Abstract
Recently we reported that the expression of the protein kinase A (PKA) regulatory subunit RIIα is dynamically regulated in human smooth muscle cells of the uterus. We showed that expression levels of mRNA/protein were substantially increased during pregnancy and decreased upon labour, changes that were mirrored by particulate type II PKA activity. This implied an important role for RIIα in maintaining uterine quiescence during pregnancy. Consequently the purpose of the present study was to identify potential mechanisms by which expression of the RIIα gene was regulated in this tissue. We indicate here that the three SpI-III (GC) binding domains within the proximal promoter region of the human RIIα gene may play important roles in modulating expression of the gene in human myometrial cells. We show that all three GC binding domains are involved in binding Sp1, Sp3, histone deacetylase (HDACs) 1/2 and RbAp48 transcriptional complexes. The functional significance of these binding domains was further analysed employing in vitro luciferase reporter assays with full-length/truncated RIIα promoter constructs. Importantly we show that treatment of primary human myometrial cell cultures with the general class I/II HDAC inhibitor trichostatin A results in an increase in mRNA/protein levels. Moreover the increase in mRNA levels appeared to be preceded by an increase in aH3, PolIIa, Sp3 and HDAC 2 binding to the three SpI-III (GC) binding sites within the RIIα promoter. These results enable us to provide a model whereby RIIα expression is epigenetically regulated in human myometrial smooth muscle cells by histone deacetylase(s) activity within the GC-rich proximal promoter region of the gene.
Keywords: myometrial, PKA, specificity proteins (Sp1-4), histone deacetylases, trichostatin A
Introduction
There is now extensive evidence to indicate that components of the cyclic adenosine monophosphate (cAMP) signalling pathway are up-regulated in the human myometrium during pregnancy so as to potentiate the maintenance of uterine quiescence until activation of smooth muscle contractions at term. These include chorionic gonadotropin/ luteinizing hormone (hCG/LH) receptors [1], calcitonin gene related peptide receptors [2], β-adrenoreceptors [3, 4], corticotrophin releasing hormone (CRH) receptor isotypes [5, 6] and the adenylyl cyclase stimulatory G-protein Gαs [7–9]. Expression levels of the latter are substantially increased within the myometrium during gestation resulting in the increased production of cAMP, which is further amplified by the progesterone-induced down-regulation of myometrial cAMP phosphodiesterase activity [10]. The effects of cAMP are mediated via protein kinase A (PKA), a heterotetrameric protein complex consisting of two regulatory (R) and two catalytic (C) subunits [11]. Two classes of PKA are expressed termed PKA type I and II, which are distinguished by differences in the R subunit RI and RII which interact with identical Ca, Cb or Cg subunit species. RI and RII each exist as two isoforms RIα/RIβ and RIIα/RIIβ. PKA containing RIa and RIb isoforms are generally present in the soluble fraction of cells, whereas PKA containing RIIα and RIIβ species are primarily associated with sub-cellular structures of the cell via interactions with specific A-kinase anchoring proteins. In this context we have recently shown that RIIα species in the human myometrium are differentially expressed during gestation and parturition [12]. This was in contrast to expression of RIα, Cα and Cβ which remained uniform whereas RIβ/RIIβ were undetected [12]. Our data indicated that expression levels of RIIα mRNA and protein were substantially increased and decreased during pregnancy and labour, respectively, which was mirrored by similar changes in particulate type II PKA activity.
Thus dynamic regulation of RIIα expression is an important means of influencing smooth muscle contractile function. Consequently the purpose of the present study was to identify potential mechanisms by which expression of the RIIα (PRKAR2A) gene is regulated in human myometrial smooth muscle cells. Although there is a paucity of information of how the RIIα gene is regulated in any tissue or cell type, the promoter region of the RIIα gene has been fully characterized [13] (see Fig. 1A). Foss et al. [13] indicated that this region is GC rich and TATA-less with a CpG island present in the proximal region of the promoter (http://linux1.softberry.com/berry.phtml) (Fig. 1A). Characteristically genes with similar features within their promoters are classified as house-keeping genes, as expression levels do not vary among different cell types [14]. However there is a growing body of evidence showing that such genes can indeed be a target of tissue/cell type specific control [15, 16]. The proximal promoter region –1206 bp upstream from the +1 transcriptional start site of the RIIα gene indicates the presence of several potential binding sites for AP1, AP2, NF1, NF-κB as well as three SpI-III GC boxes representing canonical binding sites for the specificity protein family (Sp1-4). In many instances Sp1-4 proteins have been observed to recruit HDACs to the promoter region of genes which contain GC elements to silence their expression [17–19]. We have also shown this to be the case in human myometrial cells where expression of the hCG/LH receptor gene is repressed by recruitment of HDACs 1/2 to the two SpI-II (GC) binding domains within the proximal promoter [20]. Furthermore, the presence of a high number of CpG methylation sites in the promoter region of the RIIα gene also points to a potential role for promoter DNA methylation as an important regulatory factor in controlling expression of the gene.
Fig 1.
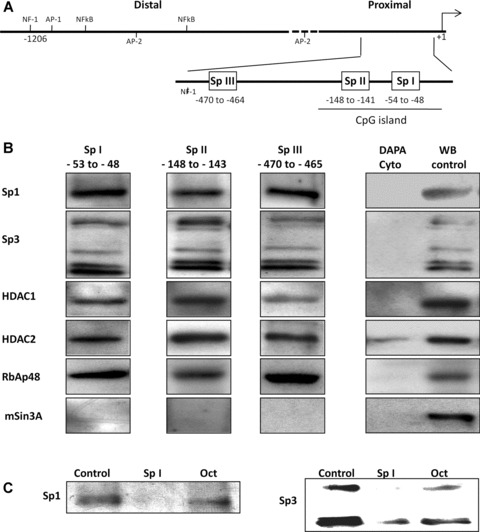
in vitro DAPA analysis indicates that the Sp I-III (GC) binding sites within the RIIα promoter bind multi-protein complexes containing Sp1, Sp3, HDACS 1 and 2 and RbAp48. (A) Schematic representation of the RIIα promoter region showing the putative transcriptional regulatory cis-elements. The transcriptional initiation site (+1) and the nucleotide position of the three Sp I-III (GC) binding elements in the proximal region are as shown. (B) DAPA analysis was performed with biotin labelled oligonucleotides for the individual Sp I-III (GC) sites and nuclear extracts from human myometrial cell cultures. Proteins which bound to the individual Sp1-III (GC) oligonucleotides were analysed by Western blotting using antibodies for Sp1, Sp3, HDAC1and 2, RbAp48 and mSin3A. All three Sp I-III (GC) elements bound protein complexes containing Sp1, Sp3, HDAC1 and 2 and RbAp48. mSin3A was not associated with the complexes, despite its presence in the myometrial cells. No binding was observed when the nuclear extracts were replaced with cytoplasmic extract (Cyto) except in the case of HDAC2 where a weak signal was observed. Western blotting of nuclear protein extracts (WB control 10 μg) were used as positive controls. Similar results were obtained in two further experiments. (C) Binding specificity of Sp1 and Sp3 to the Sp I (GC) element was determined by DAPA competition assays where 50× excess specific competitor (unlabelled Sp I oligonucleotide) or non-specific competitor (Oct element, see Table 1) was included in the DAPA binding reactions. Similar results were obtained for the Sp II and Sp III cis-elements within the RIIα promoter (data not shown).
In the present study, we assess the possible role of SpI-III GC binding domains and HDACs 1/2 in epigenetically regulating RIIα gene and protein expression in human uterine smooth muscle cells.
Materials and methods
Preparation of primary myometrial cell cultures
Primary cell cultures were established and cultured from non-pregnant myometrial tissue samples as described previously [21, 22]. Samples were taken from the uteruses removed from women undergoing hysterectomy from benign gynaecological conditions (38 to 58 years old). Written consent was obtained from all women, and ethical approval was granted by the North Tyneside Health Authority Ethics Committee.
