Abstract
Cholesterol metabolism is altered in Alzheimer's disease (AD). The nuclear hormone receptor Retinoic X Receptor a (RXRa) is a member of the nuclear ligand-activated transcription factor family. RXRs are key regulators of cholesterol synthesis and thus cholesterol metabolism. We performed a systematic screen for gene variants in the RXRA gene. The effect of these gene variants on the risk of AD was investigated in 405 AD patients (mean age: 74.27 ± 9.37 years; female 78.6%) and 347 controls (mean age: 73.26 ± 8.37 years; female 57.2%). Furthermore, the influence of RXRA gene variants on CSF and plasma levels of cholesterol, lathosterol and 24S-hydroxycholesterol were evaluated. One of the identified seven SNPs in RXRA influenced AD risk in our single marker analysis (rs3132293: P= 0.006). Haplotype analysis identified a three-marker haplotype (TGC) consisting of rs3118570, rs1536475 and rs3132293, which decreased the risk of AD (P= 0.009). The single marker rs3132293 (P= 0.026) and the TGC haplotype (P= 0.026) influenced CSF lathosterol levels in non-demented controls, and cholesterol levels in the combined sample comprising AD patients and controls (Rs3132293: P= 0.050; TGC haplotype: P= 0.035). 24S-Hydroxycholesterol CSF and plasma levels were also influenced by rs3132293 (CSF: P= 0.004; plasma: P= 0.001) and the TGC haplotype (CSF: P= 0.004; plasma: P= 0.002); this effect was most pronounced in AD patients (rs3132293: CSF: P= 0.009, plasma: P= 0.002; TGC haplotype: CSF: P= 0.019, plasma: P= 0.005). Our results suggest that RXRA gene variants might act as risk factor for AD via an influence on cerebral cholesterol metabolism.
Keywords: retinoic X receptor α, Alzheimer's disease, association, cholesterol, sequencing
Introduction
Alzheimer's disease (AD) is the most common cause of dementia world-wide. Typical pathological hallmarks are neurofibrillary tangles, amyloid angiopathy, insoluble, extracellular amyloid plaques, mainly consisting of neurotoxic 4 kD amyloid-β peptide (Aβ) and leukoaraiosis, which involves diffuse white matter changes [1]. Research of the last 10 years revealed the involvement of cholesterol metabolism in the pathogenesis of AD. Presence of the apolipoprotein E (APOE) 4 allele is the strongest genetic risk factor for late-onset AD [2]. Cholesterol metabolism is altered in AD patients in that serum levels of cholesterol metabolites such as 24S-hydroxycholesterol and 27-hydroxycholesterol are decreased [3]. Cholesterol depletion or administration of cholesterol lowering 3ß-hydroxy-3ß-methyl-glutaryl-CoA reductase (HMGCoAR) inhibitors, so called statins, inhibits the production of ß-amyloid (Aß) in vitro[4] and reduce the levels of Aßin vivo and in vitro[5]. Statins might reduce the risk of AD [6], even though this therapy is also discussed controversially for AD prevention and treatment [7, 8].
Brain cholesterol is synthesized locally and predominantly, but not entirely independent of blood cholesterol levels [9, 10]. Excess brain cholesterol derived from increased membrane turnover or neuronal loss is eliminated via cholesterol hydroxylation leading to the formation of 24S-hydroxycholesterol, the major elimination product of cerebral cholesterol.
The nuclear hormone receptor Retinoic X Receptor α (RXRα) is a member of the nuclear ligand-activated transcription factor family and belongs to the steroid hormone receptor super family. RXRs are important regulators of different pathways and are involved in cell proliferation, differentiation, and in glucose, fatty acid and cholesterol metabolism [11]. RXRs can function as homodimers or as heterodimers. Heterodimers of RXRα gene (RXRA) with the nuclear receptors liver X receptor (LXR) or peroxisome proliferator activated receptor (PPAR) have direct influence on cholesterol metabolism in that they influence expression of i.e. Apolipoprotein A1, APOE, ATP-binding cassette transporters (ABC-transporters) and sterol regulatory element-binding protein 1 (SREBP1), which are all key players in cholesterol homeostastis [12–15]. Sequence variations in RXRA might influence gene and protein function, leading to alterations in cholesterol metabolism and by this can influence the risk of AD.
RXRA is located at chromosome 9q34, nearby a putative hot spot for an AD risk gene [16, 17]. RXRA spans about 100 kilo bases and contains 10 exons. The promoter was just recently identified and characterized [18]. Several polymorphisms have been described in single-nucleotide polymorphism (SNP) databanks, but only few have been confirmed in human populations. In general, studies on RXRA polymorphisms are scarce [19] and association studies with AD are lacking. Thus, we performed a systematic screen for RXRA gene variants in AD patients and non-demented probands and investigated, if the detected polymorphisms influence the risk of AD and levels of cholesterol, the cholesterol precursor lathosterol and the cerebral cholesterol elimination product 24S-hydroxycholesterol.
Methods
Probands
AD patients (n= 405, mean age: 74.1 ± 7.9; range: 59–96 years; female: 69.1%) were recruited from the Department of Psychiatry, University of Bonn, Germany, and from the Division of Neuroradiology of the Central Institute of Mental Health, Mannheim, Germany. Patients were diagnosed according to DSM-IV, supported by clinical examination, detailed structured interviews, neuropsychological testing, cognitive screening including mini mental state examination (MMSE) [20] and neuroimaging studies. Healthy controls (n= 347, mean age: 72.8 ± 7.6; range: 64–100 years; female: 53.5%) were recruited with the support of the local Census Bureau and the regional Board of Data Protection (Nordrhein-Westfalen Germany) and from the Central Institute of Mental Health, Mannheim Germany. The cognitive status was assessed by neuropsychological testing and structured interviews. All participants of the study gave informed written consent. The study has been approved by the Ethics Committee of the Faculty of Medicine of the University of Bonn and of the institutional ethics committee, Mannheim, Germany.
Screening for variations in RXRA
Leukocyte DNA was isolated with the Qiagen® blood isolation kit according to the instructions of the manufacturer (Qiagen, Hilden, Germany). PCRs of the exons and flanking intronic regions were performed with primers designed from the sequence of chromosome 9 (GenBank acc.no.: NT_019501) as outlined in Table 1. The resulting amplification products were investigated by cycle sequencing and big dye terminator chemistry with the ABI Prism Genetic Analyzer 310A (PE Biosystems, Weiterstadt, Germany) in 40 probands. This number of probands is sufficient to detect frequent sequence variations (>5% of the population) with a likelihood of >95%.
