Abstract
Genomes of human papillomaviruses (HPV) are common in biopsies from non-melanoma skin cancers but are also found on healthy skin and it is possible that HPV positivity in tumor biopsies by PCR may merely reflect contamination of the lesion surface. To investigate this issue, 229 immunocompetent patients were tested for HPV DNA in swab samples collected on top of skin tumors and in biopsies of the same tumors, obtained after stripping with tape to remove superficial layers. HPV DNA was detected on top of 69% (159 of 229) of the lesions, and in 12% (28 of 229) of the stripped biopsies (p<0.001). The difference was seen for all four types of tumors studied. Seborrheic keratosis had 79% (34 of 43) HPV positivity on top of lesions versus 19% (eight of 43) in biopsies; actinic keratosis had 83% (38 of 46) HPV positivity on top versus 11% (five of 46) in biopsies; basal cell carcinoma had 63% (69 of 109) on top versus 8% (nine of 109) in biopsies and squamous cell carcinoma had 58% (18 of 31) on top versus 19% (six of 31) in biopsies. HPV DNA is common in superficial layers of lesions, but is not necessarily present throughout tumors.
Keywords: HPV, PCR, superficial, tumors
Certain types of human papillomaviruses (HPV) have the potential to induce malignant and benign genital tumors (Burd, 2003). The HPV types from supergroup B (HPV 5 and related HPV types originally isolated from Epidermodysplasia verruciformis) have cutaneous tropism but their possible role in the etiology of non-melanoma skin cancers is not clear. The DNA of cutaneous HPV types is found both in pre-malignant lesions and non-melanoma skin cancers, especially among immunosuppressed organ transplant patients (Pfister and Ter Schegget, 1997; Kiviat, 1999; Bouwes Bavinck et al, 2001). Renal transplant patients have up to about 100-fold increased risk to develop cutaneous squamous cell carcinoma (SCC) (Lindelof et al, 2000), and the cumulative incidence of non-melanoma skin cancer 20 y after transplantation is as much as 70% in Australia (Bouwes Bavinck et al, 1996) and 40% in The Netherlands (Hartevelt et al, 1990).
Several possible mechanisms of how HPV might act as cofactors in cutaneous cell transformation have been proposed. The promoter activity of HPV type 77 is stimulated by ultraviolet radiation and the response is mediated through a binding site for the p53 tumor suppressor protein in the upper regulatory region of the virus (Purdie et al, 1999). Moreover, ultraviolet-induced cytokines and interferons have been found to activate the promoter of the cutaneous HPV type 20 (Ruhland and de Villiers, 2001). The E6 proteins of cutaneous HPV types seem to harbor relatively low oncogenic capability, showing transformation and anchorage-independent growth of rodent cells, but without forming tumors in nude mice (Iftner et al, 1988; Kiyono et al, 1989). Recently, primary human cells were reported to be transformed by E6 of HPV 38 (Caldeira et al, 2003); however, the E6 protein of the cutaneous HPV types appear to be unable to promote degradation of p53 (Steger and Pfister, 1992; Jackson et al, 2000), in contrast to that of oncogenic genital HPV types (Tommasino et al, 2003).
The E6 protein of cutaneous human papillomaviruses might promote genomic instability of infected cells since it was demonstrated that E6 of HPV types 1 and 8 (and HPV type 16) binds to the XRCC1 protein (required for the repair of DNA single-strand breaks and genetic stability) (Iftner et al, 2002). Also, the E6 protein of cutaneous HPV 5, 10, and 77 binds the pro-apoptotic Bak protein (Jackson and Storey, 2000). This could inhibit Bak-induced apoptosis following ultraviolet radiation that could result in accumulation of deleterious mutational changes. Mutations are frequent in SCC of the skin and more than 90% of such lesions demonstrate mutations in the tumor suppressor gene p53 (Leffell, 2000).
So far, the vast majority of studies to detect cutaneous HPV DNA have used sensitive PCR methods, whereas less sensitive methods such as Southern blot are only infrequently able to detect HPV in skin lesions (Kawashima et al, 1990), indicating very low amounts of viral genomes (Bens et al, 1998; Meyer et al, 2001). But common skin warts appear to have higher amounts of viral genomes, since HPV DNA (predominantly HPV 1 and 2) is readily detectable by the Southern blot technique (Corley et al, 1988).
A significant problem for investigations of an association between cutaneous HPV and non-melanoma skin cancer is that cutaneous HPV is part of the microbiological flora of healthy human skin (Boxman et al, 1997; Astori et al, 1998; Antonsson et al, 2000, 2003a, b; Wieland et al, 2000; Forslund et al, 2003). The site for replication of cutaneous HPV DNA might be stem cells of the hair follicle (Schmitt et al, 1996; Boxman et al, 1997, 2000b) or of eccrine ducts (Egawa, 2003). These sites could also be the origin for nonmelanoma skin cancer (Egawa, 2003; Perez-Losada and Balmain, 2003); however, it is not known whether cutaneous HPV are involved in the development of skin cancers. Neither is it known if findings of HPV DNA from healthy skin represent viral particles or superficial cells containing episomal HPV genomes.
