Abstract
Studies of the genetics underlying inflammatory bowel diseases (IBD) have increased our understanding of the pathways involved in both ulcerative colitis (UC) and Crohn’s disease (CD) and focused attention on the role of the microbiome in these diseases. Full understanding of pathogenesis will require a comprehensive grasp of the delicate homeostasis between gut bacteria and the human host. In this review we present current evidence of microbiome-gene interactions in the context of other known risk factors and mechanisms, and describe the next steps necessary to pair genetic variant and microbiome sequencing data from patient cohorts. We discuss the concept of dysbiosis, proposing that the functional composition of the gut microbiome may provide a more consistent definition of dysbiosis and may more readily provide evidence of genome-microbiome interactions in future exploratory studies.
Keywords: Inflammation, inflammatory bowel disease, dysbiosis, genetics, genome-wide association studies, metagenomics, microbiome
INTRODUCTION
Family history is a well-known risk factor for developing IBD, a group of diseases that includes CD and UC. As such, the risk of developing IBD has long been recognized to have a genetic contribution. This concept has advanced considerably over the past decade as genetic studies have identified numerous loci involved in IBD susceptibility. These studies have identified key cellular pathways in IBD and enhanced our understanding of how these pathways might contribute to disease (figure 1). However, these studies have also made clear that susceptibility alleles are not sufficient on their own to trigger disease and that other genetic and non-genetic risk factors play a role in pathogenesis. Mounting evidence has indicated that among these potential factors the diversity and composition of the gut microbiota, which includes gut resident symbiotic microorganisms, are major environmental factors influencing gut homeostasis. In this review we provide a brief overview of the recent advances that have shaped our understanding of the complex interplay of the gut microbiome and genetic susceptibility to IBD.
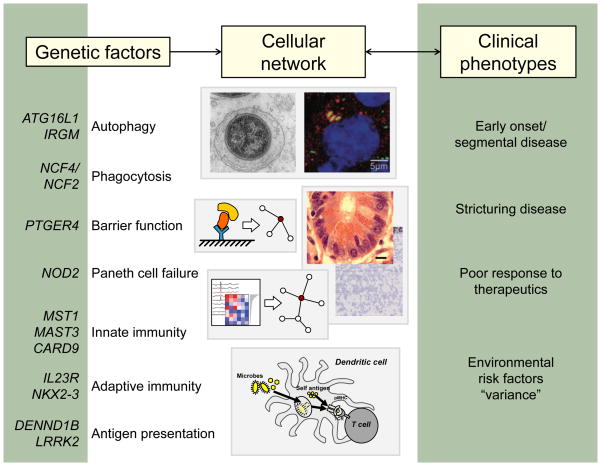
Figure 1. Genes and pathways in inflammatory bowel disease.
More than 160 loci have been associated with susceptibility to IBD. A selected list of candidate genes is shown (left) along with the cellular pathways in which these genes are thought to function. These pathways likely interact in cellular networks (center), which, when perturbed, contribute to the clinical phenotypes of IBD (right).