1.
Primers used for the screening of RXRA exonic regions
| Exon | Primer sequences | Product sizes |
|---|---|---|
| 1 | F: 5′-CAGACACAAGTAGTTTACATTGTTGG-3′ | 215 bp |
| R: 5′-GAGCGGCAGGAAATGTTTGG-3′ | ||
| 2 | F: 5′- CCTGGCCCGTAGTGAGTGTG-3′ | 753 bp |
| R: 5′-AGGCCACAGCCTCTCTCTGC-3′ | ||
| 3 + 4 | F: 5′- AGCCTCGGTGTCTGTGTGTG-3′ | 1216 bp |
| R: 5′-GCCTTGCCATGCTTCTGTC-3′ | ||
| 5 | F: 5′- TGGGGATGCTGGTGTTGTGG-3′ | 400 bp |
| R: 5′-GCAGCCCCCAGGACCTCTCT-3′ | ||
| 6 | F: 5′-GCCACACCTAATCACCTCTGC-3′ | 483 bp |
| R: 5′-GGAGGTAGGAGTGGGTCCTG-3′ | ||
| 7 | F: 5′-GAACGGGCATTCTCAGGAAC-3′ | 472 bp |
| R: 5′-TCTGAGCCTCCTCCTCATGC-3′ | ||
| 8 | F: 5′-TTGGCTTGGCCCATCTCAGC-3′ | 391 bp |
| R: 5′-CAGAAGGCAGCATGGCCACA-3′ | ||
| 9 | F: 5′-CTGGCCGTGGGTTCCACAGT-3′ | 459 bp |
| R: 5′-GGACCTCCTGCTGCCTGCTC-3′ | ||
| 10 | F: 5′-CCCTTCATCTCCGTCCTCAG-3′ | 870 bp |
| R: 5′-CTGGGCACCAGTATTCAGAGC-3′ | ||
Genotyping of SNPs
For the determination of all seven identified SNPs in RXRA in the whole study population, genotyping methods performed with restriction fragment length polymorphism (RFLP) or allele-specific PCR (Polymerase chain reaction-CTPP [21]) were established. Details are given in Table 2. As a quality measure samples were randomly analyzed by cycle sequencing with the ABI Prism genetic analyzer 310 A (PE Biosystems). The APOE genotype was studied as described by Hixson and Vernier [22].
2.
Methods for the determination of RXRA gene variants
| Variation | Primer | Method | Enzyme/primer | Alleles, fragments (Bp) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rs226677, IVS1 +33 A/G | F: 5′-CCTGGCCCGTAGTGAGTGTG-3′ | RFLP | NlaIV | G-Allel: 217, 100, 78, 73, 59 55, 35, 30, 20, 10 | |||||||
| R: 5′-AGGCCACAGCCTCTCTCTGC-3′ | A-Allele: 295, 100, 73, 59 55, 35, 30, 20, 10 | ||||||||||
| Rs1805352, IVS1-46 A/C | F: 5′-AGCCTCGGTGTCTGTGTGTG-3′ | CTPP | F-A: 5′-CGTGGGGGACATAGGGAA-3′ | A-Allele: 247, 190 | |||||||
| R: 5′-GCAGATGTGCTTGGTGAAGG -3′ | R-C: 5′-CGGGGTGTACACCAGGTTTG-3′ | C-Allele: 247, 90 | |||||||||
| Rs3118570, IVS5-99 T/G | F: 5′-GAACGGGCATTCTCAGGAAC-3′ | RFLP | HphI | G-allele: 270, 110, 90 | |||||||
| R: 5′-TCTGAGCCTCCTCCTCATGC-3′ | T-Allele: 360, 110 | ||||||||||
| Rs1536475, IVS6 + 70 A/G | F: 5′-GAACGGGCATTCTCAGGAAC-3′ | RFLP | AciI | A-Allele: 215, 140, 115 | |||||||
| R: 5′-TCTGAGCCTCCTCCTCATGC-3′ | G-Allele: 215, 140, 90, 25 | ||||||||||
| Rs3132293, IVS7-126 T/C | F: 5′-CTGGCCGTGGGTTCCACAGT-3′ | CTPP | F-C: 5′-AGGCATGTCCAGCGGCATC-3′ | C-Allele: 459, 400 | |||||||
| R: 5′-GGACCTCCTGCTGCCTGCTC-3′ | R-T: 5′-GCAGAGCAGGTGGTGGAGGA-3′ | T-Allele: 459, 94 | |||||||||
| IVS8-100 | F: 5′-CTCCACCACCTGCTCTGCAC -3′* | RFLP | DraIII | T-Allele: 350, 30 | |||||||
| R: 5′-GGACCTCCTGCTGCCTGCTC-3′ | G-Allele: 380 | ||||||||||
| Rs1805343, IVS8 -27 G/A | F: 5′-CCCTTCATCTCCGTCCTCAG-3′ | RFLP | MspI | G-Allele: 385, 228, 200, 65 | |||||||
| R: 5′-CTGGGCACCAGTATTCAGAGC-3′ | A-Allele: 385, 280, 200 | ||||||||||
The underlined letter represents a mismatch in the primer for the generation of an allele-specific enzyme restriction site.