If viral particles are produced and shed from infected healthy skin, they are likely to be contaminating large areas of the body surface, including the surface of skin tumors. If so, punch biopsies of tumors might be scored as HPV positive by PCR, although the viral DNA is not present throughout the tumor. To address this question, we investigated whether the HPV DNA prevalence on top of skin lesions was similar to that in stripped biopsies from the same tumors.
Results
Quality control
In the DNA quality/quantity test, the biopsies and the swab samples from the lesions were found positive for the β-globin gene in 100% (229 of 229) and 89% (203 of 229), respectively. The β-globin negative swab samples were found among seborrheic keratosis (SK) in 37% (16 of 43), actinic keratosis (AK) in 6% (three of 46), and basal cell carcinomas (BCC) in 6% (seven of 109). The proportions of HPV DNA positivity among these β-globin negative samples were 69% (11 of 16) for SK, 66% (two of three) for AK, and 28% (two of seven) for BCC.
Prevalence of HPV DNA
HPV DNA was detected in 69% (159 of 229) of the swab samples collected on the top of the lesions, whereas a significantly lower HPV DNA prevalence of 12% (28 of 229) was detected in the stripped biopsies of the same lesions (p<0.001). Specifically, HPV DNA was detected in 79% (34 of 43) of the swab samples vs 19% (eight of 43) in the stripped biopsies of SK, 83% (38 of 46) vs 11% (five of 46) of AK, 63% (69 of 109) vs 8% (nine of 109) of BCC, and 58% (18 of 31) vs 19% (six of 31) of SCC (Table I).
Table I. Prevalence of HPV DNA in swab samples taken from the top of lesions and in biopsies (from the same lesions previously stripped to remove superficial layers), and in swab samples from healthy skin.
| Diagnosis | Lesion swab | Lesion biopsy | Perilesion swab | Forehead swab | Buttock swab |
|---|---|---|---|---|---|
| Seborrheic keratosis | 79% (34 of 43) | 19% (eight of 43) | 69% (30 of 43) | 77% (33 of 43) | 84% (36 of 43) |
| 23% (10 of 43) | 7% (three of 43) | 37% (16 of 43) | 32% (14 of 43) | 30% (13 of 43) | |
| Actinic keratosis | 83% (38 of 46) | 11% (five of 46) | 89% (41 of 46) | 89% (41 of 46) | 69% (32 of 46) |
| 41% (19 of 46) | 2% (one of 46) | 52% (24 of 46) | 47% (22 of 46) | 32% (15 of 46) | |
| BCC | 63% (69 of 109) | 8% (nine of 109) | 76% (83 of 109) | 82% (90 of 109) | 68% (74 of 109) |
| 27% (29 of 109) | 3% (three of 109) | 38% (41 of 109) | 49% (54 of 109) | 31% (34 of 109) | |
| SCC | 58% (18 of 31) | 19% (six of 31) | 81% (25 of 31) | 94% (29 of 31) | 81% (25 of 31) |
| 13% (four of 31) | 6% (two of 31) | 30% (nine of 31) | 48% (15 of 31) | 35% (11 of 31) | |
| Totally | 69% (159 of 229) | 12% (28 of 229) | 78% (179 of 229) | 84% (193 of 229) | 73% (167 of 229) |
| 27% (62 of 229) | 4% (nine of 229) | 39% (90 of 229) | 46% (105 of 229) | 32% (73 of 229) |
Italicised figures indicate prevalence of high copy numbers of amplicons (≥+ + + on gel).
HPV DNA, human papillomavirus DNA; BCC, basal cell carcinomas; SCC, squamous cell carcinoma.
Swab samples from the top of the lesions with benign diagnoses as AK and SK showed significantly higher HPV DNA prevalences than that in swab samples of malignant lesions (SCC and BCC) (AK vs SCC, p=0.02; AK vs BCC, p=0.01; SK vs SCC, p=0.04; SK vs BCC, p=0.04). Among swab samples containing large amounts of amplicons (band intensity ≥+ + +), AK had the highest HPV DNA prevalence of 41% (19 of 46), significantly higher than 13% (four of 31) found in SCC (p<0.010) (Table I).
There was at least one common HPV type detected in 52% (13 of 25) of HPV-positive pairs of stripped biopsies and superficial lesional swabs (Table S1).
For swab samples collected at other sites, HPV DNA prevalences between 73% and 84% were observed, which decreased to between 32% and 46% for samples containing large amounts of amplicons (≥+ + +) (Table I).
HPV sequences
Among the 28 HPV-positive biopsies, 35 different HPV sequences were identified. Seven were completely characterized HPV types, 26 were designated FA types (HPV sequences of about 430 nucleotides amplified with the FAP59 and FAP64 primers, either homologous to previously described FA sequences or classified as “new” FA types), and one vs102-4 and one vs92.1 (Table S1). HPV sequences within the B1 group were most common, found in 89% (25 of 28) of the HPV positive biopsies, followed by B2 sequences found in 18% (five of 28), and one type (HPV 27) from the A4 group in one sample (patient 7) (Table S1). Multiple HPV sequences were found in 29% (eight of 28) of the HPV-positive biopsies.