Western blotting
Western blotting was carried out as described previously [20, 21]. Essentially total protein (10 μg) from untreated or treated myometrial cells were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes, blocked and then incubated with the respective antibodies for 1 hr in 1% fat free milk in PBS/Tween at room temperature (for concentrations see Table 1). After washing membranes were incubated with the secondary horse radish peroxidise (HRP)-conjugated antibodies (DAKO, Glostrup, Denmark, 1:5000) for 1 hr at room temperature and enhanced chemiluminescent reagent added and immunodetected bands scanned and quantified using the intelligent quantifier software package (BioImage, Ann Arbor, MI, USA). Membranes were re-probed with an antibody against β-actin as a loading control.
Table 1.
Concentrations of the specific antibodies used for WB, DAPA, ChIP and co-IPs and sequences of the oligonucleotide probes used in DAPAs. Positions are derived from the +1bp (AGT) transcriptional start site. Transcriptional regulatory sites are in bold. Only one of each pair of complementary oligonucleotides is shown
| Antibody | Source | DAPA/WB Dilution | ChIP (μg/ml) | co-IP (μg/reaction) |
|---|---|---|---|---|
| Sp1 (sc-59) | Santa Cruz Biotech. Inc, Santa Cruz, CA, USA | 1:300 | 30 | |
| Sp1 (ab13370) | Abcam Plc, Cambridge, UK | 2 | ||
| Sp3 (sc-644) | Santa Cruz Biotech. | 1:200 | 2 | 10 |
| HADC1 (ab7028) | Abcam | 1:2000 | 20 | 4 |
| HDAC2 (ab7029) | Abcam | 1:3000 | 20 | 20 |
| mSin3A (sc-5299) | Santa Cruz Biotech. | 1:200 | ||
| RbAp48 (ab490) | Abcam | 1:5000 | 4 | 4 |
| b-actin (ac-74) | Sigma | 1:5000 | ||
| RIIα (sc-908) | Santa Cruz Biotech. | 1:10,000 | ||
| PolIIa (05-623B) | Millipore Ltd, Dundee, UK | 5 | 5 | |
| aH3 (06-599) | Millipore | 1:10,000 | 5 | |
| GAPDH (sc-25778) | Santa Cruz Biotech. | 1:2000 | ||
| Primer | Position | Sequence | ||
| BIO Sp I | –60 to –50 | BIOTIN-5′-CGCCTGCCCCGCCCGTCCAGC-3′ | ||
| BIO Sp II | –156 to –136 | BIOTIN-5′-GCCCGAGGGGGCGGGTCATCT-3′ | ||
| BIO Sp III | –478 to –458 | BIOTIN-5′-CTTCCCCTGGGCGGGTCCTGC-3′ | ||
| Sp I | –60 to –40 | 5′-CGCCTGCCCCGCCCGTCCAGC-3′ | ||
| Sp II | –156 to –136 | 5′-GCCCGAGGGGGCGGGTCATCT-3′ | ||
| Sp III | –478 to –458 | 5′-CCTCCCCTGGGCGGGTCCTGC-3′ | ||
| Oct1 | 5′-TGTCGAATGCAAATCACTAGAA-3′ |
Preparation of myometrial cytoplasmic and nuclear extracts
Cytoplasmic and nuclear extracts were prepared using a nuclear extraction kit (Chemicon, Millipore UK Ltd, Dundee, UK) according to the manufacturer’s instructions. Briefly myometrial cells were washed and collected by scraping in PBS and centrifuged. Cell pellets were resuspended in ice cold cytoplasmic lysis buffer and incubated for 15 min. on ice and centrifuged (250 × g, 5 min., 4°C). Supernatants were discarded and cells resuspended in cytoplasmic lysis buffer with mechanical disruption and centrifuged (8000 × g, 20 min., 4°C) resulting in the cytosolic fraction. The remaining pellet was resuspended in nuclear extraction buffer and gently agitated at 4°C for 1 hr, centrifuged (16,000 × g, 5 min.) giving rise to the nuclear fraction.
DNA affinity precipitation assays (DAPAs)
DAPAs were performed as described in (16). Essentially 35 pmol of double-stranded, 5′ end biotin labelled oligonucleotides (MWG) for the respective SpI-III (GC) elements (Table 1), were incubated with either 80 μg of cytoplasmic or nuclear protein extract in 500 μl binding buffer for 1 hr at 4°C to form protein/DNA complexes. Then, 30 μl of streptavidin coated magnetic beads (Active Motif) was added for another 2 hrs to allow formation of DNA/protein/avidin complexes. Resins were then washed 5× in binding buffer. Proteins were recovered from the complex, separated by SDS-PAGE and detected by immunoblotting using specific antibodies (Table 1). To check specificity of the oligonucleotides-protein binding, control binding reactions were performed in the presence of 50× excess non-biotinylated oligonucleotide for each SpI-III (GC) element or non-specific competitor Oct1.
Co-immunoprecipitation (co-IP)
Co-IP’s of myometrial nuclear extracts were performed with the universal magnetic co-IP kit (Active Motif). Nuclear protein lysates (500 μg) were pre-cleared with the respective rabbit or mouse IgG (Upstate) and 10 μl of protein G-coated magnetic beads for 30 min. at 4°C. Primary antibodies to the individual transcriptional regulators (Table 1) were added and allowed to form complexes in a low-stringency co-IP/wash buffer supplemented with proteases, phosphatases and deacetylases inhibitors over night at 4°C. Protein/Ab complexes were then recovered with 25 μl of protein G-coated magnetic beads and washed 4× with increased stringency co-IP/wash buffer (150 mM NaCl and 0.5% detergent). Proteins were then recovered from the magnetic beads by boiling for 5 min. in 20 μl of loading buffer and subjected to Western blotting.
Chromatin immunoprecipitation (ChIP)
ChIP analysis on control and treated cell cultures was carried out using the EZ ChIP kit (Upstate, #17-371) according to the manufacturer’s instructions. In each instance the relevant rabbit or mouse IgGs were used as the negative control. Changes in the binding of the transcriptional regulators to the RIIα promoter and equal loading were investigated by amplification of the recovered DNA after ChIP or with input (I) with the primers respective to each of the three SpI-III (GC) elements (Gene Bank sequence Nb.: X99455). SpI: F5 (position 1085-1104, 5′-CCGGTGCTAAGCGGGGACG-3′) and R2 (1254-1235; 5′-CGCAACCCTACGCTACCACG-3′); SpII: F3 (992-1011; 5′-CCTGGATTCCCTCCGTGAGC-3′) and R5 (11041085; 5′GCTTAGCACCGGCCCTCAGT-3′); SpIII: F2 (679-695, 5′-ACTCCACCAGGCCTTTGCTC-3′) and R4 (808 – 78′, 5′-ATGAAGGGCAAGAGAGGGCT-3′). PCR was performed with the ready to use hot start PCR mix (VHBio), for 30 cycles and PCR products run on agarose gels and scanned and quantified using the intelligent quantifier software package (BioImage). Inputs (I) in each case were scanned to correct for loading.
Preparation of RIIα promoter luciferase reporter constructs
Four RIIα promoter luciferase reporter constructs were prepared using similar cloning methods as described in [20]. The full length promoter region construct was amplified from human genomic DNA (Roche Applied Sciences) with the following F1 primer (position: 1-20; 5′-GAGCTCCTGGGTGTGTGCCA-3′) and the above R2 primer and contained all three proximal SpI-III (GC) binding domains as well as up-stream transcriptional regulatory elements. Truncated forms of the RIIα promoter were subsequently prepared by amplification with the above F2/R2, F3/R2 and F5/R2 primers (see Fig. 4A) giving rise to the constructs containing SpIII, SpII and SpI (GC) binding elements in the promoter controlling transcription of the firefly luciferase gene.