Analysis of cholesterol, lathosterol and 24S-hydroxycholesterol in CSF and plasma
After an overnight fast, cerebro spinal fluid (CSF) samples from 149 AD patients and 86 non-demented controls were obtained by lumbar puncture. Plasma samples from 289 AD patients and 230 non-demented controls were obtained by venous puncture. AD patients were diagnosed as described above. For CSF collection, non-demented controls referring to the Department of Neurology, University of Bonn, who underwent lumbar puncture during clinical routine diagnosis of neurological disorders other than neurodegenerative or inflammatory diseases of the central nervous system were recruited. Plasma samples were obtained from non-demented controls recruited in the Department of Psychiatry, University of Bonn, Germany, and from the Division of Neuroradiology of the Central Institute of Mental Health, Mannheim, Germany. Patients with symptomatic cardiac disease, renal or hepatic dysfunction, insulin-dependent diabetes mellitus, untreated thyroidal dysfunction, blood–brain-barrier-disturbance, inflammatory disease, other neurodegenerative disorders or alcohol abuses were excluded. The cognitive status of the control group was assessed by MMSE [20]. Samples were frozen in aliquots and stored at –80°C until assay. CSF and plasma concentrations of lathosterol, 24S-hydroxycholesterol and CSF cholesterol were measured by a modified sensitive method performed with combined gas chromatographymass spectrometry as described previously [23]. Plasma concentrations of cholesterol were measured by standard enzymatic procedures (Boehringer, Mannheim, Germany). Since plasma concentrations of 24S-hydroxycholesterol or lathosterol and cholesterol are highly correlated, plasma levels of 24S-hydroxycholesterol and lathosterol were corrected for plasma cholesterol.
Statistical analysis
Allele frequencies and genotype distribution of RXRA polymorphisms in the different diagnosis groups were compared using X2-statistics. Logistic regression analysis was used to examine the simultaneous effect of RXRA polymorphisms, the APOE4 allele, age and sex on the risk of AD.
Haplotype analysis was performed with FAMHAP16 [24, 25], a software program estimating haplotype frequencies using the expectation-maximization (EM) algorithm. Haplotype frequencies in cases and controls were compared with the global likelihood ratio test as implemented in FAMHAP16. To account for multiple testing 10,000 permutations were performed. Linkage Disequilibrium (LD) plot analysis was performed with Haploview [26].
The effect of association of RXRA single SNP and haplotype with lathosterol, 24S-hydroxycholesterol and cholesterol levels was tested by MANOVA using the APOE4 allele and age as covariates. Since it is known from the literature that there are differences in the levels of cholesterol and metabolites between AD patients and controls [3, 27], separate analyses for each diagnosis group were performed. P-Values were set at two sided P < 0.05.
Results
Single locus analysis
Seven polymorphisms were identified in RXRA in our sample (Fig. 1); none of the identified polymorphisms was located in the exonic region of the gene. The Hardy-Weinberg disequilibrium was verified for AD patients and controls and all RXRA polymorphisms. Allele frequencies and genotype distributions are given in Table 3. Due to a frequency of less than 5% of the minor allele of IVS8-100, this polymorphism was excluded from further analyses. X2-Statistics revealed differences in genotype distribution between AD patients and controls for SNPs rs1805352, rs3118570, rs1536475 and rs3132293.
1.
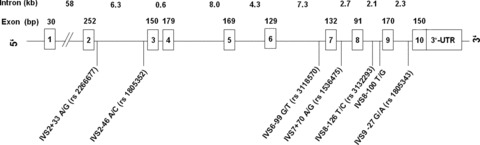
Schematic representation of the RXRA gene and localization of the identified gene variants.
3.
Genotype distribution and allele frequencies of RXRA polymorphisms in AD patients and controls
| Diagnosis | n | Genotypes [%] | Allele frequencies | X2-test | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IVS1 + 33 A/G, rs2266677 | AA | AG | GG | A | G | X2 | df | P | |||||||||||||||
| Controls | 347 | 217 (62.5) | 116 (33.4) | 14 (4.1) | 0.79 | 0.21 | 0.373 | 2 | 0.83 | ||||||||||||||
| AD | 405 | 246 (60.7) | 140 (34.6) | 19 (4.7) | 0.78 | 0.22 | |||||||||||||||||
| IVS1-46 A/C, rs1805352 | AA | AC | CC | A | C | X2 | df | P | |||||||||||||||
| Controls | 347 | 175 (50.4) | 128 (36.9) | 44 (12.7) | 0.69 | 0.31 | 6.36 | 2 | 0.042* | ||||||||||||||
| AD | 405 | 179 (44.2) | 186 (46.0) | 40 (9.8) | 0.67 | 0.33 | |||||||||||||||||
| IVS5-99 T/G, rs3118570 | TT | TG | GG | T | G | X2 | df | P | |||||||||||||||
| Controls | 347 | 241 (69.5) | 90 (25.9) | 16 (4.6) | 0.82 | 0.18 | 8.61 | 2 | 0.014* | ||||||||||||||
| AD | 405 | 261 (64.4) | 136 (33.6) | 8 (2.0) | 0.81 | 0.19 | |||||||||||||||||
| IVS6 + 70 A/G, rs1536475 | GG | GA | AA | G | A | X2 | df | P | |||||||||||||||
| Controls | 347 | 246 (70.9) | 82 (23.6) | 19 (5.5) | 0.83 | 0.17 | 6.54 | 2 | 0.038* | ||||||||||||||
| AD | 405 | 257 (63.5) | 130 (32.1) | 18 (4.4) | 0.80 | 0.20 | |||||||||||||||||
| IVS7 -126 T/C, rs3132293 | CC | CT | TT | C | T | X2 | df | P | |||||||||||||||
| Controls | 347 | 216 (62.2) | 107 (30.8) | 24 (7.0) | 0.78 | 0.22 | 13.60 | 2 | 0.001* | ||||||||||||||
| AD | 405 | 203 (50.1) | 177 (43.7) | 25 (6.2) | 0.72 | 0.28 | |||||||||||||||||
| IVS8-100 T/G | TT | TG | GG | T | G | X2 | df | P | |||||||||||||||
| Controls | 347 | 333 (96.0) | 5 (1.4) | 9 (2.6) | 0.97 | 0.03 | 1.48 | 2 | 0.48 | ||||||||||||||
| AD | 405 | 392 (96.8) | 8 (2.0) | 5 (1.2) | 0.98 | 0.02 | |||||||||||||||||
| IVS8 -27 G/A, rs1805343 | AA | AG | GG | A | G | X2 | df | P | |||||||||||||||
| Controls | 347 | 166 (47.8) | 129 (37.2) | 52 (15.0) | 0.66 | 0.34 | 4.96 | 2 | 0.084 | ||||||||||||||
| AD | 405 | 162 (40.0) | 179 (44.2) | 64 (15.8) | 0.62 | 0.38 | |||||||||||||||||
p-values < 0.05.