Among the 44 HPV-positive swab samples from lesions, 51 different HPV sequences were identified. Nine were completely characterized HPV types, 39 were designated FA types and the two isolates vs102-4 and vs92.1 were also detected (Table S1). HPV sequences within the B1 group were found in 84% (37 of 44) of the HPV-positive swab samples from lesions, followed by B2 types found in 41% (18 of 44) (Table S1). Multiple HPV sequences were found in 57% (25 of 44) of the HPV-typed swab samples. No distinct HPV type predominated, but HPV type 23 was detected in five samples (three SCC and two AK) and HPV 5 in four samples (Table S1).
Among the HPV-typed samples, nine new putative HPV types, 14 new putative subtype, and five new putative variants were identified (Table S2, on-line supplemental material).
Detection of HPV particles in a swab sample
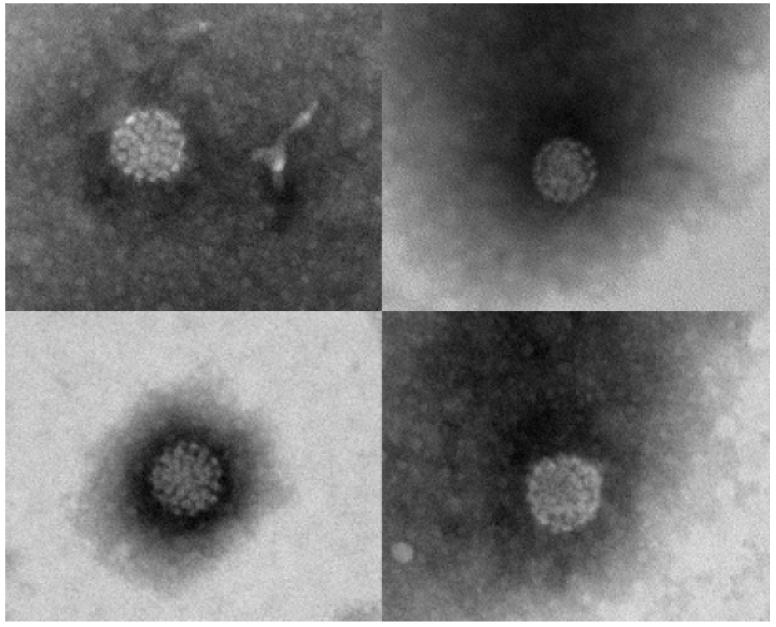
To investigate if the presence of viral DNA in swab samples may be due to viral particles, a swab sample from perilesional healthy skin of a patient with SCC (patient 20 in the reference Forslund et al, 2003) was analyzed by electron microscopy. Viral particles of typical HPV morphology were detected (Fig 1).
Figure 1. Human papillomavirus (HPV) particles from perilesional healthy skin of an Australian patient with squamous cell carcinoma.
The HPV particles were visualized by electron microscopy from a sample collected with a cotton swab. DNA from HPV types 23, 24, and 92 were identified.
Effect of the stripping on the stratum corneum
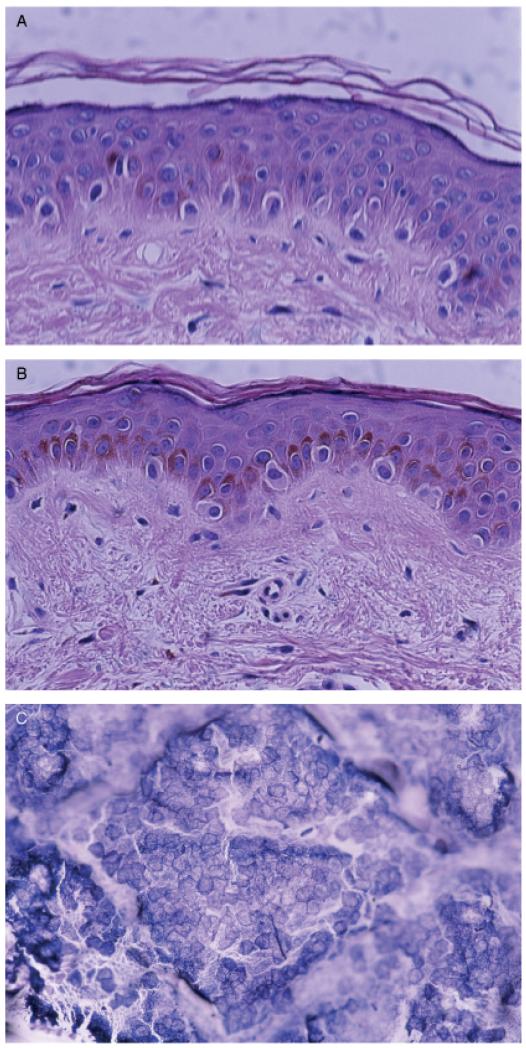
In order to investigate the stripping effect on stratum corneum, biopsies from non-stripped healthy skin and from the same healthy skin after stripping were collected from one of the investigators. No difference in the thickness of the stratum corneum could be observed by histopathological examination (Fig 2A and B). But on the adhesive surface of the tape a dense layer of corneocytes were present (Fig 2C).