Fig 4.
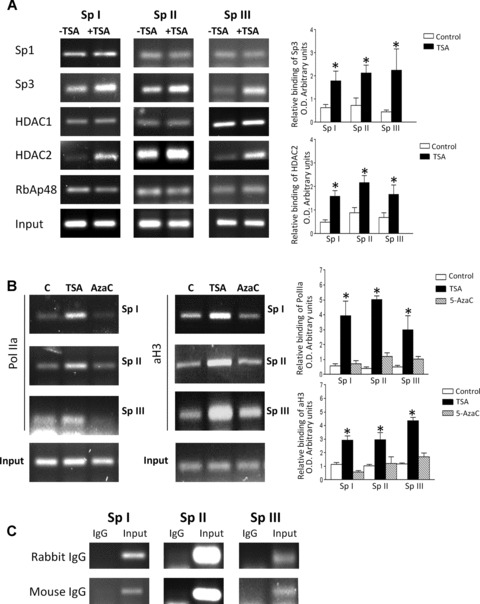
ChIP analysis confirms that the SpI-III (GC) binding sites within the RIIα promoter bind multi-protein complexes containing Sp1, Sp3, HDACS 1 and 2 and RbAp48 in vivo in human myometrial cells. Human myometrial cells were treated in the absence or presence of TSA (330 nM) or 5-AzaC (100 μM) for 6 hrs and ChIP analysis carried out as described in Materials and Methods. (A) Sp1, Sp3, HDAC1 and 2 and RbAp48 antibodies specifically precipitated PCR amplicons associated with the three Sp I-III (GC) cis-elements within the RIIα promoter. Incubation of myometrial cells with TSA resulted in increased binding of Sp3 and HDAC2 to the three Sp I-III (GC) sites. Data were obtained from three individual myometrial cell preparations analysed in triplicate and are expressed as mean ± S.E.M. of the relative binding after correcting for Input (I) as a loading control. *P < 0.05 Student’s t-test (n = 9) for TSA treated compared to control. (B) Incubation of myometrial cells with TSA resulted in increased binding of acetylated Histone 3 (aH3) and PolIIa to the three Sp I-III (GC) sites; no increase in binding was observed for 5-AzaC. Data were obtained from three individual myometrial cell preparations analysed in triplicate (n = 9) and are expressed as mean ± S.E.M. of the relative binding after correcting for Input. In each instance treatment with TSA resulted in a significant increase in binding compared to control or 5-AzaC treatment. *P < 0.05 Student’s t-test. (C) Rabbit and Mouse IgG controls indicated the specificity of all pull-downs.
Transient DNA transfection of human myometrial cells
Cells were grown in 35-mm culture plates to 70% confluence. For each plate, 8 μl of TransIT-LT-1 (Mirus, Bio LLC, Madison, WI, USA) were mixed with 250 μl D-MEM (Invitrogen Limited, Paisley, UK) and incubated for 20 min. at room temperature. Then, 2 μg of sterile, highly purified plasmid DNA containing the respective RIIα promoter luciferase construct and 1 μg of control Renilla luciferase plasmid used as internal transfection control (Promega, Southampton, UK) were added and incubated 30 min. at room temperature. Subsequently DMEM/LT-1/plasmid mix was added to cells in pen/strep-free, DMEM/10% foetal calf serum (FCS). Cultures were incubated for 24 hrs at 37°C to allow transfection to occur. Then the transfection mix was replaced with DMEM/10%FCS, pen/strep free medium and cells were treated for 6 hrs with Trichostatin A (TSA). Data from three independent transfections, each in triplicate, were collected using the dual-luciferase reporter assay system (Promega). Transfection efficiency was determined (10–15% for each experiment) using LT-1 and the β-galactosidase-encoding plasmid/β-galactosidase staining Kit (Mirus).
RIIα promoter luciferase reporter construct methylation
To investigate the effect of promoter methylation on expression of the RIIα gene, the promoter luciferase reporter construct containing the three SpI-III (GC) binding elements was methylated in vitro using CpG methyltransferase m.SssI (New England Biolabs Ltd, Hitchin, UK) in the presence of S-adenosylmethionine for 3 hrs at 37°C. After the methylation reaction, transfection of myometrial cells was carried out as described above. To check the specificity of methylation, 0.5 μg of methylated, mock treated and un-treated plasmid constructs were digested with the CpG methylation sensitive restriction enzyme XhoI.
siRNA-mediated gene silencing
Primary human myometrial cells were prepared and cultured as above. For siRNA transfection, cells were transfected at 70–80% confluence. Dharmacon siGENOME™ SMARTpools (Thermo Fisher Scientific, Lafayette, CO, USA) of four specific siRNA duplexes (targeted to individual Sp1 and Sp3 species, 100 nM siRNA each) were used to transfect myometrial cells using the Dharmafect™ 1 lipid reagent (Thermo Fisher). After 48 hrs cells were harvested and proteins extracted and resolved on SDS-PAGE. Knockdown of Sp1, Sp3 and RIIα was monitored employing specific antibodies detailed in Table 1. A GAPDH antibody (see Table 1) was used to confirm specificity of the siRNA knockdowns.
RNA isolation and real time PCR
Total RNA was isolated from myometrial cell cultures harvested in 1 μl Tri Reagent (Sigma-Aldrich, St. Louis, MO, USA)/10 cm2 culture plates according to the manufacturer’s instructions. 1 μg of total RNA was reverse transcribed using MMLV reverse transcriptase (Promega). Primers for RIIα (F: 679-700, 5′-GCATTCCGGTACTGTTGGTAA-3′, R: 829 to 811, 5′-TCAAGTTCTCGATGCCATGTTT-3′) and the internal GAPDH control (F: 275-295, 5′-GCACCGTCAAGGCTGAGAAC-3′; R: 425-406, 5′-GCCTTCTCCATGGTGGTGAA-3′) were designed using the AbiPrism primer design software, based on reported human sequences in the GeneBank (RIIα: BC002763, GAPDH: NM002046.3). Real time PCR reactions were performed with an Abi Prism 7000 cycler (Applied Biosystems, Europe BV, Warrington, UK) and a SYBRGreen PCR kit (Qiagen Ltd., Crawley, UK). The PCR reaction mix contained 1μ QuantiTect SYBRGreen PCR Mix, 0.3 μM of the forward and reverse primers each, and 2 μl of cDNA template, or 2 μl of standard, respectively. The initial 10 min. denaturation was followed by 40 denaturation amplification cycles of 30 sec. at 95°C, 1 min. annealing and elongation at 60°C. To ensure the specificity of the PCR amplicons, a temperature-controlled melting curve analysis was performed as a last step of the PCR reaction. The experiment was carried out with four independent tissue cultures, each in triplicate. Expression levels of RIIα mRNA for each individual sample were calculated by the ΔΔCt method [23].
Myometrial contractility studies
Longitudinal myometrial strips (measuring approximately 2 × 2 × 10 mm) were mounted for isometric recording under 2 mN of tension in organ baths as previously described [24]. The tissue baths contained 10 ml of Krebs-Henseleit physiological salt solution (PSS) maintained at 37°C, pH 7.4 were gassed continuously with a mixture of 95% oxygen / 5% carbon dioxide. Myometrial strips were allowed to equilibrate for at least 1 hr during which time the Krebs-Henseleit physiological salt solution was changed every 20 min. After equilibration, contractions were stimulated by bath exposure of the strips to oxytocin (10 nM). TSA was then added to the tissue bath at 3.3 μM. Control experiments were performed with strips exposed to 10 nM oxytocin and vehicle. The effects of TSA and the respective controls were assessed by calculation of the integral from minimum of selected areas for each 20 min. intervals and expressed as a percentage of the integral obtained in the 20 min. period prior to TSA addition using the PowerLab hardware unit and Chart v4.2 software (AD Instruments, Hastings, UK). Myometrial strips were frozen in liquid nitrogen at either 1 hr (after contractility measurements) or 6 hrs (maintained in buffer ± TSA) and proteins extracted and resolved on PAGE-SDS with subsequent Western blotting for expression of RIIα and aH3.