We performed a logistic regression analysis including the remaining six RXRA polymorphisms and the co-variates, APOE4 allele, age and sex. Rs3132293 (IVS7-126 T/C) significantly influenced the risk of AD in that carriers of an rs3132293 CC genotype presented with a reduced risk of AD (rs3132293: X2= 7.60; df = 1; P= 0.006; OR = 0.64; 95% CI = 0.46–0.88). As expected, age (P= 0.037), female gender (P= 0.001) and the presence of the APOE4 allele (P < 0.001) were associated with an increased risk of AD. The other RXRA polymorphisms did not influence the risk of AD in this model (carrier of the minor allele versus non-carriers: rs226677: P= 0.34; rs1805352: P= 0.96; rs3118570: P= 0.39, rs1536475: P= 0.71; rs1805343: P= 0.42).
Haplotype analysis
LD plot analysis performed with Haploview revealed high-linkage disequilibrium D' scores between all RXRA polymorphisms investigated in this study (Fig. 2). For haplotype analysis performed with FAMHAP16, only haplotypes with a frequency of more than 5% were included. Famhap16 identified a three-marker haplotype consisting of rs3118570 (IVS5-99 T/G), rs1536475 (IVS6 + 70 A/G) and rs3132293 (IVS7-126 T/C) to influence the risk of AD (P= 0.009, global statistics, 10,000 permutations). Homozygote carriers of the TGC haplotype were significantly less frequent in the group of AD patients compared to controls (X2= 10.09; df = 1; P= 0.001, Tables 4 and 5), suggesting that this haplotype might act as a protective factor of AD.
2.
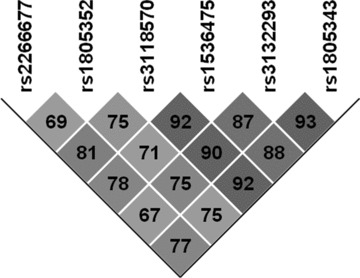
Linkage Disequilibrium (LD) structure of RXRA gene variations. The number at the intersection of each pair of SNPs represents the pairwise D' values between two SNPs.
4.
RXRA haplotypes in AD patients and controls
| Haplotype | Controls (n = 346) | AD-patients (n = 400) | Single values | Global statistics | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3118570 | Rs1536475 | rs3132293 | n | % | n | % | x2 | df | P | x2 | df | P | |||||||||||||||||||||||||
| T | G | C | 266 | 74.9 | 282 | 69.0 | 8.18 | 1 | 0.004* | 13.20 | 4 | 0.004* | |||||||||||||||||||||||||
| G | A | T | 56 | 15.8 | 71 | 17.4 | 0.96 | 1 | 0.33 | ||||||||||||||||||||||||||||
| T | G | T | 22 | 6.1 | 39 | 9.5 | 5.82 | 1 | 0.02* | ||||||||||||||||||||||||||||
| T | A | C | 2 | 0.6 | 8 | 1.9 | 4.91 | 1 | 0.03* | ||||||||||||||||||||||||||||
p-values < 0.05.
5.
Distribution of RXRA TGC haplotype including SNPs rs3118570, rs1536475 and rs3132293 in AD patients and controls
| Diagnosis | n | TGC Homocygotes | Carrier versus non-carrier | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carrier | Non-carrier | X2 | df | P | ||||||||||||||||||||
| Controls | 347 | 145 (41.7) | 202 (58.3) | 7.14 | 1 | 0.001* | ||||||||||||||||||
| AD | 405 | 216 (53.3) | 189 (46.7) | |||||||||||||||||||||
p-values < 0.05.
To consider for co-variates, we performed an additional logistic regression analysis by including the co-variates APOE4 allele, age and gender. Homozygote carriers of the TGC haplotype presented with a reduced risk of AD (X2= 7.32; df = 1; P= 0.007; OR = 0.648; 95% CI = 0.47–0.89). As expected, female gender (P= 0.001), age (P= 0.032) and the presence of the APOE4 allele (P < 0.001) were associated with an increased risk of AD.
CSF parameters
To verify if RXRA single marker, rs3132293 and the RXRA TGC haplotype, which both influenced the risk of AD, might also influence CSF levels of cholesterol, the cholesterol precursor lathosterol and the cerebral cholesterol elimination product 24S-hydroxycholesterol, we performed MANOVAs independently for the RXRA single marker and the haplotype.
MANOVAs in the whole sample comprising AD patients and controls detected that CSF levels of 24S-hydroxycholesterol and cholesterol were lower in carriers of the rs3132293 C allele (24S-hydroxycholesterol: P= 0.004; cholesterol: P= 0.050; Table 6) or the TGC haplotype (24S-hydroxycholesterol: P= 0.004; cholesterol: P= 0.035; Table 6), while levels of lathosterol were not altered (P= 0.2). In both analyses presence of the APOE4 allele influenced 24S-hydroxycholesterol levels (P < 0.001) but not those of the other cholesterol parameters (P > 0.5). Age influenced levels of cholesterol and 24S-hydroxycholesterol (each P < 0.001), but not of lathosterol. Levels of cholesterol and lathosterol were lower in AD patients compared to controls (cholesterol: P < 0.001; lathosterol: P < 0.05).
6.