Untangling pathways from genetics
More than a decade ago, nucleotide oligomerization domain 2 (NOD2) was identified as the first susceptibility gene for CD.1, 2 In the last five years, population-based genome-wide association studies (GWAS), followed by subsequent meta-analysis of GWAS and Immunochip data, have greatly expanded the number of IBD-associated loci to more than 160.3
These types of genetic studies have reinforced the importance of genes and pathways previously known to be involved in IBD pathogenesis, such as barrier function, the role of T cell subsets, and cytokine-cytokine receptor signaling. In addition, these studies have helped uncover unanticipated new genes and pathways, including autophagy, regulation of interleukin 23 (IL23) signaling, and others (table 1). Recent studies have also highlighted the importance of host defense pathways, specifically those involved in the handling of mycobacteria, as important in balancing inflammatory responses as discussed further below.3 IBD GWAS have also demonstrated a significant degree of overlap between loci for UC and CD, as well as a high degree of overlap between susceptibility genes for IBD and for other complex diseases. Given this overlap, it is likely that similar pathways underlie IBD and that small differences result in diverse phenotypic presentations of UC and CD. Potentially the same polymorphism can have different cell- or tissue type-specific effects. Also, additional sequencing studies in carefully phenotyped patients may identify additional variants within the same loci that result in different phenotypes. Many genetic factors underlying early onset and adult IBD also appear to be the same, although it remains unknown whether the genes/genomic regions are entirely the same regardless of the age of onset. In patients 0–2 years of age, clinical presentation of IBD is atypical, with CD cases affecting the colon and UC patients often presenting with pancolitis. The great degree of overlap between IBD and other autoimmune diseases can largely be explained by the involvement of pathways such as antigen presentation and pathways that contribute to intestinal mucosal homeostasis. However, the identification of pathways that involve Th17 cells, CARD9, NOD2, reactive oxygen species, and IL1β suggests that IBD can also be defined as an autoinflammatory disease. The diagnostic value of these genetic associations with disease remains unclear. Although some studies have begun to find clinical associations, these approaches need to be confirmed in large well-phenotyped cohorts.
Table 1. Pathways identified by IBD GWAS.
Genetic studies have identified a number of genes and pathways as potential players in the initiation and progression of IBD. Some of these pathways were previously known to be involved in IBD pathogenesis (left), while other pathways had been previously unknown (right).
| Known IBD pathways confirmed by GWAS | Novel pathways implicated by GWAS |
|---|---|
| Barrier function | Autophagy |
| Role of T cell subsets | Regulation of IL23 signaling; Th17 development |
| Cytokine-cytokine receptor signaling | ROS signaling; IL1β |
| CARD9, NOD2 in innate immunity | |
| Innate lymphoid cells | |
| Enrichment for genes implicated in immunodeficiencies | |
| Shared risk genes associated with susceptibility to Mycobacterium tuberculosis |
Deciphering phenotype from pathways
Although IBD-associated susceptibility genes and pathways offer great promise for researchers, limiting our investigations to the identification of these risk factors is likely to lead to an incomplete understanding of pathogenesis. Given that IBD-associated genetic variants are present in many individuals who do not develop disease, as well as the suggestion that classic loss-of-function variants play only a minor role in pathogenesis, a full explanation of disease complexity will require significantly more knowledge.4, 5 For example, it will be imperative to understand the cell types involved in disease initiation and progression, how IBD pathways are regulated in these cell types, how a single pathway can exert different biological phenotypes in discrete cell and tissue types, and how multiple disease variants interact.
Many of the pathways in IBD are known to have heterogeneous effects when activated in different cell types, and these cellular outcomes may be compounded to affect disease. For example, in epithelial cells, autophagy pathways play a key role in bacterial clearance; however, in macrophages, the same autophagy genes affect the ability of cells to secrete IL1β, a key inflammatory mediator involved in host defense. Therefore the same pathway can affect disease pathogenesis through different actions in discrete cell types. Furthermore, IL1β can act through both innate lymphoid cells and CD4 T cells to stimulate IL17 secretion and chronic intestinal inflammation,6 demonstrating that the same cytokine can act on multiple arms of the immune system to promote inflammation.
The IL23-Th17 pathway is a key IBD pathway with well-characterized roles in microbial defense and intestinal immune homeostasis, and a number of genes within this pathway have been found within risk loci. In addition to its effects on classical inflammatory monocytes, myeloid cells, and stromal cells, this pathway is highly influenced by environment. In recent studies of murine models of IL23-dependent colitis, inflammatory cytokines have been shown to affect hematopoietic stem and progenitor cells, resulting in the overproduction and accumulation of granulocyte-monocyte progenitor cells in the intestine that are sufficient to aggravate the colitis phenotype.7 Additional studies will help define the heterogeneous spectrum of cellular phenotypes associated with each IBD pathway and contribute to our understanding of IBD pathogenesis.
