 1.25 ±0.72 |
| Bladder | Transitional cell carcinoma | 18 | 1.03 ±0.20 |
| Brain | Glioblastoma | 3 | 1.00 ±0.00 |
| Colon | Adenocarcinoma | 18 | 0.81 ±0.17 |
| Pancreas | Adenocarcinoma | 12 | 0.75 ±0.20 |
| Liver | Adenocarcinoma | 10 | 0.75 ±0.40 |
| Lung | Adenocarcinoma | 18 | 0.78 ±0.13 |
| Gallbladder | Squamous cell carcinoma | 3 | 0.67 ±0.44 |
| Cervix | Squamous cell carcinoma | 11 | 0.64 ±0.32 |
| Gastric | Adenocarcinoma | 13 | 0.62 ±0.18 |
| Head and neck | Squamous cell carcinoma | 44 | 0.51 ±0.11 |
| Oesophagus | Squamous cell carcinoma | 12 | 0.38 ±0.16 |
| Kidney | Adenocarcinoma; renal cell carcinoma; clear cell carcinoma | 13 | 0.27 ±0.12 |
HTm4 staining Intensity Criteria:
0: absent; 0.5: trace; 1: weak; 1.5: weak plus; 2: moderate;
2.5: moderate plus; 3: strong; 3.5: strong plus.
Discussion
HTm4 is a member of a newly defined, family of human and murine proteins [1–5]. Each member of this group has a distinctive four-transmembrane structure and is thus referred to as the MS4 family of proteins. To date, few members of this family are well understood, but a diverse functionality is beginning to emerge. These functions include roles as cell surface signalling receptors and intracellular adapter proteins [6, 9–11, 17]. HTm4, in human beings, is a 214 amino acid protein whose expression initially appeared restricted to mature and precursor cells of the haematopoietic lineage [1]. We have previously demonstrated that HTm4 is an adapter molecule that regulates the function of the KAP phosphatase, cyclin A and CDK2, key regulators of cell cycle progression [3–5]. Thus, the restricted expression pattern of HTm4 may reflect a functional role in driving the differentiation and/or regulating the proliferation of haematopoietic lineage-specific stem cells.
A second important feature of human HTm4 biology relates is its chromosomal localization. HTm4 maps to the chromosomal locus 11q13.1, in close proximity to the FcɛRI antigen receptor β chain gene. Both of these genes are MS4 family members. Interestingly, the 11q13.1 region has been significantly linked to human atopic phenotypes. Taq I restriction fragment length polymorphism in the third intron of the human HTm4 showed a strong association with atopic asthma [2]. This variant showed similar odds ratios comparable with that which intron 2 of the FcɛRIβ gene has shown on severe asthma, as well as on severe atopy phenotypes. Therefore, HTm4 is considered an atopy gene candidate on locus 11q13.1 [2].
In this report, we define the HTm4 expression patterns in normal blood cells, human tissues and in a wide variety of cancers. Our previous work had shown that HTm4 appeared to be predominantly expressed in human haematopoietic cells [1, 4]. In addition, we showed the lineage specific expression of HTm4 in murine haematopoietic cells [5]. In this report we provide an extensive description of the expression pattern of HTm4 in normal cell types and tissues. The pattern of HTm4 expression in human haematopoietic cells was very similar to the pattern seen in the murine system. Among the haematopoietic cells examined, HTm4 expression was generally absent, although weak expression could be found in B cells, granulocytes and monocytes. The highest expression level of HTm4 was found in basophils as was shown in prior studies [16]. The reliability of the test was verified by the specific expression of two other MS4A family member-MS4A1 (CD20) [7] and MS4A2 (FcɛRIβ) [8]. As noted in Fig. 1, high levels of CD20 expression was noted in B lymphoid cells and high FcɛRIb expression was confirmed in basophils and mast cells, in accord with current knowledge regarding those two molecules [8, 16, 18]. High HTm4 basophil expression underscored its role as a potential allergy-related molecule located on chromosome 11q13.1 [2].