Influence of single marker rs3132293 and the three-marker TGC haplotype on CSF lathosterol, 24S-hydroxycholesterol and cholesterol levels in AD patients and controls
| Sample | Genotype | n | Mean ± SE | F | df | P | Mean ± SE | F | df | P | Mean ± SE | F | df | P | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3132293 | Lathosterol [μg/dl] | 24S-Hydroxycholesterol [ng/ml] | Cholesterol [mg/dl] | ||||||||||||||||||||||||||||||||||
| All | C-allele carrier | 214 | 0.33 ± 0.02 | 1.53 | 1 | 0.22 | 2.61 ± 0.07 | 8.347 | 1 | 0.004* | 0.49 ± 0.01 | 3.88 | 1 | 0.050* | |||||||||||||||||||||||
| Non-carrier | 21 | 0.39 ±0.05 | 3.27 ± 0.22 | 0.55 ± 0.03 | |||||||||||||||||||||||||||||||||
| Controls | C-allele carrier | 77 | 0.34 ± 0.03 | 5.15 | 1 | 0.026* | 2.56 ± 0.12 | 1.57 | 1 | 0.16 | 0.53 ± 0.02 | 2.41 | 1 | 0.13 | |||||||||||||||||||||||
| Non-carrier | 9 | 0.54 ± 0.09 | 3.02 ± 0.32 | 0.61 ± 0.05 | |||||||||||||||||||||||||||||||||
| AD | C-allele carrier | 137 | 0.30 ± 0.02 | 0.24 | 1 | 0.63 | 2.58 ± 0.09 | 6.97 | 1 | 0.009* | 0.44 ± 0.01 | 1.93 | 1 | 0.17 | |||||||||||||||||||||||
| Non-carrier | 12 | 0.27 ± 0.06 | 3.4 ± 0.3 | 0.51 ± 0.04 | |||||||||||||||||||||||||||||||||
| Three-marker TGC haplotype | Lathosterol [μg/dl] | 24S-Hydroxycholesterol [ng/ml] | Cholesterol [mg/dl] | ||||||||||||||||||||||||||||||||||
| All | TGC-haplotype carrier | 193 | 0.32 ± 0.02 | 0.75 | 1 | 0.39 | 2.57 ± 0.07 | 8.53 | 1 | 0.004* | 0.48 ± 0.01 | 4.49 | 1 | 0.035* | |||||||||||||||||||||||
| Non-carrier | 25 | 0.35 ± 0.04 | 3.16 ± 0.19 | 0.54 ± 0.03 | |||||||||||||||||||||||||||||||||
| Controls | TGC-haplotype carrier | 67 | 0.32 ± 0.03 | 5.19 | 1 | 0.026* | 2.53 ± 0.12 | 3.92 | 1 | 0.052 | 0.53 ± 0.02 | 1.97 | 1 | 0.16 | |||||||||||||||||||||||
| Non-carrier | 11 | 0.49 ± 0.07 | 3.12 ± 0.29 | 0.59 ± 0.05 | |||||||||||||||||||||||||||||||||
| AD | TGC-haplotype carrier | 126 | 0.30 ± 0.02 | 0.78 | 1 | 0.38 | 2.54 ± 0.09 | 5.66 | 1 | 0.019* | 0.43 ± 0.01 | 2.90 | 1 | 0.09 | |||||||||||||||||||||||
| Non-carrier | 14 | 0.25 ± 0.05 | 3.22 ± 0.27 | 0.49 ± 0.04 | |||||||||||||||||||||||||||||||||
p-values < 0.05.
In agreement with the literature, we found differences in the levels of cholesterol parameters between AD patients and controls; thus we performed exploratory analyses in the subgroups of AD patients and controls.
In controls levels of lathosterol were lower in carriers of the rs3132293 C allele (P= 0.026) or the TGC haplotype (P= 0.026). Levels of 24S-hydroxycholesterol and cholesterol were also lower in carriers of the C allele or the TGC haplotype, but this did not reach statistical significance (Table 6).
In AD patients, levels of 24S-hydroxycholesterol were lower in carriers of the rs3132293 C allele (P= 0.009) or the TGC haplotype (P= 0.019), also cholesterol levels were altered in these patients, but this did not reach significance (P < 0.17). We did not find an effect of gene variants on the CSF levels of lathosterol in AD patients.
Plasma parameters
To investigate if RXRA single marker, rs3132293 and the RXRA TGC haplotype also influenced peripheral cholesterol metabolism, we performed MANOVAs independently for the RXRA single marker and the haplotype and investigated their effect on plasma levels of cholesterol, lathosterol/cholesterol and 24S-hydroxycholesterol/cholesterol.
MANOVAs in the whole sample comprising AD patients and controls detected that only levels of 24S-hydroxycholesterol/cholesterol were lower in carriers of the rs3132293 C allele (P= 0.001; Table 7) or the TGC haplotype (P= 0.002; Table 7). While levels of lathosterol/cholesterol or cholesterol were not altered (P > 0.2). Presence of the APOE4 allele influenced levels of all cholesterol parameters (P < 0.01), age influenced levels of 24S-hydroxycholesterol/cholesterol (P= 0.022) and lathosterol/cholesterol (P= 0.008).
7.
Influence of single marker rs3132293 and the three-marker TGC haplotype on serum lathosterol, 24S-hydroxycholesterol and cholesterol levels in AD patients and controls
| Sample | Genotype | n | Mean±SE | F | df | P | Mean±SE | F | df | P | Mean±SE | F | df | P | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3132293 | Lathosterol/Cholesterol [μg/mg] | 24S-Hydroxycholesterol/Cholesterol[ng/mg] | [spanname“12to15”]Cholesterol [mg/dl] | ||||||||||||||||||||||||||||||||||
| All | C-allele carrier | 512 | 1.14 ± 0.02 | 0.01 | 1 | 0.95 | 36.27±0.44 | 12.0 | 1 | 0.001* | 237.6±2.1 | 0.09 | 1 | 0.76 | |||||||||||||||||||||||
| Non-carrier | 28 | 1.14±0.1 | 29.64±1.87 | 240.3±8.7 | |||||||||||||||||||||||||||||||||
| Controls | C-allele carrier | 231 | 1.18±0.03 | 0.31 | 1 | 0.58 | 36.77±0.76 | 2.75 | 1 | 0.099 | 238.5±3.4 | 1.43 | 1 | 0.23 | |||||||||||||||||||||||
| Non-carrier | 12 | 1.26±0.14 | 31.58±3.09 | 255.0±13.6 | |||||||||||||||||||||||||||||||||
| AD | C-allele carrier | 281 | 1.11±0.03 | 0.29 | 1 | 0.59 | 35.43±0.56 | 10.32 | 1 | 0.001* | 237.7±2.8 | 0.48 | 1 | 0.49 | |||||||||||||||||||||||
| Non-carrier | 16 | 1.04±0.13 | 27.96±2.26 | 229.6±11.4 | |||||||||||||||||||||||||||||||||
| Three-marker TGC haplotype | Lathosterol/Cholesterol [μg/mg] | 24S-Hydroxycholesterol/Cholesterol [ng/ml] | [spanname“12to15”]Cholesterol [mg/dl] | ||||||||||||||||||||||||||||||||||
| All | TGC-haplotype carrier | 482 | 1.14±0.02 | 0.49 | 1 | 0.48 | 36.29±0.45 | 9.49 | 1 | 0.002* | 238.9±2.1 | 0.06 | 1 | 0.81 | |||||||||||||||||||||||
| Non-carrier | 37 | 1.08±0.08 | 31.13±1.62 | 237.1±7.5 | |||||||||||||||||||||||||||||||||
| Controls | TGC-haplotype carrier | 214 | 1.17±0.04 | 0.12 | 1 | 0.73 | 36.76±0.78 | 2.69 | 1 | 0.102 | 239.9±3.3 | 0.73 | 1 | 0.39 | |||||||||||||||||||||||
| Non-carrier | 16 | 1.22±0.12 | 32.28±2.67 | 249.9±11.5 | |||||||||||||||||||||||||||||||||
| AD | TGC-haplotype carrier | 268 | 1.12±0.03 | 1.27 | 1 | 0.26 | 35.56±0.54 | 8.12 | 1 | 0.005* | 238.3±2.8 | 1.06 | 1 | 0.30 | |||||||||||||||||||||||
| Non-carrier | 21 | 0.99±0.12 | 29.69±1.98 | 227.7±9.9 | |||||||||||||||||||||||||||||||||
We also performed separate analyses in the subgroups of AD patients and controls. In controls, levels of 24S-hydroxycholesterol/cholesterol were higher in carriers of the C allele or the TGC haplotype, but this did not reach statistical significance (Table 7). Plasma cholesterol and lathosterol/cholesterol levels were neither influenced by the C allele nor by the TGC haplotype in non-demented persons (Table 7).