Statistical analysis
Data were compared using an unpaired two-tailed t-test; P < 0.05 was considered statistically significant. All experiments were performed from myometrial cell preparations from individual patient biopsies (n = 3–6) and experiments were carried out in triplicate and results expressed as the mean ± S.E.M. For contractility studies n = 3 myometrial strips were used.
Results
Assessment of transcriptional regulator recruitment to the SpI-III (GC) binding elements within the RIIα promoter region in human myometrial cells
DAPA analysis indicated that both Sp1 and Sp3 specifically bound the three SpI-III (GC) boxes within the proximal promoter region of the RIIα gene (Fig. 1B). Note that Sp3 has several well documented isoforms which are detected with the Sp3 antibody used here in DAPAs and Western blotting. No binding was observed for either Sp2 or Sp4 (data not shown). In each instance Sp1 and Sp3 binding was only observed in nuclear fractions whereas no signal was observed after incubation of the biotinylated oligonucleotides with cytoplasmic fractions (Fig. 1B). Specificity of DNA protein binding proteins was determined by co-incubation with specific and non-specific competitors. Here incubation of 50× excess of specific non-labelled oligonucleotides for the respective SpI element showed reductions in the detected signal for both Sp1 and Sp3 whereas the non-specific competitor Oct1 consensus element had no effect (Fig. 1C), similar results were obtained for the SpII and III elements (data not shown). HDAC1 and 2 were also specifically pulled down by all three SpI-III (GC) promoter elements in nuclear fractions, however a weak signal for HDAC2 was also observed in the cytoplasmic fraction (Fig. 1B). DAPAs were also performed to determine whether mSin3A and RbAp48 also formed complexes with Sp1/Sp3 and HDACs 1/2 as observed for the hCG/LH receptor gene [19, 20]. Although mSin3A was expressed in human myometrial cells as determined by Western blotting only RbAp48 was found to form transcriptional complexes with Sp1/Sp3 and HDACs 1/2 on the three SpI-III (GC) binding domains (Fig. 1B).
Control DAPAs using the same mSin3A antibody with GC oligonucleotide sequences within the Gαs promoter indicated that mSin3A could be captured in the complexes with the above proteins (data not shown) thus confirming that this can occur in myometrial cells but not in the context of RIIα.
To further determine that binding of Sp1-4 proteins were involved in regulating expression of RIIαin vivo in human myometrial cells, cells were treated with Mithramycin A. In this instance Mithramycin A interacts non-covalently with G/C-rich duplex DNA and therefore competes with proteins such as Sp1 and Sp3, as well as other zinc finger family proteins, which bind to regulatory elements containing GC-rich sequences [25, 26]. This appeared to be the case in myometrial cells where 6 hrs treatment with 2 μM Mithramycin A significantly decreased basal RIIα protein levels (Fig. 2A). Note that 100–200 nM concentrations of Mithramycin A that are normally used elicited smaller effects at 6 hrs and thus μM concentrations were used as reported by Hirasawa [27], which were observed to specifically inhibit Sp1 DNA binding in the macrophage cell line RAW264. However when myometrial cells were treated with 200 nM Mithramycin A for 24 hrs a significant decrease in basal RIIα protein expression was also observed (Fig. 2B) further indicating specificity of Sp1/Sp3 in regulating expression of the RIIα gene. Moreover employing siRNA oligonucleotides directed against Sp1 and Sp3 resulted in substantial decreases of both protein species as well as RIIα as monitored by Western blotting (Fig. 2C) also indicating a role for Sp1/Sp3 in regulating expression of RIIα in myometrial cells.
Fig 2.
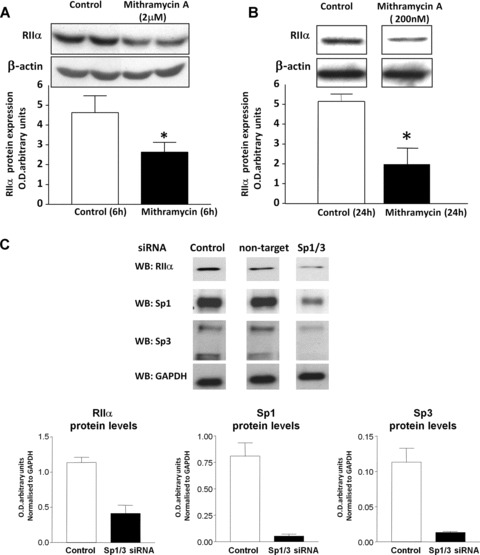
Incubation with Mithramycin A and siRNA knockdown of Sp1 and Sp3 inhibit basal RIIα protein expression in human myometrial cells. (A) Decrease in RIIα protein levels monitored by Western blotting after 6 hrs treatment of human myometrial cells with 2 μM Mithramycin. Data are expressed as mean ± S.E.M. Data were obtained from six myometrial cell preparations. *P, 0.05 Student’s t-test (n 5 6) treated versus untreated cells. (B) Mithramycin A at 200 νM also decreased RIIα protein levels as monitored by Western blotting after 24 hrs treatment of human myometrial cells indicating further specificity of the effect. Data are expressed as mean ± S.E.M. *P < 0.05 Student’s t-test (n = 4) treated versus untreated cells. Equal loading in each case (A and B) was confirmed using a β-actin antibody. (C) Effect of Sp1/Sp3 siRNA transfection of human myometrial cells on RIIα protein expression. Myometrial cells were transfected with siRNAs directed against both Sp1 and Sp3 as well as control non-target siRNAs. Expression of Sp1, Sp3 and RIIα protein levels after 48 hrs transfection were monitored by Western blotting. A substantial decrease in Sp1, Sp3 and RIIα protein levels was observed with Sp1/Sp3 siRNAs compared to control (no siRNAs) or non-target siRNAs. A GAPDH antibody was used to confirm specificity of the siRNA transfection. Data are means ± S.E.M. (n 5 5) from two myometrial cell preparations.
HDACs 1/2 have been previously reported to form complexes with Sp1 on the hCG/LH receptor promoter in placental choriocarcinoma cells; whereas binding to Sp3 occurred via binding to RbAp48 [19]. Co-immunoprecipitations assays were therefore employed to determine if similar events occurred on the RIIα promoter in human myometrial cells. In this instance interactions between Sp1 and HDACs1/2 was detected (Fig. 3), as per the hCG/LH receptor promoter [19], whereas in contrast Sp3 only co-precipitated with HDAC2 but not with HDAC1 (Fig. 3). These results were confirmed by a second co-IP with HDAC1 and 2 with subsequent Western blotting with the anti-Sp3 Antibody (data not shown). In these assays RbAp48 was observed to be precipitated with both HDACs1/2 (Fig. 3).
Fig 3.
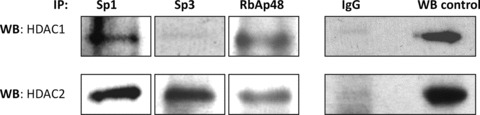
Determination of the protein interactions within the transcriptional complexes binding the SpI-III (GC) sites in the RIIα promoter. Nuclear extracts prepared from human myometrial primary cells were subjected to co-immunoprecipitation assays with anti-rabbit antibodies for Sp1, Sp3, and anti-mouse antibody for RbAp48, followed by immunoblotting with antibodies for HDAC1 or 2, or RbAp48. Respective, mouse and rabbit, IgGs were used as a negative control and Western blotting of nuclear protein extracts (10 μg) were used as positive controls. Similar results were obtained in two further experiments.