Decoding non-coding DNA
To date, the bulk of the research on IBD genetics has focused on the impact of mutations in coding regions of genes and on the placement of these genes into discrete pathways. While these studies have been valuable in helping to characterize IBD susceptibility genes, approximately 70% of known IBD susceptibility loci are not coding variants. Understanding the implication of this finding has been aided by recent results published by the ENCODE (Encyclopedia of DNA Elements) consortium. This consortium has provided an unprecedented view of the genome that goes beyond cataloguing human sequence variation and begins to provide an integrated view of the role of functional elements in gene regulation.8 Importantly, the ENCODE project found that more than 80% of the genome is involved in at least one biochemical process, although this number is likely overestimated.9
Data from the ENCODE project and similar studies could help characterize largely unstudied IBD susceptibility elements and reveal important insights into key genes and pathways. To address how these non-coding DNA elements might have an impact on disease, the ENCODE project evaluated thousands of reported single-nucleotide polymorphisms (SNPs) curated in the National Human Genome Research Institute GWAS catalogue, finding that the majority of disease-associated SNPs reside in ENCODE-defined regulatory regions.8, 10 This observation is consistent with data suggesting that most disease-associated SNPs reside in regions that affect gene expression.11, 12 For IBD, 64 of the associated SNPs are in linkage disequilibrium with variants that are known to regulate gene expression.3 In T helper cells, CD-associated variants are sensitive to DNase, a phenotype generally associated with cis-regulatory modules such as promoters and enhancers.11 This finding suggests that fine mapping studies of the regulatory landscape surrounding IBD SNPs in specific cell types and tissues could yield important phenotypic insights into pathway heterogeneity.
Gene regulation goes far beyond the presence of promoters or enhancers. Non-coding risk variants could act at the level of DNA, through modification of transcription factor binding sites or epigenetic modification of regulatory regions that control the expression level of a given gene, as well as at the RNA level, through long intergenic non-coding RNAs (lincRNA) or microRNAs (miRNAs). Mounting data suggest clear roles for epigenetic modifications in maintaining immune homeostasis. For example, studies have revealed that altering the metabolic rate of intracellular transmethylation reactions is sufficient to ameliorate autoimmunity in mouse models of lupus.13 However, the precise role of non-coding RNAs in IBD pathogenesis is still being defined. Non-coding RNAs are not transcribed into protein products, although they can interact with chromatin regulators to adjust the expression of other genes in cis or, more commonly, in trans.14 Treatment of cells with pro-inflammatory molecules such as muramyl dipeptide, an activator of the NOD2 pathway, causes upregulation of a specific group of miRNAs, implying that these miRNAs could help fine-tune the inflammatory response and act synergistically with IBD variants in these inflammatory pathways.15 Recent studies have also catalogued changes in miRNA expression in IBD and suggested a role for miRNAs in IBD pathogenesis (reviewed in 16).
Demystifying dysbiosis
Much of the genetic susceptibility data from IBD studies suggest impaired handling of bacteria as well as an improper immune response to potential pathogens. Consistent with this hypothesis, dramatic shifts in the gut microbiota have been associated with IBD risk. These include alterations in the relative abundances of approximately one dozen bacterial taxa, as well as a decrease in the diversity of the community.17, 18 It remains largely unknown whether the severity of gut dysbiosis is the cause of, or the response to, the severity of the disease. Although certain opportunistic pathogens such as Enterobacteriaceae have increased relative abundance in IBD patients19 and in mouse models of intestinal inflammation,20 in most cases causal connections remain elusive, and the possibility remains that alterations in the abundance of gut commensal bacteria play a role in IBD pathogenesis. Plausible causal mechanisms have been proposed for certain taxa, such as the noted decrease in the genera Roseburia and Phascolarctobacterium associated with CD. Based on studies of related taxa, these bacteria are expected to produce butyrate21 and propionate,22 respectively; Roseburia is also expected to increase production of T regulatory cells.23 A reduction in the relative abundance of these members could therefore cause a decrease in anti-inflammatory agents. In the cases of alterations of other common gut commensals such as Ruminococcaceae and Leuconostocaceae, the direction of causality remains unclear.