Figure 2. Illustration of stratum corneum.
(A) Non-stripped healthy skin, (B) stripped healthy skin, and (C) corneocytes on tape after stripping.
Discussion
This is a study aimed to investigate specifically whether the HPV DNA prevalence on top of skin tumors differs from the prevalence in biopsies from the same tumors. The prevalence of HPV DNA was indeed much higher (69%) in the swab samples collected from lesions than in biopsies taken after removal of superficial cell layers by repeated tape stripping (12%). The result demonstrates that cutaneous HPV DNA is commonly present on top of skin lesions but to a lesser extent throughout the lesions. The fact that swab samples from healthy skin sites were found to be HPV positive to about the same extent as swab samples from the top of tumors, suggests that presence of virus is not specifically related to the tumor below. Whether the presence of viral DNA in superficial skin is due to viral particles or to episomal HPV genomes of infected cells is not known. We could demonstrate viral particles in one swab sample from perilesional skin of a patient with SCC, suggesting that presence of viral particles is at least contributing to the presence of HPV DNA in such samples.
In a previous study of Australian immunocompetent patients with skin tumors, 71% (29 of 41) and 36% (15 of 41) of paired perilesional swab samples and biopsies (collected under standard surgical sterile conditions), respectively, were found HPV positive by PCR (Forslund et al, 2003). The proportion of samples with at least one HPV type in common between paired biopsies and HPV-positive perilesional swab samples was 38% (11 of 29). But in this study, the corresponding proportion of at least one HPV type in common between the stripped biopsies and HPV-positive superficially collected samples from the same lesions was only 8% (13 of 159).
Multiple HPV types were more commonly detected among swab samples (57%) compared with that of biopsies (31%) (p=0.03). But since the determination of HPV types was limited to analysis of only three clones from each sample, it is possible that higher proportions of mixed infections could have been demonstrated if more clones had been analyzed.
The low prevalence of HPV in stripped biopsies suggests that stripping is rather effective for removal of superficial layers harboring HPV DNA from the lesions. The method of tape stripping has previously been used in atopy patch tests on non-lesional skin (Langeveld-Wildschut et al, 1995; Tengvall Linder et al, 2000) and is expected to reduce the superficial layers of the normal epidermis (predominantly the stratum corneum). But we were not able to detect any obvious effect of the tape stripping in histopathological morphology after tape stripping of healthy epidermis. The presence of a dense layer of corneocytes on the tape indicates that the outermost loose part of stratum corneum is removed by the stripping.
Overall, the HPV DNA prevalence on top of the tumors (69%) was similar to that of swab samples from healthy skin, taken at perilesional sites (78%), foreheads (84%), and from the buttock (73%). It should be noted that the area for collection of top of tumor samples was limited to the size of the tumor (usually 1 and 2 cm in diameter) whereas a larger area of about 5 × 5 cm was used for collection of superficial swab samples from the healthy skin.
Among three swab samples from top of lesions, no HPV DNA was detected although HPV DNA was found in the stripped biopsies. Possibly, the larger amount of cells in the biopsy may have resulted in improved ability to detect the HPV DNA.
In the previous study of Australian skin tumor patients, a relatively high HPV DNA prevalence was reported in AK by the FAP-PCR method (60%, six of ten) (Forslund et al, 2003). This HPV DNA prevalence, is considerably higher than the finding of 13% (six of 45) detected among the stripped AK in this study (p=0.004). Although population differences cannot be excluded, it seems likely that the approach of stripping the lesion before taking the biopsy is the main reason for the reduced prevalence. Overall, the HPV DNA prevalences of stripped punch biopsies in our study are considerably lower than those found in other studies using fresh biopsies, collected under standard sterile conditions from immunocompetent patients, and analyzed with various PCR protocols, e.g., among our biopsies with the diagnosis of BCC, 8% (nine of 109) demonstrated HPV DNA. The HPV DNA prevalences in BCC in five other studies ranged from 21% to 55% (Shamanin et al, 1996; Harwood et al, 2000; Meyer et al, 2000, 2001; Wieland et al, 2000). For SCC, we detected HPV DNA in 19% (six of 31) of the biopsies compared with prevalences between 27% and 54% in four other studies (Shamanin et al, 1996; Harwood et al, 2000; Meyer et al, 2000, 2001). For AK, we observed an HPV DNA prevalence of 11% (five of 46) compared with 36% (five of 14) by Meyer et al (2001), and to 85% (46 of 54) by Pfister et al (2003). Except for the differences in the sampling procedure (tape stripping), there are also differences in the PCR methods used; however, the FAP PCR method used in this study is established as having high sensitivity for detection of HPV DNA on the skin of almost all healthy adult individuals (Forslund et al, 1999; Antonsson et al, 2000).