Assessment of in situ recruitment of transcriptional regulators to the RIIα promoter in human myometrial cells
ChIP assays were performed to confirm that the data accrued via in vitro DAPA analyses also occurred in vivo. In this instance ChIP analysis of all three RIIα SpI-III (GC) binding domains indicated that Sp1, Sp3, HDAC1/2 and RbAp48 were all recruited to these cis-elements whereas mSin3A appeared not to be. Moreover, TSA treatment for 6 hrs increased the binding of both Sp3 and HDAC2 approximately 2-fold to the three SpI-III (GC) elements (Fig. 4A); whereas no change in binding was observed for Sp1, RbAp48 and HDAC1. Additionally a 3–4-fold increase in binding of acetylated Histone 3 (aH3) after 6 hrs treatment with TSA (330 nM) was also seen (Fig. 4B) which correlated with an increase in acetylated Histone 3 (aH3) as monitored by Western analysis (data not shown). Because real time RT-PCR indicated that RIIα mRNA levels increased after 6 hrs treatment with TSA (see below) a specific Polymerase IIa (PolIIa) antibody was used in ChIP to ascertain whether increased mRNA expression was associated with increased PolIIa binding to these RIIα promoter regions. Indeed for all three binding sites a 3–4-fold increase in binding of PolIIa was observed for cells treated with TSA whereas levels of binding did not significantly change for cells treated with 5-AzaC (100 μM) (Fig. 4B). All pull-downs were specific as monitored by Rabbit/Mouse IgG controls compared to DNA inputs (Fig. 4C).
Regulation of transcriptional activation of RIIα in human myometrial cells by acetylation or methylation
Full and truncated forms of the RIIα promoter region, detailed in Fig. 4, were used in transient transfection of human primary myometrial cell cultures to investigate the involvement of the three SpI-III (GC) consensus elements in transcriptional activity of the RIIα gene. Initially, basal luciferase activity was measured for the full length promoter and the three constructs with sequential deletion of the SpI-III (GC) elements. The highest expression as monitored by luciferase activity was observed for the promoter construct containing all three SpI-III (GC) elements (Fig. 4A) but lacking the distal part of the promoter, indicating that all three SpI-III (GC) elements are necessary for maximal expression of the RIIα gene. Because transfection of the full-length construct resulted in lower luciferase activities than that of the construct containing the three SpI-III (GC) elements this may indicate the presence of inhibitory elements up-stream of these GC binding domains. Sequential deletions of the SpIII and II elements resulted in decreasing luciferase activity compared to the construct containing all three elements (Fig. 5A). However the presence of the single SpI element was still enough to increase luciferase activity compared to the empty vector control (Fig. 5A). To elucidate whether inhibition of HDACs1/2 could enhance basic expression of the constructs; myometrial cells were transiently transfected with full-length and truncated luciferase constructs and treated for 6 hrs with TSA (330 nM). Here luciferase activity was significantly increased in all constructs compared to the empty vector control (Fig. 5B). No significant difference was seen between full-length and truncated constructs indicating that TSA maximally activated all constructs to the same level. This was similar to that found by us for the hCG/LH receptor gene [20]. To determine if DNA methylation might influence transcriptional activity of the RIIα gene, the promoter construct containing all three SpI-III (GC) elements was methylated in vitro with the CpG methyltransferase m.SssI. This methylated construct was then transfected into myometrial cells cultures and luciferase activity measured. Methylation of the promoter region significantly decreased basal luciferase activity compared to the non-methylated ‘mock’ construct (Fig. 5C). However methylation did not attenuate the effect of TSA treatment on transcriptional activity of this RIIα construct, as the luciferase signal was increased to the same extent in both the methylated and mock treated constructs (Fig. 5C).
Fig 5.
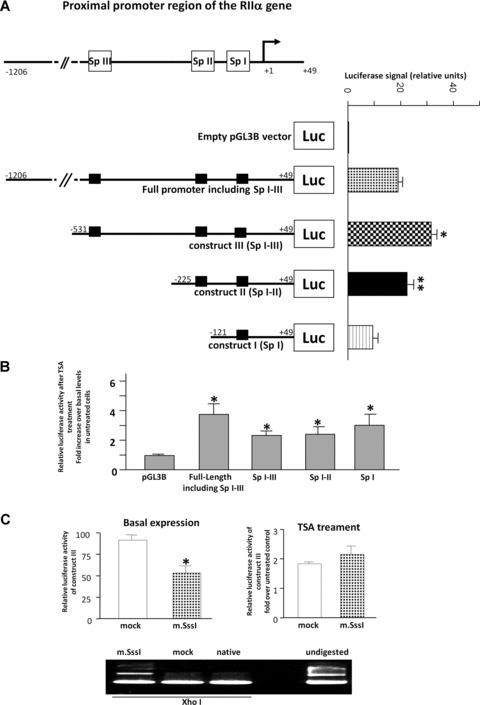
in vitro transcriptional activation of RIIα by TSA. (A) Diagrams of the 5′-flanking proximal promoter region of the RIIα gene containing the three Sp I-III (GC) binding domains and their sequential deletions. All numbers of constructs refer to the upstream sequence from the +1 (AGT) transcriptional start site. The closed LUC box refers to the coding region of the luciferase gene in the pGLB3 vector into which the varying lengths of the 59-flanking region of the RIIα promoter were cloned. Full-length and deleted constructs were transiently transfected into human myometrial cells and basal luciferase activity compared to the empty vector control measured with respect to a Renilla control. Data are expressed as mean ± S.E.M. Results were obtained from six myometrial cell preparations assayed in triplicate (n = 18). Construct III was significantly higher compared to the full length construct, construct II and construct I (*P < 0.05 Student’s t-test). Construct II was significantly higher compared to construct I (**P < 0.05 Student’s t-test) (B) The effect of 6 hrs TSA (330 νM) treatment on myometrial cells transfected with the RIIα full-length and Sp I-III (GC) deleted luciferase constructs in comparison to the empty pGL3B vector control. Results were obtained from six myometrial cell preparations assayed in triplicate (n = 18). Data are expressed as mean ± S.E.M. Results are expressed as a fold increase over basal levels in untreated cells. *P= 0.05 Student’s t-test compared to pGL3B empty vector control. (C) in vitro methylation with m.SssI methyltransferase of the RIIα luciferase construct III containing all three Sp I-III (GC) sites. Basal and 6 hrs treatment with TSA (330 νM) data were obtained from three myometrial cell preparations assayed in triplicate (n = 9). *P < 0.05 Student’s t-test basal m.Sss1 compared to mock. Restriction analysis with methylation site sensitive XhoI was used to show the effectiveness of CpG site methylation compared to mock treated and untreated native plasmid. Undigested plasmid is also shown as a control.
Regulation of RIIα protein levels in human myometrial cells following TSA or 5-AzαC treatment
To define whether inhibition of HDACs1/2 and DNA methylation status could affect RIIα protein expression levels in vivo in human myometrial cells; total protein extracts were prepared from myometrial cell cultures treated with TSA and 5-AzaC and subjected to Western blotting with the RIIα antibody. To determine the optimal treatment times, cells were incubated with TSA (330 nM) and 5-AzaC (100 μM) for 6, 24 and 48 hrs. TSA increased RIIα protein levels after 6 hrs which then slightly declined after 24 and 48 hrs but remained higher than control levels (Fig. 5A). 5-AzaC also increased RIIα protein levels after 6 hrs treatment, which was maintained up to 24 and 48 hrs (Fig. 6A). On the basis of these initial results 6 hrs treatment with TSA and 5-AzaC were chosen for further protein and mRNA analysis (as well as the above ChIP analyses). Note that a 6 hrs treatment of GH3 cells with TSA has been shown to significantly increase GH mRNA [28]. Similarly an 80% decrease in DNA methyltransferase activity has been observed in cell lines treated for 4 hrs with the demethylating agent 5-AzaC [29]. Employing this incubation time significant increases in RIIα protein levels were observed for both TSA and 5-AzaC treatment (Fig. 6B).