A severe imbalance in the composition of the gut microbiome is often referred to as dysbiosis, but the term is poorly defined. A balance of healthy gut commensal bacteria is required for suppression of pathogenic infections,24 with increasing evidence that restoration of normal commensals via transplant is more effective at fighting Clostridium difficile infection than antibiotics.25 Transplants are especially relevant for IBD patients, where recurrent C. difficile infections increase morbidity and mortality and are increasing in prevalence.26 Given recent findings of high variability of relative abundances of constituent taxa both between healthy individuals and within a single healthy individual over time,27, 28 we may continue to find dysbiosis challenging to define in terms of taxonomic or phylogenetic composition. It has been proposed that human gut microbial communities may be partitioned into three discrete clusters.29 If this were true it would greatly simplify the definition of dysbiosis and could have important implications for disease diagnosis and treatment, but subsequent analysis of a substantially larger population revealed that gut microbial community composition follows a relatively smooth distribution across the global human population, with primary variation largely driven by continuous gradients of dominant taxa.30 In contrast, the functional repertoire of the gut microbiota appears to be relatively stable both within and between individuals.27 Changes in functional composition have been observed in subjects with IBD, including enrichment of genes in sulfur-metabolism pathways, and a decrease in butanoate and propanoate metabolism specifically in subjects with ileal CD. Bacterial proteases, from both pathogens and commensals, have also been implicated in intestinal inflammation.31 Given its relative stability, the functional composition of the gut microbiome may provide a more consistent definition of dysbiosis and may more readily provide evidence of genome-microbiome interactions in future exploratory studies.
Altered immune response to bacterial products
IBD-associated genes in host cells indicate altered response to gut microbiota as a primary determinant of disease risk and a likely mechanism for the disease. A number of host biological functions related to protection from and management of gut bacteria are susceptible to deleterious genetic mutations in constituent genes (figure 2). These include NOD2, which stimulates the immune system to respond to the presence of certain bacteria-produced peptidoglycans. Several NOD2 mutations are known to be pathogenic in CD.3, 32 Although NOD2-deficient mice are more susceptible to infection by specific bacterial pathogens,33 it is not known the extent to which NOD2 deficiencies alter host immune response to gut commensal bacteria. The IL23 receptor (IL23R) also plays an important role in response to pathogens34 and mutations of IL23R associate with increased IBD risk.3 Elevated levels of IL23 have been found in the epithelial mucosal barrier in subjects with IBD, further indicating the role of IL23 in the chronic inflammatory response to luminal bacteria.
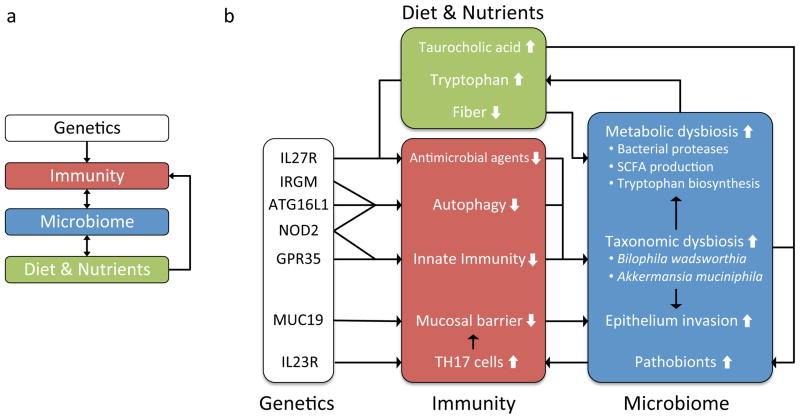
Figure 2. Interaction network of host genetics, the gut microbiome and diet in overview (a) and in detail (b).