In AD patients, levels of 24S-hydroxycholesterol/cholesterol were higher in carriers of the rs3132293 C allele (P= 0.001) or the TGC haplotype (0.005). Levels of cholesterol or lathosterol/cholesterol were not influenced by any RXRA gene variant (Table 7).
Discussion
Alterations of cholesterol metabolism contribute to AD risk and pathology [3, 28]. RXRA is a key regulator of cholesterol metabolism and the gene is located next a region on chromosome 9 comprising increased LOD scores in AD linkage analyses [17, 29], making RXRA an ideal candidate for an AD risk gene.
We screened exons and flanking introns of RXRA and detected six polymorphisms with a frequency of >5% of the minor allele, which were included in our analyses. We selected only gene variants with a frequency of >5% for our study, and by this followed the common disease-common variant hypothesis [30, 31]. One might argue, that due to an other hypothesis, the common disease-multiple rare variants hypothesis, also multiple gene variants presenting with a low frequency might act as risk factors of AD. However, since it was the aim of our study to investigate the effect of one specific gene we focussed only on common variants in RXRA.
One RXRA polymorphism rs3132293 influenced AD risk in the single marker analysis, in that carriers of the CC genotype presented with a reduced risk of AD. Haplotype analysis identified a three-marker haplotype TGC, combining rs3118570, rs1536475 and rs3132293, which also reduced the risk of AD. Thus it might be concluded that gene variants in RXRA act as risk factor of AD.
All SNPs included in the identified haplotype are in strong linkage disequilibrium. The single marker rs3132293, which influenced AD risk might act as a taq SNP in our analysis. This also explains why the effects of the rs3132293 single marker and of the TGC haplotype on AD risk and on cholesterol parameters were in comparable ranges.
The rs3132293 C-allele and also the TGC haplotype influenced cholesterol metabolism of the brain as measured as reduced CSF levels of lathosterol, 24S-hydroxycholesterol and cholesterol in carriers of these gene variants; however, these effects differed between AD patients and controls. We found RXRA gene variants to influence levels of the cholesterol precursor lathosterol only in non-demented controls. However, influences on 24S-hydroxycholesterol were detected in the whole sample comprising AD patients and controls, but were most pronounced in AD patients. We also found CSF cholesterol levels in carriers of the rs3118570 C allele or the TGC haplotype to be reduced in the whole sample, but statistical analysis of the subgroups of AD patients or controls revealed only marginal effects. Influences of RXRA gene variants on peripheral cholesterol metabolism were also identified by altered levels of 24S-hydroxycholesterol/cholesterol, especially in AD patients, while the other plasma parameters were not changed. Our data suggest, that RXRA gene variants might influence AD risk mainly via an influence on cerebral cholesterol metabolism as measured by altered levels of 24S-hydroxycholesterol in CSF and plasma.
24S-Hydroxycholesterol is the major cholesterol elimination product of the brain [32, 33]. The detailed mechanism for the transport of 24S-hydroxycholesterol from brain via the blood–brain barrier into the periphery is not fully clear. ABCA1 transporters or organic anion transporting peptide 2 has been shown to be involved in this mechanism [34, 35] and ABCA1 expression is regulated by heterodimers of RXRA with the nuclear receptor LXR [15]. Previous publications report on reduced levels of 24S-hydroxycholesterol in plasma of AD patients compared to controls [3, 36]. In line with this observation, we found AD patients who were non-carrier of the protective RXRA C allele or the TGC haplotype to present with lower plasma levels; however, these probands showed increased CSF levels of 24S-hydroxycholesterol. These findings suggest, that in non-carriers of the protective RXRA gene variants, the elimination of 24S-hydroxycholesterol might be less efficient, possibly via a reduced activation of ABCA1 transporters, leading to an accumulation of CSF 24S-hydroxycholesterol.
In non-demented controls, who were non-carriers of the protective RXRA gene variants, we found increased CSF levels of lathosterol, which might be a sign of increased cholesterol de novo synthesis. Cholesterol synthesis and thus levels of lathosterol increase if the transcription factor LXR is activated by 24S-hydroxycholesterol [37]. It might be speculated that the higher CSF levels of 24S-hydroxycholesterol in non-demented persons who are homozygote carriers of the RXRA risk genotype might induce an increased synthesis of lathosterol, although the changes in the 24S-hydroxycholesterol levels were only marginally significant in these probands.
The lack of an effect of RXRA gene variants on CSF levels of lathosterol in AD patients might be due to the observation that alterations of cholesterol metabolism are a pathological process in AD [3, 27, 36]. Due to the neurodegenerative processes cholesterol synthesis in the brain of AD patients might be reduced. Comparable diagnosis-specific effects of gene variations on cholesterol metabolism have also been described for the APOE4 allele. It is generally accepted that the APOE4 allele influences plasma cholesterol in non-demented probands and that increased plasma cholesterol levels are observed in carriers of the APOE4 allele [38]. However, in AD patients it has been shown that serum cholesterol levels are not influenced by the presence of the APOE4 allele [39, 40]. It can be assumed that pathological alterations may cover gene effects especially in manifest AD, and thus gene effects might be seen mainly in non-demented probands.