Fig 6.
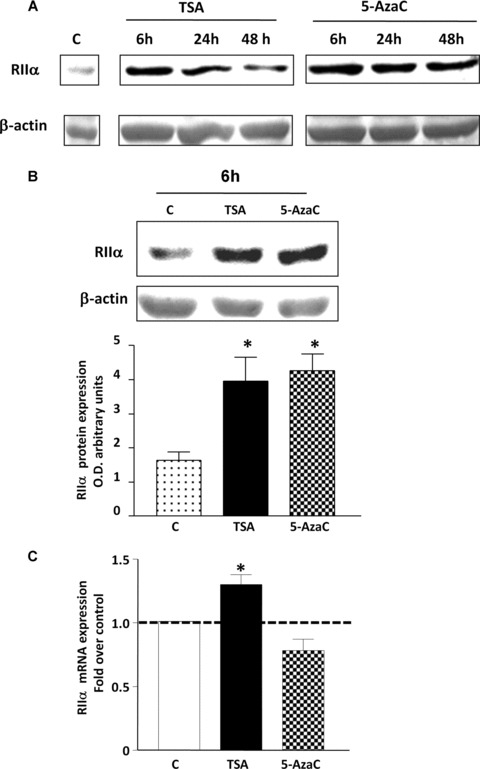
The effect of TSA and 5-AzaC treatment on protein and mRNA RIIα levels in human myometrial cell cultures. (A). Time course of treatment of myometrial cells with TSA (330 νM) and 5-AzaC (100 μM) for 6, 24 and 48 hrs. (B) 6 hrs treatment of human myometrial cell cultures with TSA and 5-AzaC as monitored by Western blotting. Data are expressed as mean ± S.E.M. Results were obtained from six myometrial cell preparations. *P, 0.05 Student’s t-test (n = 6) TSA and 5-AzaC treated compared to untreated control. In (A) and (B), equal loading was confirmed using a b-actin antibody. (C) Quantitative real time RT-PCR analysis of myometrial cells treated for 6 hrs with TSA (330 νM) and 5-AzaC (100 μM). Data are expressed as mean ± S.E.M. Results were obtained from four myometrial cell preparations each analysed in triplicate. *P < 0.05 Student’s t-test (n = 4) TSA treated compared to untreated control.
TSA, but not 5-AzaC, increases RIIα mRNA levels in human myometrial cells
Because Western blotting analysis revealed significant increases in protein levels of RIIα after 6 hrs treatment with TSA or 5-AzaC; we subsequently investigated if these changes were associated with an increase in mRNA levels. GAPDH was chosen as an internal control as no changes in gene expression were observed after TSA and 5-AzaC treatment (data not shown). ΔΔCt analysis showed a significant increase (31%) in mRNA levels (P < 0.05) in cells stimulated for 6 hrs with TSA (330 nM) compared to control untreated cells whereas no change was observed with 5-AzaC (100 mM) (Fig. 6C). No change in RIIα mRNA levels was observed in cells treated with 5-AzaC for either 12 or 24 hrs (data not shown).
TSA inhibits myometrial contractions
Because TSA treatment increases RIIα protein expression in cell cultures we investigated whether this also occurred in myometrial strips and defined the effect on myometrial contractility. Here, TSA caused a significant decrease in oxytocin-induced contractions (Fig. 7A–D) similar to that described in [30] and this was associated with an increase in RIIα protein expression and an increase in Histone 3 acetylation at 1 and 6 hrs (Fig. 7E).
Fig 7.
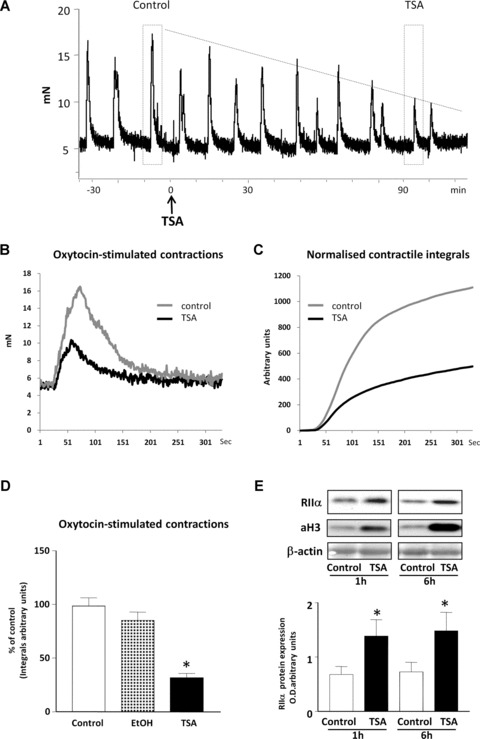
The effect of TSA on myometrial contractions. (A) Representative isometric tension recordings of oxytocin (10 μM) -induced contractions in human myometrial strips after TSA treatment (3.3 μM). Tension is represented in mN with time in minutes. Dashed boxes mark representative peaks of contraction for the myometrial tissue before treatment (control) and after 90 min. of TSA treatment (TSA). Comparison of the representative recordings and integrated area under the tension curve (integrals) for these peaks are presented in (B) and (C). (D) Effect of vehicle (EtOH) and TSA treatment on myometrial contractions measured as percentage of contraction of untreated control. Data are expressed as mean ± S.E.M. Results were obtained from three myometrial strips. *P < 0.05 Student’s t-test, TSA treated compared to untreated controls. (E) Effect of TSA treatment on expression of RIIα and acetylated Histone3 (aH3) levels in myometrial tissue strips treated for 1 and 6 hrs, respectively. Equal loading was confirmed using a β-actin antibody. Data are expressed as mean ± S.E.M. Results were obtained from three myometrial strips for 1 and 6 hrs, respectively. *P= 0.05 Student’s t-test, TSA treated compared to untreated control.
Discussion
Our data have established important features of the molecular regulation of the RIIα gene and protein expression in human smooth muscle cells. The human RIIα proximal promoter has the potential to recruit the Sp1-4 family of transcriptional factors to the multiple GC cis-elements within its proximal promoter to regulate its expression. Two such family members are Sp1 and Sp3 which are closely related zinc finger DNA binding transcription factors that are ubiquitously expressed in mammalian tissues [31–33]. In this context our results from in vitro DAPA and in vivo ChIP analysis indicate that all three SpI, II and III (GC) domains within the RIIα promoter are capable of specifically binding Sp1 and Sp3 which in turn recruit HDACs 1/2 and RbAp48 to regulate expression of the gene in human myometrial cells. A role for Sp1 and Sp3 in regulating RIIα gene expression in these cells is demonstrated by incubation with Mithramycin A which results in a decrease in basal levels of RIIα protein. Although it is realised that Mithramycin A may also potentially perturb the binding of other zinc finger family members within the KLF family which may also bind these GC-rich elements. However, siRNA targeted against Sp1 and Sp3 resulted in a substantial decrease in RIIα protein expression providing further evidence for the regulatory involvement of Sp1/Sp3 species. From co-IP experiments we show that, for each of the SpI-III (GC) binding sites within the RIIα promoter, Sp1 interacts with HDACs1/2 whereas Sp3 only interacts with HDAC2. No evidence was found for any interaction with the putative co-repressor mSin3A.