The fact that swab samples from the top of benign lesions were somewhat more commonly HPV positive (81%) than swab samples taken from the top of malignant tumors (61%) may have several possible explanations, e.g., keratinization or morphological features resulting in an increased tendency to trap environmental HPV. High amounts of amplicons were detected among swab samples from AK (42% had band intensity of ≥+ + +). This suggests that a large amount of HPV DNA is present superficially among these lesions. But the intensity of a PCR band is dependent of the amplification efficiency of different HPV genotypes with the general primers used and the scored band intensities should not be regarded as absolute, quantitative values. Furthermore, the variation of band intensities between swab samples could also depend on the amount of collected cells.
Interestingly, HPV DNA has been detected in 100% (11 of 11) of a benign neoplasm (Trichilemmomas) of the hair follicle (Rohwedder et al, 1997). But in non-melanoma skin cancer HPV DNA is reported to be lost after in vitro passage (Purdie et al, 1993; Boxman et al, 2000a), suggesting that the virus is not needed for maintenance of the malignant phenotype or may even have been an environmental contaminant of the specimen.
In conclusion, we have shown that cutaneous HPV DNA is commonly present on the top of both skin lesions and healthy skin, but is less common in the skin lesions themselves. Consideration of the difference between viral presence throughout the lesion and presence only on the top of the lesions may be important in further studies aiming to elucidate a putative role of HPV in non-melanoma skin cancers.
Materials and Methods
Patients and sample collection
Two hundred and twenty-nine immunocompetent patients attending dermatology clinics in Sweden (Stockholm, 107 patients; Gothenburg, 44 patients; Malmö, 69 patients) and Austria (Vienna, nine patients) were, following informed consent, providing swab and biopsy samples for HPV DNA analysis. Subjects included were restricted to those with the diagnoses of SCC, BCC, AK, and SK. The mean age was 74 y (range, 34–95 y), and 128 males and 101 women participated.
For collection of superficial cells, a cotton-tipped swab was pre-wetted in saline (0.9% NaCl), rolled on the lesion (within margins of the lesion), and suspended in 1 mL of saline.
From each patient, samples were also collected from healthy perilesional skin, foreheads and buttock skin by cotton-tipped swabs that were drawn to and fro 15 times within an area of 5 × 5 cm, and suspended in 1 mL of saline.
Before taking a biopsy sample, the skin was anesthetized (without cleaning with ethanol) and stripped with tape (Art no 1527-1, Transpore, 3M Health Care, St Paul Minnesota) that was attached five times, then new tape was attached for another five times. Thereafter, a 2 mm diameter punch biopsy was taken from the tumor and the remaining tumor was excised and sent for histopathological analysis as per normal procedures. AK and SK were in most cases only diagnosed clinically. Samples were stored at −20°C and shipped frozen to the laboratory, except samples from Gothenburg that were delivered unfrozen by overnight express mail. The DNA from each punch biopsy was extracted and dissolved in 200 μL TE buffer using a simple phenol-free method (Forslund et al, 1999). For each swab sample, 5 μL of the 1 mL suspension was added directly to the PCR without DNA extraction.
Consensus primer PCR
The 25 μL PCR solution contained 5 μL of the sample and 0.75 μM of each primer FAP59 and FAP64 (Forslund et al, 1999), 0.2 mM of each dNTP (Roche, Mannheim, Germany), 0.2% BSA (Fraction V, Sigma, St. Louis, Missouri), 0.625 U AmpliTaq Gold DNA polymerase, GeneAmp 1 × PCR buffer II and 3.5 mM MgCl2 (Perkin-Elmer, Foster City, California). The PCR was performed in a thermal cycler (Hybaid Omnigene, Middlesex, UK), programmed for block temperature, using the parameters: 10 min at 94°C followed by 45 cycles of 1.5 min at 94°C, 1.5 min at 50°C, and 1.5 min at 72°C. In each test panel, H2O without DNA was included as a negative control and 2000 copies of recombinant HPV 5 genomes served as positive control. Five μL aliquots of the PCR products were separated by electrophoresis in 2% agarose gel (Type 1-A, Sigma-Aldrich, St Louis, Missouri) with ethidium bromide (20 ng per mL, Sigma-Aldrich) in 1 × TBE buffer (Sambrook et al, 1989). The HPV-specific amplicons were identified by size determination in ultraviolet light. The intensities of the amplicons varied between samples and were therefore graded arbitrarily from one to four (most intensive) in comparisons with the positive control of HPV 5.
HPV typing
Amplicons were cloned (three clones from each sample were usually obtained) by the TOPO TA CloningKit (Invitrogen, Leek, The Netherlands) and sequenced. Owing to limited resources, not all amplicons were sequenced. All positive “pairs” of HPV-positive biopsies and swabs were sequenced. In one case, the “positive pair” was not confirmed (the “positive” band was found to be human DNA in the sequencing). In addition, the first 16 of the 45 swab samples from top of tumors that had amplicons intensities of ≥+ + + and the two first-collected HPV-positive swab samples (<+ +) from top of tumors were sequenced.
The nucleotide sequences were generated via cycle sequencing (ABI Prism BigDye Terminator Cycle Sequencing, Applied Biosystems, California) and an automated DNA sequencer (3730 DNA Sequencer, Applied Biosystems). The sequences were compared with the GenBank database by the Blast program (http://www.ncbi.nlm.nih.gov/BLAST/).