Chronic inflammation in the intestinal epithelium has been associated with increased production of Th17 cells, impaired innate immune response, decreased mucosal barrier, impaired autophagy, and a decrease in antimicrobial agents. There is a complex network of potential interactions, in some cases involving feedback, between impaired host immune functions, diet, and the taxonomic and functional dysbiosis of the gut microbiome. For example, deleterious mutations in NOD2, GPR35, ATG16L1, or IRGM may lead to impaired immune response to commensal bacteria, and subsequently to taxonomic dysbiosis, an imbalance in the taxonomic composition of the gut microbiota; taxonomic dysbiosis may cause metabolic dysbiosis, an imbalance in the metabolic capabilities of the gut microbiome; metabolic dysbiosis may include increased biosynthesis of tryptophan; increased tryptophan is expected to lead to decreased antimicrobial activity through several pathways (see text); impaired antimicrobial activity may lead to further taxonomic and metabolic dysbiosis. A similar feedback system may be proposed for the physical integrity of the epithelial barrier: impaired innate immune response and increased production of Th17 cells may lead to decreased integrity of the mucosal barrier; altered or impaired mucus production due to MUC19 deficiency may compound this effect; subsequent invasion of pathobionts, or opportunistic pathogens, may increase inflammation, leading to further breakdown of the epithelial barrier.
The association of genes SLC22A5, GPR35, and GPR65 with IBD pathogenesis suggests an impaired immune response to bacteria-derived ligands and metabolites.35 Although it is known that gut commensals can produce pathogen-associated molecular patterns similar to pathogenic species, in healthy subjects toll-like receptors in dendritic cells respond selectively to pathogenic bacteria while largely ignoring gut commensals.36 Due to the high degree of selectivity required by this task, even subtle defects in microbial product sensing might be expected to contribute to a chronic inflammatory response. Bacterial proteases are also expected to play a role in intestinal inflammation, including those proteases from commensals. The gut commensal Enterococcus faecalis produces gelatinase, a metalloproteinase that disrupts the epithelial barrier and increases inflammation in mice.31 This disruption occurs only when the host has genetic susceptibility to inflammation, for example via IL10 or NOD2 deficiency, thereby associating genetic risk of IBD with increased sensitivity to by-products of commensal bacteria.
The role of dietary nutrients and metabolites
In addition to short-chain fatty acids, a number of other metabolites are expected to be involved in host-microbiome interactions. Tryptophan provides an important intermediate metabolite for the action of aryl hydrocarbon receptor (AHR) in suppressing immune responses in dendritic cells. Specifically, AHR induces indoleamine 2,3-dioxygenase (IDO), which catabolizes tryptophan into kynurenine (Kyn).37 A deficiency in AHR causes reduced Kyn and subsequently increased production of pro-inflammatory Th17 cells. In a similar case, a synthase acting to reduce tryptophan levels, tryptophan hydroxylase-1 (Tph-1), is required for immune suppression in several inflammation models.38 Direct interaction of tryptophan levels with the microbiome has been demonstrated through the tryptophan-induced production of anti-microbial peptides via the mammalian target of rapamycin (mTOR) pathway.39 Recent findings implicate cytokine IL27 in maintaining epithelial barrier protection against normal intestinal bacteria, and IL27 has been associated through GWAS with increased risk of CD.3 Transcriptional analysis indicates that IL27 activates several members of the signal transducers and activators of transcription family (STAT1, STAT3, STAT6), and depends on STAT1 to activate IDO.40 In this study IDO was found to exert antibacterial effects on luminal bacteria specifically by depletion of tryptophan independent of the presence of Kyn. Thus modulation of gut tryptophan levels by diet or by microbial biosynthesis is likely to have differential effects in individuals with altered or impaired function in AHR, IL17/IL23, or IL27 pathways.
Long-term dietary habits are associated not only with the overall structure of the gut microbiome in humans41 but also with expansion of a particular pathobiont, Bilophila wadsworthia, in mice.42 Increased levels of taurocholic acid induced by consumption of certain saturated fats causes an increase in organic sulfur in hepatic bile. This leads to the subsequent blooming of sulphite-reducing B. wadsworthia, which then leads to higher rates of colitis in IL10-deficient mice. Together these interactions provide a possible mechanism by which shifts in dietary nutrients could have inflammation-inducing interactions with the gut microbiota in individuals with certain genetic mutations (figure 2).