in vivo ChIP analysis on myometrial cells treated with the HDAC inhibitor TSA indicated that Sp3 and HDAC2 binding to the three SpI-III (GC) cis-elements within the RIIα promoter were significantly increased. Generally Sp1 is reported to be a transactivator [34] whereas Sp3 is strongly promoter dependent in that it can act as transcriptional activator [35] or a transcriptional repressor [36, 37]. A similar increase in Sp3 binding within the Major Vault Protein gene promoter due to TSA treatment of cells has also been reported [38]. This appeared to precede an increase in expression of the gene and was suggested to arise due to increased acetylation of the lysine residue in the inhibitory domain of Sp3 which enhanced its transcriptional activity [37]. Consequently the observed increase in binding of HDAC2 to the three GC cis-elements within the RIIα promoter due to TSA treatment may arise as a result of its interaction with Sp3 as determined by co-IPs. Results from myometrial ChIP assays also indicated a significant increase in binding of aH3 and PolIIa to the three SpI-III (GC) cis-elements within the RIIα promoter upon TSA treatment. Consequently the increase in RIIα mRNA levels brought about inhibition of HDAC1/2 activity by TSA would appear to involve a combination of increased H3 histone acetylation, changes in the ratios of transcriptional regulators that bind the three SpI-III (GC) cis-elements and a substantial increase in RNA polymerase II binding with its subsequent effect on transcription of the gene.
Results from reporter luciferase assays indicated that inhibition of HDAC1/2 activities by TSA resulted in a similar increase in luciferase signal, compared to control empty vector, for each of the RIIα constructs transfected. These data suggest that in the context of this in vitro experimental protocol each SpI-III (GC) binding domain within the RIIα gene is capable of maximal transcriptional activity. However under non-stimulated basal conditions the presence of all three SpI-III (GC) sites appeared to be essential for maximal transcriptional activity. These experiments also implicated the distal region of the promoter as having inhibitory properties. This was demonstrated by a decrease in the level of promoter luciferase activity with the full length construct compared to the construct containing only the three SpI-III (GC) sites. In this context the distal promoter region of the RIIα gene contains several AP-2, AP-1, NF-1 and NFkB regulatory elements that may have a repressive affect on transcription of the RIIα gene as monitored by these luciferase assays [39, 40].
Treatment of myometrial cells with TSA resulted in a significant 31% and 100% increase in RIIα mRNA and protein levels, respectively, and provides further evidence that HDAC1/2 activity may potentially regulate RIIα gene expression in vivo. In many cell types TSA administration results in a large rise in mRNA levels of the genes being studied. However, there are instances where small increases have also been detected as observed in this investigation. An example of this is the 50% increase in GH mRNA levels in GH3 cells treated with TSA [28]. The small increase in RIIα mRNA expression coupled with the large increase in protein expression induced by TSA may indicate that these mRNA species are highly stable and thus are not rapidly decayed. Indeed homology searches for the presence of AU-rich elements in the 3′-untranslated region (3′-UTR) of the human RIIα mRNA indicates the presence of only one copy of the pentamer AUUUA which may be involved in inhibiting (or enhancing) mRNA decay. Furthermore the larger increase in RIIα protein expression with respect to mRNA levels could arise as a result of inhibition by TSA of proteosome activity, which has been observed in colon cancer cell lines [40], this would thus increase the half-life of myometrial RIIα protein species. Importantly, data presented here also indicate that TSA treatment of myometrial strips under tension also causes a substantial increase in RIIα protein expression which is associated with an increase aH3 levels and results in a significant decrease in oxytocin-induced contractions.
Promoters with a high concentration of GC-rich sequences, known as CpG islands are often unmethylated and therefore transcriptionally active. However, our data from in vitro methylation of the RIIα promoter reporter luciferase construct containing all three SpI-III (GC) domains resulted in a decrease in basal luciferase activity compared to the mock treated control. This suggested that RIIα promoter methylation may potentially affect expression of the gene. However methylation of this construct did not alter the effect of TSA as similar luciferase activities were seen for both control mock and treated plasmids. If DNA methylation of the CpG island within the RIIα promoter played a role in transcriptional regulation of the gene in vivo then incubation of myometrial cells with 5-AzaC would result in demethylation of cytidine residues within this region leading to increased transcription with subsequent increased mRNA expression. This appears not to occur and thus negates a role for DNA methylation in regulating expression of RIIα mRNA in myometrial cells. Although no increase in RIIα mRNA levels was observed in cells treated with 5-AzaC a substantial increase in RIIα protein levels was detected. The mechanism(s) by which this occurs remains to be elucidated.
In conclusion we provide evidence to indicate that the RIIα gene in human smooth muscle cells is regulated by epigenetic mechanisms involving recruitment of HDAC 1/2 to multiple GC cis-elements within the proximal promoter of the gene. Increased expression of RIIα would be expected to limit contraction of human myometrial cells in vivo, which is reflected by the decrease in myometrial contractility observed here. Consequently, these studies indicate that specific HDAC 1/2 inhibitors could possibly be employed to maintain levels of RIIα with subsequent effects on myometrial quiescence during pregnancy. More generally, the data elucidate epigenetic mechanisms by which dynamic modulation of RIIα expression may occur in human smooth muscles.
Acknowledgments
The study was funded by a grant made available from Action Medical Research (SP3972). We wish to thank Prof. M.J. Taggart for helpful comments regarding the manuscript.
Conflict of interest
The authors have nothing to disclose.
References
- 1.Zuo J, Lei ZM, Rao CV. Human myometrial chorionic gonadotropin/luteinizing hormone receptors in preterm and term deliveries. J Clin Endocrinol Metab. 1994;79:907–11. doi: 10.1210/jcem.79.3.8077381. [DOI] [PubMed] [Google Scholar]
- 2.Dong YL, Fang L, Kondapaka S, et al. Involvement of calcitonin gene-related peptide in the modulation of human myometrial contractility during pregnancy. J Clin Invest. 1999;104:559–65. doi: 10.1172/JCI6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottari SP, Severne Y, Kaivez E, et al. Myometrial beta 1-adrenoreceptors are detectable only in the midfollicular phase. J Clin Endocrinol Metab. 1986;62:1220–6. doi: 10.1210/jcem-62-6-1220. [DOI] [PubMed] [Google Scholar]
- 4.Hayashida DN, Leung R, Goldfien A, et al. Human myometrial adrenergic receptors: identification of the beta-adrenergic receptor by [3H]dihydroalprenolol binding. Am J Obstet Gynecol. 1982;142:389–93. doi: 10.1016/s0002-9378(16)32378-x. [DOI] [PubMed] [Google Scholar]
- 5.Grammatopoulos DK. The role of CRH receptors and their agonists in myometrial contractility and quiescence during pregnancy and labour. Front Biosci. 2007;12:561–71. doi: 10.2741/2082. [DOI] [PubMed] [Google Scholar]
- 6.Grammatopoulos DK, Hillhouse EW. Role of corticotropin-releasing hormone in onset of labour. Lancet. 1999;354:1546–9. doi: 10.1016/S0140-6736(99)03418-2. [DOI] [PubMed] [Google Scholar]