Interestingly, despite the observation that HTm4 expression in resting lymphoid cells was limited, HTm4 expression could be unregulated following PHA stimulation in normal human CD4+ T-cell subsets (Fig. 3). Antigenic stimulation, which is mimicked by PHA stimulation, triggers T-cell apoptosis after an initial phase of lymphoid expansion [19, 20]. In light of HTm4’s known function in cell cycle regulation [1, 3–5], further examination of the role of HTm4 in activation-induced cell death is warranted.
The high levels of expression of HTm4 in foetal liver haematopoietic cells underscored the potential importance of HTm4 in human haematopoietic development. Also of note, was the strong staining of lymphoblasts in the cortical regions of the thymus, which is known to contain the earliest lymphoid precursors; further evidence for a role of HTm4 in haematolymphoid development. As we reported in murine developing brain, HTm4 is strongly expressed [5]. In this report, we demonstrated that HTm4 is strongly expressed within the glial cells and neurons of the foetal brain (Figs 4 and 5) but in modulating patterns across development, suggesting a role in cell differentiation. The expression in ‘pioneer’ neurons (Cajal-Retzius and subplate-like cells) further hints at an important role in normal cortical organization.
Additionally, small numbers of cell types in some non-haematopoietic or neural tissues demonstrated HTm4 expression including ductal epithelium of the breast, testis (seminiferous tubules and rete), pancreas, prostate, thymus and stomach (Table I and Fig. 6). Importantly, human malignancies derived from these tissue types also show high levels of HTm4 expression (Table 2 and Fig. 6K and L). Given the role of HTm4 as an adapter molecule that regulates the function of the cell cycle regulators KAP phosphatase, cyclin A and CDK2, HTm4 may be a rational therapeutic target in a wide variety of cancers. Additional studies of the effects of HTm4 disruption in cancer cell lines will help to further elucidate the role of HTm4 in tumour cell survival.
Acknowledgments
This work was supported by NIH Grant AI 43663 from the National Institute of Allergy and Infectious Diseases and by Grant RSG-01–241-01-LIB from the American Cancer Society (To C.A.); NIH-P30CA6516 (Specialized Histopathology Lab/Dana Farber Cancer Center). This work was also supported by the Adra family and is dedicated to the memories of Dr. Ramzi Cotran and Dr. Stephen H. Robinson. We would like to acknowledge the technical assistance of Ms. Yvonne Nguyen.
References
- 1.Adra CN, Lelias JM, Kobayashi H, et al. Cloning of the cDNA for a hematopoietic cell-specific protein related to CD20 and the beta subunit of the high-affinity IgE receptor: evidence for a family of proteins with four membrane-spanning regions. Proc Natl Acad Sci USA. 1994;91:10178–82. doi: 10.1073/pnas.91.21.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adra CN, Mao XQ, Kawada H, et al. Chromosome 11q13 and atopic asthma. Clin Genet. 1999;55:431–7. doi: 10.1034/j.1399-0004.1999.550606.x. [DOI] [PubMed] [Google Scholar]
- 3.Chinami M, Yano Y, Yang X, et al. Binding of HTm4 to cyclin-dependent kinase (Cdk)-associated phosphatase (KAP).Cdk2.cyclin A complex enhances the phosphatase activity of KAP, dissociates cyclin A, and facilitates KAP dephosphorylation of Cdk2. J Biol Chem. 2005;280:17235–42. doi: 10.1074/jbc.M413437200. [DOI] [PubMed] [Google Scholar]
- 4.Donato JL, Ko J, Kutok JL, et al. Human HTm4 is a hematopoietic cell cycle regulator. J Clin Invest. 2002;109:51–8. doi: 10.1172/JCI14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kutok JL, Yang X, Folkerth RD, et al. The cell cycle associated protein, HTm4, is expressed in differentiating cells of the hematopoietic and central nervous system in mice. J Mol Histol. 2005;36:77–87. doi: 10.1007/s10735-004-3913-8. [DOI] [PubMed] [Google Scholar]