A new putative HPV type was designated FA and a consecutive number, if the sequence showed <90% sequence homology with any of previously known HPV sequences. FA isolates showing sequence homology between 90% and <100% (90%–97% homology=subtype, 98% and <100% homology=variant) to an earlier identified FA isolate were given the same FA number plus an extra succession number. Subtypes with a sequence homology between greater than 90% and 97% to any completely characterized HPV type were also designated FA and a consecutive number.
Following sequence comparisons, the detected HPV types/candidates were grouped according to phylogenetic classification described by Chan et al (1995).
Quality control
The quality/quantity of DNA of biopsy samples and swab samples from lesions were analyzed in separate tubes by using a PCR amplifying the human β-globin gene. The 25 μL PCR solution contained 2.5 μL of the sample and 0.5 μM of each primer for the human β-globin gene PC03 and PC05 (de Roda Husman et al, 1995), 0.2 mM of each dNTP (Roche), 0.2% BSA (Fraction V, Sigma), 0.625 U AmpliTaq Gold DNA polymerase, GeneAmp 1 × PCR buffer II, and 3.5 mM MgCl2 (Perkin-Elmer).The PCR was performed in a thermal cycler (Mastercycler, Eppendorf, Hamburg, Germany), programmed for block temperature, using the parameters: 10 min at 94°C followed by 45 cycles of 1.5 min at 94°C, 1.5 min at 45°C, and 1.5 min at 72°C. In each batch of tests, H2O without DNA was included as a negative control. Five μL aliquots from each PCR were then analyzed by PCR-EIA (Forslund et al, 2002).
Statistical analysis
The Fisher exact probability test was used to compare subgroups with regard to the prevalences of HPV DNA. p-values below 0.05 were regarded statistically significant.
The study adheres to the Declaration of Helsinki Guidelines and was approved by the Ethics Committees of Karolinska Institute and of Lund University, Sweden, and of Medical University of Vienna, Austria.
Acknowledgments
Supported by grants from the European Commission (“Viraskin” QLK2-CT-2002-01500), the Science Council of Sweden, the County Council of the Scania Region and the Cancer Foundation of the University Hospital, Malmö. Kjell-Olof Hedlund, the Swedish Institute for Infectious Disease Control is acknowledged for the electron microscopy analysis. Mari-Anne Hedblad, Department of Dermatology, Karolinska Hospital, Stockholm, Sweden, is acknowledged for the illustrations and histological analysis of “stripped” stratum corneum. The laboratory assistance of Christina Gerrouda, Kristin Olsson and Tobias Hey is gratefully acknowledged.
Abbreviations
- AK
actinic keratosis
- BCC
basal cell carcinomas
- HPV
human papillomavirus
- SCC
squamous cell carcinoma
- SK
seborrheic keratosis
Footnotes
Supplementary Material The following material is available from http://www.blackwellpublishing.com/products/journals/suppmat/JID/JID23205/JID23205sm.htm
References
- Antonsson A, Erfurt C, Hazard H, et al. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol. 2003a;84:1881–1886. doi: 10.1099/vir.0.18836-0. [DOI] [PubMed] [Google Scholar]
- Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J Virol. 2000;74:11636–11641. doi: 10.1128/jvi.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Karanfilovska S, Lindqvist PG, Hansson BG. General acquisition of human papillomavirus infections of skin occurs in early infancy. J Clin Microbiol. 2003b;41:2509–2514. doi: 10.1128/JCM.41.6.2509-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astori G, Lavergne D, Benton C, Hockmayr B, Egawa K, Garbe C, de Villiers EM. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J Invest Dermatol. 1998;110:752–755. doi: 10.1046/j.1523-1747.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- Bens G, Wieland U, Hofmann A, Hopfl R, Pfister H. Detection of new human papillomavirus sequences in skin lesions of a renal transplant recipient and characterization of one complete genome related to epidermodysplasia verruciformis-associated types. J Gen Virol. 1998;79:779–787. doi: 10.1099/0022-1317-79-4-779. [DOI] [PubMed] [Google Scholar]
- Bouwes Bavinck JN, Feltkarmp M, Struijk L, ter Scheggett J. Human papillomavirus infection and skin cancer risk in organ transplant recipients. J Investig Dermatol Symp Proc. 2001;6:207–211. doi: 10.1046/j.0022-202x.2001.00048.x. [DOI] [PubMed] [Google Scholar]
- Bouwes Bavinck JN, Hardie DR, Green A, et al. The risk of skin cancer in renal transplant recipients in Queensland, Australia. A follow-up study. Transplantation. 1996;61:715–721. doi: 10.1097/00007890-199603150-00008. [DOI] [PubMed] [Google Scholar]