Genetic risk of infection
IBD genetic risk loci have substantial overlap with risk loci for primary immunodeficiencies, especially with susceptibility to mycobacterium infection.3 IBD risk loci potentially involved in IL-17 production and therefore in response to bacterial infection are enriched for balancing selection, often seen in genes related to anti-parasite immunity. Given this and other recent evidence of genetic predisposition to acquisition of certain pathogens or parasites in humans, we must consider the possibility of such effects in IBD pathogenesis. There is suggestive evidence that 13 genetic loci are associated with colonization by periodontal pathogens,43 although the mechanism of action is not understood. Similarly, mutations in two genetic loci increase susceptibility to severe forms of falciparum malaria.44 Here the loci, found through GWAS, led directly to likely disease mechanisms when coupled with a thorough understanding of the complex stages of infection. There are a number of plausible mechanisms for a parallel effect in IBD. For example, impaired autophagy of invasive bacteria is a plausible point for genome-microbe interaction in which defenses against otherwise commensal bacteria allow them to become pathogenic. Mutations in loci containing immunity-related GTPase family M (IRGM) and autophagy-related 16-like 1 (ATG16L1) are both found to increase CD risk (odds ratio 1.3 and 1.2, respectively).3 NOD2, one of the strongest GWAS associations with CD risk, induces autophagy in dendritic cells and requires proper function of ATG16L1.45
Altered mucosal protection and bacterial invasion
The epithelial mucosal barrier that normally helps isolate the lamina propia from luminal bacteria is reduced in patients with IBD.46 The possible association of MUC19 with IBD risk3 could indicate an additional mechanism for host-microbe interaction. It is known that certain bacterial species such as Akkermansia muciniphila and Enterorhabdus mucosicola degrade mucus and can thrive on the mucus layer.47 Therefore inherited alterations in mucosal composition or production have the potential to alter the composition of the luminal bacteria, especially those most proximal to the host epithelial cells (figure 2).
Multistage triggers for chronic inflammation
Given the large number of genetic loci associated both with IBD risk and with host-microbiome interactions, we may wish to consider pathogenic models that include multiple stages of disease development (figure 2). This implies bi-directional causality between altered host immune function and altered bacterial community functions, features, or by-products. A number of simple mechanisms may be considered that involve multiple stages of triggers. For example, a host genetic variant in NOD2 or IL23R may lead to elevated inflammatory response to the presence of a pathogen; this excessive response may damage the epithelial barrier, leading to colonization by an opportunistic pathogen or an imbalance in normal gut commensal bacteria; increased exposure to these bacteria may cause further inflammation, leading to a chronic state of dysregulation. Genetic variants causing impaired mucosal barrier production may accelerate the process. Another such mechanism begins with impaired host sensing of bacterial byproducts and metabolites via defects in SLC22A5A, GPR35, or GPR65, again leading to dysbiosis and eventually a chronic inflammation state. In contrast, a healthy immune system would respond appropriately to transient infections by opportunistic pathogens without entering the over-inflamed state, thereby avoiding the development of chronic dysbiosis.
Bi-directional causality with multistage triggers is supported by mouse experiments in which disease phenotypes can be caused not only by a genetic mutation but also by transplant of the microbiota from the mutant mouse to a wild-type mouse. This has been observed for colitis in mice deficient in NOD-like receptor family, pyrin domain containing 6 (NLRP6)48 and in malnutrition-related intestinal inflammation in angiotensin-converting enzyme 2 (ACE2) knockout mice.39 In the former case, mutant mice had altered fecal microbiota and increased susceptibility to chemically induced colitis; wild-type mice acquired the increased susceptibility after transplant. In the latter case, a detailed mechanism was determined involving impaired production of antimicrobial peptides via the mTOR pathway. Mice with the knock-out developed inflammation, whereas wild-type mice developed the same inflammation after transplant of the “inflamed” microbiome. In closer relation to IBD, NOD2 deficiency has been found to induce colitis-causing dysbiosis in mice, with phenotype conferred by either genetic inheritance or inheritance through maternally transmitted microbiota.49 Host-microbiome feedback in chronic inflammation is further supported by evidence that immune response to transient infection can lead to long-term adaptive immune response to gut commensals.50 During mucosal infection by Toxoplasma gondii, some T cells differentiate into memory cells specific to gut commensals. Because increased CD4 T cell activation is associated with IBD,3, 51, 52 it is possible that improper sensing of commensal bacteria leads to chronic inflammation, but only after exposure to a bacterial infection.