Since studies on gene variations in RXRA are scarce, there is only one study investigating RXRA polymorphisms in psoriasis [41], it is unknown, if the identified polymorphisms might influence RXRA function or gene expression. The possible effect of RXRA SNPs on RNA splicing was evaluated using automated online splice site analysis (https://splice.cmh.edu) [42]. This program revealed that the rs3132293 C allele might cause a five to ninefold increased activity of a SRp40 site, while the rs3118570 T allele might cause a 4–25-fold increased activity of the donor site. It is unknown, if these effect are also present in human beings. Thus, these results have to be considered carefully and putative functions of the SNPs might be speculative. However, these analyses might help to gain a theoretical insight into the putative function of the identified RXRA gene variants.
We cannot exclude, that the relevant polymorphisms are in linkage disequilibrium with another yet unknown gene variant in the RXRA promoter. The promoter of RXRA was recently identified and described [18]; however, this study did not screen for gene variations in this region. It was reported, that the GC base content of the RXRA promoter is especially high, >80% and that there are several shorter regions presenting with 100% GC base content, making amplification of this region extremely difficult. Due to these limitations we were only able to screen a short region of the promoter, and it cannot be excluded, that other functionally relevant variations are located in the other promoter-regions of RXRA.
In conclusion, our results suggest that RXRA gene variants are risk factors of AD in that they influence cholesterol metabolism of the brain. However, the detailed mechanisms will have to be explored in additional studies.
Acknowledgments
These data were supported by grants from the Alzheimer Forschungs Initiative (AFI #03802), the Deutsche Forschungsgemeinschaft (He 2318/1-2 and KO2327/2-1) and by the German Federal Ministry for Education and Research within the framework of the Competence Network Dementia (grant: 01GI0422). We thank Anne Fiedler, Christine Frahnert-Ledschbor, Sandra Schmitz, Anja Kerksiek and Sandra Schulz for technical assistance.
References
- 1.Szolnoki Z. Pathomechanism of leukoaraiosis: a molecular bridge between the genetic, biochemical, and clinical processes (a mitochondrial hypothesis) Neuromolecular Med. 2007;9:21–34. doi: 10.1385/nmm:9:1:21. [DOI] [PubMed] [Google Scholar]
- 2.Strittmatter WJ, Weisgraber KH, Huang DY, Dong L-M, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgraber D, Roses AD. Binding of human apolipoprotein E to synthetic amyloid (i peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:8098–102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kölsch H, Heun R, Kerksiek A, Bergmann KV, Maier W, Lütjohann D. Altered levels of plasma 24S- and 27-hydroxycholesterol in demented patients. Neurosci Lett. 2004;368:303–8. doi: 10.1016/j.neulet.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, Simons K. Cholesterol depletion inhibits the generation of (J-amyloid in hippocampal neurons. Proc Natl Acad Sci USA. 1998;95:6460–4. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fassbender K, Simons M, Bergmann C, Stroick M, Lütjohann D, Keller P, Runz H, Kuhl S, Bertsch T, Von Bergmann K, Hennerici M, Beyreuther K, Hartmann T. Simvastatin strongly reduces levels of Alzheimer's disease beta – amyloid pep-tides Abeta 42 and Abeta 40 in vitro and in vivo. Proc Natl Acad Sci USA. 2001;98:5856–61. doi: 10.1073/pnas.081620098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolozin B, Kellman W, Ruosseau P, Celesia GG, Siegel G. Decreased prevalence of Alzheimer disease associated with 3-hydroxy-3- methyglutaryl coenzyme A reductase inhibitors. Arch Neurol. 2000;57:1439–43. doi: 10.1001/archneur.57.10.1439. [DOI] [PubMed] [Google Scholar]
- 7.Mielke MM, Zandi PP, Sjogren M, Gustafson D, Ostling S, Steen B, Skoog I. High total cholesterol levels in late life associated with a reduced risk of dementia. Neurology. 2005;64:1689–95. doi: 10.1212/01.WNL.0000161870.78572.A5. [DOI] [PubMed] [Google Scholar]
- 8.Li G, Higdon R, Kukull WA, Peskind E, Van Valen MK, Tsuang D, Van Belle G, McCormick W, Bowen JD, Teri L, Schellenberg GD, Larson EB. Statin therapy and risk of dementia in the elderly: a community-based prospective cohort study. Neurology. 2004;63:1624–8. doi: 10.1212/01.wnl.0000142963.90204.58. [DOI] [PubMed] [Google Scholar]
- 9.Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Curr Opin Lipidol. 2001;12:105–12. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Jurevics H, Morell P. Cholesterol for synthesis of myelin is made locally, not imported into the brain. J Neurochem. 1995;64:895–901. doi: 10.1046/j.1471-4159.1995.64020895.x. [DOI] [PubMed] [Google Scholar]
- 11.Ahuja HS, Szanto A, Nagy L, Davies PJ. The retinoid X receptor and its ligands: versatile regulators of metabolic function, cell differentiation and cell death. J Biol Regul Homeost Agents. 2003;17:29–45. [PubMed] [Google Scholar]
- 12.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laffitte BA, Repa JJ, Joseph SB, Wilpitz DC, Kast HR, Mangelsdorf DJ, Tontonoz P. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc Natl Acad Sci USA. 2001;98:507–12. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentili C, Tutolo G, Pianezzi A, Cancedda R, Descalzi CF. Cholesterol secretion and homeostasis in chondrocytes: a liver X receptor and retinoid X receptor heterodimer mediates apolipoprotein A1 expression. Matrix Biol. 2005;24:35–44. doi: 10.1016/j.matbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Uehara Y, Miura S, Von EA, Abe S, Fujii A, Matsuo Y, Rust S, Lorkowski S, Assmann G, Yamada T, Saku K. Unsaturated fatty acids suppress the expression of the ATP-binding cassette transporter G1 (ABCG1) and ABCA1 genes via an LXR/RXR responsive element. Atherosclerosis. 2007;191:11–21. doi: 10.1016/j.atherosclerosis.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer's disease. Exp Gerontol. 2000;35:1343–52. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 17.Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, Wavrant DF, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O'Donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2005;135:24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Yin W, Chamberlain R, Hewett-Emmett D, Roberts JN, Yang X, Lippman SM, Clifford JL. Identification and characterization of the human retinoid X receptor alpha gene promoter. Gene. 2006;372:118–27. doi: 10.1016/j.gene.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 19.Hegele RA, Cao H. Single nucleotide polymorphisms of RXRA encoding retinoid X receptor alpha. J Hum Genet. 2001;46:423–5. doi: 10.1007/s100380170061. [DOI] [PubMed] [Google Scholar]