- Boxman IL, Berkhout RJ, Mulder LH, Wolkers MC, Bouwes Bavinck JN, Vermeer BJ, ter Schegget J. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J Invest Dermatol. 1997;108:712–715. doi: 10.1111/1523-1747.ep12292090. [DOI] [PubMed] [Google Scholar]
- Boxman IL, Mulder LH, Vermeer BJ, Bavinck JN, ter Schegget J, Ponec M. HPV-DNA is not detectable in outgrowing cells from explant cultures of skin lesions established at the air–liquid–interface. J Med Virol. 2000a;61:281–288. doi: 10.1002/1096-9071(200007)61:3<281::aid-jmv1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Boxman IL, Russell A, Mulder LH, Bavinck JN, Schegget JT, Green A. Case–control study in a subtropical Australian population to assess the relation between non-melanoma skin cancer and epidermodysplasia verruciformis human papillomavirus DNA in plucked eyebrow hairs. The Nambour Skin Cancer Prevention Study Group. Int J Cancer. 2000b;86:118–121. doi: 10.1002/(sici)1097-0215(20000401)86:1<118::aid-ijc18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldeira S, Zehbe I, Accardi R, et al. The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties. J Virol. 2003;77:2195–2206. doi: 10.1128/JVI.77.3.2195-2206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Delius H, Halpern AL, Bernard HU. Analysis of genomic sequences of 95 papillomavirus types: Uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley E, Pueyo S, Goc B, Diaz A, Zorzopulos J. Papillomaviruses in human skin warts and their incidence in an Argentine population. Diagn Microbiol Infect Dis. 1988;10:93–101. doi: 10.1016/0732-8893(88)90046-6. [DOI] [PubMed] [Google Scholar]
- de Roda Husman AM, Snijders PJ, Stel HV, van den Brule AJ, Meijer CJ, Walboomers JM. Processing of long-stored archival cervical smears for human papillomavirus detection by the polymerase chain reaction. Br J Cancer. 1995;72:412–417. doi: 10.1038/bjc.1995.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa K. Do human papillomaviruses target epidermal stem cells? Dermatology. 2003;207:251–254. doi: 10.1159/000073085. [DOI] [PubMed] [Google Scholar]
- Forslund O, Antonsson A, Edlund K, et al. Population-based type-specific prevalence of high-risk human papillomavirus infection in middle-aged Swedish Women. J Med Virol. 2002;66:535–541. doi: 10.1002/jmv.2178. [DOI] [PubMed] [Google Scholar]
- Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80:2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- Forslund O, Ly H, Reid C, Higgins G. A broad spectrum of human papillomavirus types is present in the skin of Australian patients with non-melanoma skin cancers and solar keratosis. Br J Dermatol. 2003;149:64–73. doi: 10.1046/j.1365-2133.2003.05376.x. [DOI] [PubMed] [Google Scholar]
- Hartevelt MM, Bavinck JN, Kootte AM, Vermeer BJ, Vandenbroucke JP. Incidence of skin cancer after renal transplantation in The Netherlands. Transplantation. 1990;49:506–509. doi: 10.1097/00007890-199003000-00006. [DOI] [PubMed] [Google Scholar]
- Harwood CA, Surentheran T, McGregor JM, Spink PJ, Leigh IM, Breuer J, Proby CM. Human papillomavirus infection and non-melanoma skin cancer in immunosuppressed and immunocompetent individuals. J Med Virol. 2000;61:289–297. doi: 10.1002/1096-9071(200007)61:3<289::aid-jmv2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Iftner T, Bierfelder S, Csapo Z, Pfister H. Involvement of human papillomavirus type 8 genes E6 and E7 in transformation and replication. J Virol. 1988;62:3655–3661. doi: 10.1128/jvi.62.10.3655-3661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftner T, Elbel M, Schopp B, Hiller T, Loizou JI, Caldecott KW, Stubenrauch F. Interference of papillomavirus E6 protein with single-strand break repair by interaction with XRCC1. EMBO J. 2002;21:4741–4748. doi: 10.1093/emboj/cdf443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Harwood C, Thomas M, Banks L, Storey A. Role of bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 2000;14:3065–3073. doi: 10.1101/gad.182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Storey A. E6 proteins from diverse cutaneous HPV types inhibit apoptosis in response to UV damage. Oncogene. 2000;19:592–598. doi: 10.1038/sj.onc.1203339. [DOI] [PubMed] [Google Scholar]
- Kawashima M, Favre M, Obalek S, Jablonska S, Orth G. Premalignant lesions and cancers of the skin in the general population: Evaluation of the role of human papillomaviruses. J Invest Dermatol. 1990;95:537–542. doi: 10.1111/1523-1747.ep12504887. [DOI] [PubMed] [Google Scholar]