- 6.Liang Y, Buckley TR, Tu L, et al. Structural organization of the human MS4A gene cluster on Chromosome 11q12. Immunogenetics. 2001;53:357–68. doi: 10.1007/s002510100339. [DOI] [PubMed] [Google Scholar]
- 7.Tedder TF, Streuli M, Schlossman SF, et al. Isolation and structure of a cDNA encoding the B1 (CD20) cell-surface antigen of human B lymphocytes. Proc Natl Acad Sci USA. 1988;85:208–12. doi: 10.1073/pnas.85.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ra C, Jouvin MH, Kinet JP. Complete structure of the mouse mast cell receptor for IgE (Fc epsilon RI) and surface expression of chimeric receptors (rat-mouse-human) on transfected cells. J Biol Chem. 1989;264:15323–7. [PubMed] [Google Scholar]
- 9.Hulett MD, Pagler E, Hornby JR, et al. Isolation, tissue distribution, and chromosomal localization of a novel testis-specific human four-transmembrane gene related to CD20 and FcepsilonRI-beta. Biochem Biophys Res Commun. 2001;280:374–9. doi: 10.1006/bbrc.2000.4088. [DOI] [PubMed] [Google Scholar]
- 10.Ishibashi K, Suzuki M, Sasaki S, et al. Identification of a new multigene four-transmembrane family (MS4A) related to CD20, HTm4 and beta subunit of the high-affinity IgE receptor. Gene. 2001;264:87–93. doi: 10.1016/s0378-1119(00)00598-9. [DOI] [PubMed] [Google Scholar]
- 11.Liang Y, Tedder TF. Identification of a CD20-, FcEpsilonRIbeta-, and HTm4-related gene family: sixteen new MS4A family members expressed in human and mouse. Genomics. 2001;72:119–27. doi: 10.1006/geno.2000.6472. [DOI] [PubMed] [Google Scholar]
- 12.Metcalf D. The roles of stem cell self-renewal and autocrine growth factor production in the biology of myeloid leukemia. Cancer Res. 1989;49:2305–11. [PubMed] [Google Scholar]
- 13.Weissman D, Dybul M, Daucher MB, et al. Interleukin-2 up-regulates expression of the human immunodeficiency virus fusion coreceptor CCR5 by CD41 lymphocytes in vivo. J Infect Dis. 2000;181:933–8. doi: 10.1086/315303. [DOI] [PubMed] [Google Scholar]
- 14.Ellisen LW, Carlesso N, Cheng T, et al. The Wilms tumor suppressor WT1 directs stage-specific quiescence and differentiation of human hematopoietic progenitor cells. EMBO J. 2001;20:1897–99. doi: 10.1093/emboj/20.8.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima T, Iikura M, Okayama Y, et al. Identification of granulocyte subtype receptors and ion channels by using a high-density oligonucleotide probe array. J Allergy Clin Immunol. 2004;113:58–35. doi: 10.1016/j.jaci.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 17.Hulett MD, Pagler E, Hornby JR. Cloning and characterization of a mouse homologue of the human haematopoietic cell-specific four-transmembrane gene HTm4. Immunol Cell Biol. 2001;79:345–9. doi: 10.1046/j.1440-1711.2001.01017.x. [DOI] [PubMed] [Google Scholar]
- 18.Tedder TF, Engel P. CD20: a regulator of cell-cycle progression of B lymphocytes. Immunol Today. 1994;15:450–4. doi: 10.1016/0167-5699(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 19.Lenardo M, Chan KM, Hornung F, et al. Mature T lymphocyte apoptosis–immune regulation in a dynamic and unpredictable antigenic environment. Annu Rev Immunol. 1999;17:221–53. doi: 10.1146/annurev.immunol.17.1.221. [DOI] [PubMed] [Google Scholar]
- 20.Michallet MC, Saltel F, Flacher M, et al. Cathepsin-dependent apoptosis triggered by supraoptimal activation of T lymphocytes: a possible mechanism of high dose tolerance. J Immunol. 2004;172:5405–14. doi: 10.4049/jimmunol.172.9.5405. [DOI] [PubMed] [Google Scholar]