- 20.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 21.Hamajima N, Saito T, Matsuo K, Kozaki K, Takahashi T, Tajima K. Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Jpn J Cancer Res. 2000;91:865–8. doi: 10.1111/j.1349-7006.2000.tb01026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–8. [PubMed] [Google Scholar]
- 23.Teunissen CE, Lutjohann D, Von BK, Verhey F, Vreeling F, Wauters A, Bosmans E, Bosma H, Van Boxtel MP, Maes M, Delanghe J, Blom HJ, Verbeek MM, Rieckmann P, De BC, Steinbusch HW, De VJ. Combination of serum markers related to several mechanisms in Alzheimer's disease. Neurobiol Aging. 2003;24:893–902. doi: 10.1016/s0197-4580(03)00005-8. [DOI] [PubMed] [Google Scholar]
- 24.Becker T, Knapp M. A powerful strategy to account for multiple testing in the context of haplotype analysis. Am J Hum Genet. 2004;75:561–70. doi: 10.1086/424390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker T, Knapp M. Maximum-likelihood estimation of haplotype frequencies in nuclear families. Genet Epidemiol. 2004;27:21–32. doi: 10.1002/gepi.10323. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Mailer J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Mulder M, Ravld R, Swaab DF, De Kloet ER, Haasdljk ED, Julk J, Van Der Boom J, Havekes LM. Reduced levels of cholesterol, phospholipids, and fatty acids in cerebrospinal fluid of Alzheimer disease patients are not related to apolipoprotein E4. Alzheimer Dis Assoc Disord. 1998;12:198–203. doi: 10.1097/00002093-199809000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Simons M, Keller P, De Strooper B, Beyreuther K, Dottl CG, Simons K. Cholesterol depletion inhibits the generation of (β-amyloid in hippocampal neurons. Proc Natl Acad Sci USA. 1998;95:6460–4. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blacker D, Bertram L, Saunders AJ, Moscarlllo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folsteln MF, Mclnnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer's Disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 30.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–9. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 31.Peng B, Klmmel M. Simulations provide support for the common disease-common variant hypothesis. Genetics. 2007;175:763–76. doi: 10.1534/genetics.106.058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Björkhem I, Lütjohann D, Breuer O, Saklnls A, Wennmalm A. Importance of a novel oxidative mechanism for elimination of brain cholesterol. J Biol Chem. 1997;48:30178–84. doi: 10.1074/jbc.272.48.30178. [DOI] [PubMed] [Google Scholar]
- 33.Lütjohann D, Breuer O, Ahlborg G, Nennesmo I, Sidén A, Diczfalusy U, Björkhem I. Cholesterol homeostasis in human brain: Evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc Natl Acad Sci USA. 1996;93:9799–804. doi: 10.1073/pnas.93.18.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panzenboeck U, Balazs Z, Sovic A, Hrzenjak A, Levak-Frank S, Wintersperger A, Malle E, Sattler W. ABCA1 and scavenger receptor class B, type I, are modulators of reverse sterol transport at an in vitro blood-brain barrier constituted of porcine brain capillary endothelial cells. J Biol Chem. 2002;277:42781–9. doi: 10.1074/jbc.M207601200. [DOI] [PubMed] [Google Scholar]
- 35.Solomon A, Kareholt I, Ngandu T, Winblad B, Nissinen A, Tuomilehto J, Soininen H, Kivipelto M. Serum cholesterol changes after midlife and latelife cognition: twenty-one-year follow-up study. Neurology. 2007;68:751–6. doi: 10.1212/01.wnl.0000256368.57375.b7. [DOI] [PubMed] [Google Scholar]
- 36.Bretillon L, Siden A, Wahlund LO, Lütjohann D, Minthon L, Crisby M, Hillert J, Groth CG, Diczfalusy U, Björkhem I. Plasma levels of 24S-hydroxycholesterol in patients with neurological diseases. Neurosci Lett. 2000;293:87–90. doi: 10.1016/s0304-3940(00)01466-x. [DOI] [PubMed] [Google Scholar]
- 37.Bose-Boyd RA, Ou J, Goldstein JL, Brown MS. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc Natl Acad Sci USA. 2001;98:1477–82. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dufouil C, Richard F, Fievet N, Dartigues JF, Ritchie K, Tzourio C, Amouyel P, Alperovitch A. APOE genotype, cholesterol level, lipid-lowering treatment, and dementia: the Three-City Study. Neurology. 2005;64:1531–8. doi: 10.1212/01.WNL.0000160114.42643.31. [DOI] [PubMed] [Google Scholar]
- 39.Dupuy AM, Mas E, Ritchie K, Descomps B, Badiou S, Cristol JP, Touchon J. The relationship between apolipoprotein E4 and lipid metabolism is impaired in Alzheimer's disease. Gerontology. 2001;47:213–8. doi: 10.1159/000052801. [DOI] [PubMed] [Google Scholar]
- 40.Von Trotha KT, Heun R, Schmitz S, Lutjohann D, Maier W, Kolsch H. Influence of lysosomal acid lipase polymorphisms on chromosome 10 on the risk of Alzheimer's disease and cholesterol metabolism. Neurosci Lett. 2006;402:262–6. doi: 10.1016/j.neulet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 41.Vasku V, Bienertova VJ, Pavkova GM, Vasku A. Three retinoid X receptor gene polymorphisms in plaque psoriasis and psoriasis guttata. Dermatology. 2007;214:118–24. doi: 10.1159/000098569. [DOI] [PubMed] [Google Scholar]
- 42.Nalla VK, Rogan PK. Automated splicing mutation analysis by information theory. Hum. Mutat. 2005;25:334–42. doi: 10.1002/humu.20151. [DOI] [PubMed] [Google Scholar]