- Kiviat NB. Papillomaviruses in non-melanoma skin cancer: Epidemiological aspects. Semin Cancer Biol. 1999;9:397–403. doi: 10.1006/scbi.1999.0143. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Nagashima K, Ishibashi M. The primary structure of major viral RNA in a rat cell line transfected with type 47 human papillomavirus DNA and the transforming activity of its cDNA and E6 gene. Virology. 1989;173:551–565. doi: 10.1016/0042-6822(89)90567-9. [DOI] [PubMed] [Google Scholar]
- Langeveld-Wildschut EG, van Marion AM, Thepen T, Mudde GC, Bruijnzeel PL, Bruijnzeel-Koomen CA. Evaluation of variables influencing the outcome of the atopy patch test. J Allergy Clin Immunol. 1995;96:66–73. doi: 10.1016/s0091-6749(95)70034-x. [DOI] [PubMed] [Google Scholar]
- Leffell DJ. The scientific basis of skin cancer. J Am Acad Dermatol. 2000;42:18–22. doi: 10.1067/mjd.2000.103340. [DOI] [PubMed] [Google Scholar]
- Lindelof B, Sigurgeirsson B, Gabel H, Stern RS. Incidence of skin cancer in 5356 patients following organ transplantation. Br J Dermatol. 2000;143:513–519. [PubMed] [Google Scholar]
- Meyer T, Arndt R, Christophers E, Nindl I, Stockfleth E. Importance of human papillomaviruses for the development of skin cancer. Cancer Detect Prev. 2001;25:533–547. [PubMed] [Google Scholar]
- Meyer T, Arndt R, Christophers E, Stockfleth E. Frequency and spectrum of HPV types detected in cutaneous squamous-cell carcinomas depend on the HPV detection system: A comparison of four PCR assays. Dermatology. 2000;201:204–211. doi: 10.1159/000018489. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Balmain A. Stem-cell hierarchy in skin cancer. Nat Rev Cancer. 2003;3:434–443. doi: 10.1038/nrc1095. [DOI] [PubMed] [Google Scholar]
- Pfister H, Fuchs PG, Majewski S, Jablonska S, Pniewska I, Malejczyk M. High prevalence of epidermodysplasia verruciformis-associated human papillomavirus DNA in actinic keratoses of the immunocompetent population. Arch Dermatol Res. 2003;295:273–279. doi: 10.1007/s00403-003-0435-2. [DOI] [PubMed] [Google Scholar]
- Pfister H, Ter Schegget J. Role of HPV in cutaneous premalignant and malignant tumors. Clin Dermatol. 1997;15:335–347. doi: 10.1016/s0738-081x(96)00162-9. [DOI] [PubMed] [Google Scholar]
- Purdie KJ, Pennington J, Proby CM, Khalaf S, de Villiers EM, Leigh IM, Storey A. The promoter of a novel human papillomavirus (HPV77) associated with skin cancer displays UV responsiveness, which is mediated through a consensus p53 binding sequence. EMBO J. 1999;18:5359–5369. doi: 10.1093/emboj/18.19.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie KJ, Sexton CJ, Proby CM, Glover MT, Williams AT, Stables JN, Leigh IM. Malignant transformation of cutaneous lesions in renal allograft patients: A role for human papillomavirus. Cancer Res. 1993;53:5328–5333. [PubMed] [Google Scholar]
- Rohwedder A, Keminer O, Hendricks C, Schaller J. Detection of HPV DNA in Trichilemmomas by polymerase chain reaction. J Med Virol. 1997;51:119–125. doi: 10.1002/(sici)1096-9071(199702)51:2<119::aid-jmv6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Ruhland A, de Villiers EM. Opposite regulation of the HPV 20-URR and HPV 27-URR promoters by ultraviolet irradiation and cytokines. Int J Cancer. 2001;91:828–834. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1129>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Sambrook JE, Fritsch F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schmitt A, Rochat A, Zeltner R, Borenstein L, Barrandon Y, Wettstein FO, Iftner T. The primary target cells of the high-risk cottontail rabbit papillomavirus colocalize with hair follicle stem cells. J Virol. 1996;70:1912–1922. doi: 10.1128/jvi.70.3.1912-1922.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanin V, zur Hausen H, Lavergne D, et al. Human papillomavirus infections in nonmelanoma skin cancers from renal transplant recipients and nonimmunosuppressed patients. J Natl Cancer Inst. 1996;88:802–811. doi: 10.1093/jnci/88.12.802. [DOI] [PubMed] [Google Scholar]
- Steger G, Pfister H. In vitro expressed HPV 8 E6 protein does not bind p53. Arch Virol. 1992;125:355–360. doi: 10.1007/BF01309654. [DOI] [PubMed] [Google Scholar]
- Tengvall Linder M, Johansson C, Scheynius A, Wahlgren C. Positive atopy patch test reactions to Pityrosporum orbiculare in atopic dermatitis patients. Clin Exp Allergy. 2000;30:122–131. doi: 10.1046/j.1365-2222.2000.00702.x. [DOI] [PubMed] [Google Scholar]
- Tommasino M, Accardi R, Caldeira S, Dong W, Malanchi I, Smet A, Zehbe I. The role of TP53 in cervical carcinogenesis. Hum Mutat. 2003;21:307–312. doi: 10.1002/humu.10178. [DOI] [PubMed] [Google Scholar]
- Wieland U, Ritzkowsky A, Stoltidis M, et al. Communication: Papillomavirus DNA in basal cell carcinomas of immunocompetent patients: an accidental association. J Invest Dermatol. 2000;115:124–128. doi: 10.1046/j.1523-1747.2000.00015.x. [DOI] [PubMed] [Google Scholar]