Pursuing microbiome-wide association studies (MWAS)-GWAS
Each of the above models requires both genetic predisposition to IBD and exposure to certain types of bacteria or bacterial products. Although the details of such interactions are largely suppositional, given the large environmental component of disease risk in IBD, the strong associations of genetic risk loci with response to microbial symbionts, and the associations of a number of bacterial taxa with IBD, genome-microbiome interaction is a likely candidate for further study. No such connections have been demonstrated to date in a diseased cohort. Discovery of such interactions is complicated by a number of factors including the multiple stages of pathogenesis, the large number of interactions to be tested, and high inter- and intra-subject variability in the gut microbiota. Depending on the strength of the true associations, these limitations may be overcome by careful treatment of cohort selection and data analysis. The feedback-based models described above involve cascades of several trigger events that lead to eventual establishment of chronic inflammation. Therefore the ideal analysis would involve observation of the gut microbiota longitudinally before and after disease presentation. It is likely that exploration of interactions between host genetics and the functional, rather than taxonomic, composition of the gut microbiome will provide both stronger association signals and more direct insights into the mechanisms of the disease.
Key messages.
More than 160 genetic loci have been associated with susceptibility to inflammatory bowel disease (IBD). These genetic findings have led to the identification of several known and novel pathways that are involved in IBD, but understanding the cell types involved in these pathways remains an important unresolved goal.
IBD-associated genes in host cells indicate that altered responses to gut microbiota may be a primary determinant of disease risk and a likely mechanism for the disease.
The diversity and composition of the gut microbiota are major environmental factors influencing gut homeostasis. A severe imbalance in the composition of the gut microbiome, often referred to as dysbiosis, has been associated with IBD.
The concept of dysbiosis remains poorly defined. Describing dysbiosis in terms of taxonomic or phylogenetic composition is likely to remain challenging due to high intra- and inter-individual variation. In contrast, defining dysbiosis in terms of the relatively stable functional composition of the gut microbiome may be a more promising approach.
Particular dietary nutrients and metabolites likely interact with host genetics to influence host-microbiome interactions and thereby contribute to inflammation.
Microbiome interactions with host genetics may be best understood as a bi-directional relationship between altered host immune function and altered bacterial community functions, features, or by-products.
Acknowledgments
We thank Natalia Nedelsky for editorial assistance. This work was supported by funding from the Crohn’s and Colitis Foundation of America Genetics Initiative as well as N.I.H. grants DK097485 and DK062432 to R.J.X.
Abbreviations
- AHR
aryl hydrocarbon receptor
- CD
Crohn’s disease
- ENCODE
Encyclopedia of DNA Elements
- GWAS
genome-wide association study
- IBD
inflammatory bowel disease
- IDO
indoleamine 2,3-dioxygenase
- IL
interleukin
- Kyn
kynurenine
- lincRNA
long intergenic non-coding RNA
- miRNA
microRNA
- SNP
single-nucleotide polymorphisms
- UC
ulcerative colitis
Footnotes
Licensing statement:
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive license (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Gut editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our license.
Competing Interest: All authors declare that the answer to the questions on your competing interest form are all No and therefore have nothing to declare.