- 7.Europe-Finner GN, Phaneuf S, Watson SP, et al. Identification and expression of G-proteins in human myometrium: up-regulation of G alpha s in pregnancy. Endocrinology. 1993;132:2484–90. doi: 10.1210/endo.132.6.8504751. [DOI] [PubMed] [Google Scholar]
- 8.Europe-Finner GN, Phaneuf S, Tolkovsky AM, et al. Down-regulation of G alpha s in human myometrium in term and preterm labor: a mechanism for parturition. J Clin Endocrinol Metab. 1994;79:1835–9. doi: 10.1210/jcem.79.6.7989491. [DOI] [PubMed] [Google Scholar]
- 9.Lopez Bernal A, Europe-Finner GN, Phaneuf S, et al. Preterm labour: a pharmacological challenge. Trends Pharmacol Sci. 1995;16:129–33. doi: 10.1016/s0165-6147(00)89000-8. [DOI] [PubMed] [Google Scholar]
- 10.Kofinas AD, Rose JC, Koritnik DR, et al. Progesterone and estradiol concentrations in nonpregnant and pregnant human myometrium. Effect of progesterone and estradiol on cyclic adenosine monophosphate-phosphodiesterase activity. J Reprod Med. 1990;35:1045–50. [PubMed] [Google Scholar]
- 11.Tasken K, Skalhegg BS, Tasken KA, et al. Structure, function, and regulation of human cAMP-dependent protein kinases. Adv Second Messenger Phosphoprotein Res. 1997;31:191–204. doi: 10.1016/s1040-7952(97)80019-5. [DOI] [PubMed] [Google Scholar]
- 12.MacDougall MW, Europe-Finner GN, Robson SC. Human myometrial quiescence and activation during gestation and parturition involve dramatic changes in expression and activity of particulate type II (RII alpha) protein kinase A holoenzyme. J Clin Endocrinol Metab. 2003;88:2194–205. doi: 10.1210/jc.2002-021862. [DOI] [PubMed] [Google Scholar]
- 13.Foss KB, Solberg R, Simard J, et al. Molecular cloning, upstream sequence and promoter studies of the human gene for the regulatory subunit RII[alpha] of cAMP-dependent protein kinase. Biochimica et Biophysica Acta (BBA) – Gene Struct Expr. 1997;1350:98–108. doi: 10.1016/s0167-4781(96)00152-2. [DOI] [PubMed] [Google Scholar]
- 14.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–58. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funk CD, Hoshiko S, Matsumoto T, et al. Characterization of the human 5-lipoxygenase gene. Proc Natl Acad Sci USA. 1989;86:2587–91. doi: 10.1073/pnas.86.8.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauerwald A, Hoesche C, Oschwald R, et al. The 5’-flanking region of the synapsin I gene. A G+C-rich, TATA- and CAAT-less, phylogenetically conserved sequence with cell type-specific promoter function. J Biol Chem. 1990;265:14932–7. [PubMed] [Google Scholar]
- 17.Feng D, Kan YW. The binding of the ubiquitous transcription factor Sp1 at the locus control region represses the expression of beta-like globin genes. Proc Natl Acad Sci USA. 2005;102:9896–900. doi: 10.1073/pnas.0502041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Dufau ML. Repression of the luteinizing hormone receptor gene promoter by cross talk among EAR3/COUP-TFI, Sp1/Sp3, and TFIIB. Mol Cell Biol. 2003;23:6958–72. doi: 10.1128/MCB.23.19.6958-6972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Dufau ML. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J Biol Chem. 2002;277:33431–8. doi: 10.1074/jbc.M204417200. [DOI] [PubMed] [Google Scholar]
- 20.Phillips RJ, Tyson-Capper Nee PollardAJ, Bailey J, et al. Regulation of expression of the chorionic gonadotropin/ luteinizing hormone receptor gene in the human myometrium: involvement of specificity protein-1 (Sp1), Sp3, Sp4, Sp-like proteins, and histone deacetylases. J Clin Endocrinol Metab. 2005;90:3479–90. doi: 10.1210/jc.2004-1962. [DOI] [PubMed] [Google Scholar]
- 21.Phillips RJ, Bailey J, Robson SC, et al. Differential expression of the adenylyl cyclase-stimulatory guanosine triphosphate-binding protein G(s)alpha in the human myometrium during pregnancy and labor involves transcriptional regulation by cyclic adenosine 3’,5’-monophosphate and binding of phosphorylated nuclear proteins to multiple GC boxes within the promoter. J Clin Endocrinol Metab. 2002;87:5675–85. doi: 10.1210/jc.2002-020640. [DOI] [PubMed] [Google Scholar]
- 22.Phaneuf S, Europe-Finner GN, Varney M, et al. Oxytocin-stimulated phosphoinositide hydrolysis in human myometrial cells: involvement of pertussis toxin-sensitive and -insensitive G-proteins. J Endocrinol. 1993;136:497–509. doi: 10.1677/joe.0.1360497. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bugg GJ, Riley MJ, Johnston TA, et al. Hypoxic inhibition of human myometrial contractions in vitro: implications for the regulation of parturition. Eur J Clin Invest. 2006;36:133–40. doi: 10.1111/j.1365-2362.2006.01600.x. [DOI] [PubMed] [Google Scholar]
- 25.Demicheli C, Albertini JP, Garnier-Suillerot A. Interaction of mithramycin with DNA. Evidence that mithramycin binds to DNA as a dimer in a right-handed screw conformation. Eur J Biochem. 1991;198:333–8. doi: 10.1111/j.1432-1033.1991.tb16020.x. [DOI] [PubMed] [Google Scholar]
- 26.Nehls MC, Brenner DA, Gruss HJ, et al. Mithramycin selectively inhibits collagen-alpha 1(I) gene expression in human fibroblast. J Clin Invest. 1993;92:2916–21. doi: 10.1172/JCI116914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Hirasawa N, Torigoe M, Kano K, et al. Involvement of Sp1 in lipopolysaccharide-induced expression of HDC mRNA in RAW 264 cells. Biochem Biophys Res Commun. 2006;349:833–7. doi: 10.1016/j.bbrc.2006.08.104. [DOI] [PubMed] [Google Scholar]
- 28.Lizcano F, Koibuchi N, Fukuda H, et al. Cell type-specific roles of histone deacetylase in TR ligand-independent transcriptional repression. Mol Cell Endocrinol. 2001;172:13–20. doi: 10.1016/s0303-7207(00)00400-7. [DOI] [PubMed] [Google Scholar]
- 29.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2’-deoxycytidine. J Biol Chem. 1982;257:2041–8. [PubMed] [Google Scholar]
- 30.Moynihan AT, Hehir MP, Sharkey AM, et al. Histone deacetylase inhibitors and a functional potent inhibitory effect on human uterine contractility. Am J Obstet Gynecol. 2008;199:167 e1–7. doi: 10.1016/j.ajog.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Saffer JD, Jackson SP, Annarella MB. Developmental expression of Sp1 in the mouse. Mol Cell Biol. 1991;11:2189–99. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagen G, Muller S, Beato M, et al. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–25. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supp DM, Witte DP, Branford WW, et al. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev Biol. 1996;176:284–99. doi: 10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- 34.Mastrangelo IA, Courey AJ, Wall JS, et al. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci USA. 1991;88:5670–4. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennett SB, Udvadia AJ, Horowitz JM. Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res. 1997;25:3110–7. doi: 10.1093/nar/25.15.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagen G, Muller S, Beato M, et al. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–51. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ammanamanchi S, Freeman JW, Brattain MG. Acetylated sp3 is a transcriptional activator. J Biol Chem. 2003;278:35775–80. doi: 10.1074/jbc.M305961200. [DOI] [PubMed] [Google Scholar]
- 38.Steiner E, Holzmann K, Pirker C, et al. SP-transcription factors are involved in basal MVP promoter activity and its stimulation by HDAC inhibitors. Biochem Biophys Res Commun. 2004;317:235–43. doi: 10.1016/j.bbrc.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 39.Lee SK, Kim JH, Lee YC, et al. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappaB, and serum response factor. J Biol Chem. 2000;275:12470–4. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- 40.Place RF, Noonan EJ, Giardina C. HDAC inhibition prevents NF-kappa B activation by suppressing proteasome activity: down-regulation of proteasome subunit expression stabilizes I kappa B alpha. Biochem Pharmacol. 2005;70:394–406. doi: 10.1016/j.bcp.2005.04.030. [DOI] [PubMed] [Google Scholar]