References
- 1.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 2.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 3.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. Genomes Project C. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–8. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, et al. IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. J Exp Med. 2012;209:1595–609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griseri T, McKenzie BS, Schiering C, Powrie F. Dysregulated hematopoietic stem and progenitor cell activity promotes interleukin-23-driven chronic intestinal inflammation. Immunity. 2012;37:1116–29. doi: 10.1016/j.immuni.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium EP, Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013 doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardison RC. Genome-wide epigenetic data facilitate understanding of disease susceptibility association studies. J Biol Chem. 2012;287:30932–40. doi: 10.1074/jbc.R112.352427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang ML, Gee AJ, Gee RJ, Zurita-Lopez CI, Khare S, Clarke SG, et al. Lupus autoimmunity altered by cellular methylation metabolism. Autoimmunity. 2013;46:21–31. doi: 10.3109/08916934.2012.732133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasler R, Jacobs G, Till A, Grabe N, Cordes C, Nikolaus S, et al. Microbial pattern recognition causes distinct functional micro-RNA signatures in primary human monocytes. PLoS One. 2012;7:e31151. doi: 10.1371/journal.pone.0031151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pekow JR, Kwon JH. MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:187–93. doi: 10.1002/ibd.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroenterol. 2010;26:327–31. doi: 10.1097/MOG.0b013e328339536b. [DOI] [PubMed] [Google Scholar]
- 19.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–54. e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 20.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–29. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov. a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–20. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe Y, Nagai F, Morotomi M. Characterization of Phascolarctobacterium succinatutens sp. nov. an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol. 2012;78:511–8. doi: 10.1128/AEM.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagano Y, Itoh K, Honda K. The induction of Treg cells by gut-indigenous Clostridium. Curr Opin Immunol. 2012;24:392–7. doi: 10.1016/j.coi.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–9. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N Engl J Med. 2013 doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 26.Issa M, Ananthakrishnan AN, Binion DG. Clostridium difficile and inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1432–42. doi: 10.1002/ibd.20500. [DOI] [PubMed] [Google Scholar]
- 27.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, et al. A Guide to Enterotypes across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steck N, Mueller K, Schemann M, Haller D. Bacterial proteases in IBD and IBS. Gut. 2012;61:1610–8. doi: 10.1136/gutjnl-2011-300775. [DOI] [PubMed] [Google Scholar]
- 32.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshmukh HS, Hamburger JB, Ahn SH, McCafferty DG, Yang SR, Fowler VG., Jr Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect Immun. 2009;77:1376–82. doi: 10.1128/IAI.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riol-Blanco L, Lazarevic V, Awasthi A, Mitsdoerffer M, Wilson BS, Croxford A, et al. IL-23 receptor regulates unconventional IL-17-producing T cells that control bacterial infections. J Immunol. 2010;184:1710–20. doi: 10.4049/jimmunol.0902796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujiya M, Inaba Y, Musch MW, Hu S, Kohgo Y, Chang EB. Cytokine regulation of OCTN2 expression and activity in small and large intestine. Inflamm Bowel Dis. 2011;17:907–16. doi: 10.1002/ibd.21444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–95. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, et al. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–6. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowak EC, de Vries VC, Wasiuk A, Ahonen C, Bennett KA, Le Mercier I, et al. Tryptophan hydroxylase-1 regulates immune tolerance and inflammation. J Exp Med. 2012;209:2127–35. doi: 10.1084/jem.20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–81. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diegelmann J, Olszak T, Goke B, Blumberg RS, Brand S. A novel role for interleukin-27 (IL-27) as mediator of intestinal epithelial barrier protection mediated via differential signal transducer and activator of transcription (STAT) protein signaling and induction of antibacterial and anti-inflammatory proteins. J Biol Chem. 2012;287:286–98. doi: 10.1074/jbc.M111.294355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Divaris K, Monda KL, North KE, Olshan AF, Lange EM, Moss K, et al. Genome-wide association study of periodontal pathogen colonization. J Dent Res. 2012;91:21S–8S. doi: 10.1177/0022034512447951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timmann C, Thye T, Vens M, Evans J, May J, Ehmen C, et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489:443–6. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- 45.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 46.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–68. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013 doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–6. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]